Abstract
Understanding the energetics of peripheral protein/membrane interactions is important to many areas of biophysical chemistry and cell biology. Estimating free energy landscapes by molecular dynamics (MD) simulation is challenging for such systems, especially when membrane recognition involves complex lipids, e.g. phosphatidylinositol phosphates (PIPs). We combined coarse-grained MD simulations with umbrella sampling to quantify the binding of the well-explored GRP1 pleckstrin homology (PH) domain to model membranes containing PIP molecules. The experimentally observed preference of GRP1-PH for PIP3 over PIP2 was reproduced. Mutation of a key residue (K273A) within the canonical PIP-binding site significantly reduced the free energy of PIP binding. The presence of a non-canonical PIP-interaction site, observed experimentally in other PH domains but not previously in GRP1-PH, was also revealed. These studies demonstrate how combining coarse-grained simulations and umbrella sampling can unmask the molecular basis of the energetics of interactions between peripheral membrane proteins and complex cellular membranes.
Keywords: lipid bilayer, peripheral membrane protein, molecular dynamics, umbrella sampling, coarse-grained
The binding of lipid-recognizing peripheral proteins to cell membranes is essential for many cellular processes. Targeting of proteins to specific lipid molecules and/or to bilayers of particular lipid compositions, mediated by lipid-binding domains, allows their recruitment to be regulated in both a temporal and spatial fashion.1 Perhaps the most intensively studied class of lipid-binding domains is the Pleckstrin Homology (PH) domain, which has been shown in many cases to recognise and bind to phosphatidylinositol phosphate (PIP) lipids.2,3 PIPs are a family of lipids characterized by different phosphorylation patterns of a common inositol head-group; interconversion between different PIP species allows them to act as second messengers in a variety of signalling and regulatory pathways.3,4 Variations in the sequence of individual PH domains allows them to recognise PIP species with differing selectivities and affinities.2 The PH domain of GRP1, a protein involved in cytoskeletal dynamics5, is of note due its ability to bind phosphatidyl inositol (3,4,5)-trisphosphate (PI(3,4,5)P3 or more briefly PIP3) with high affinity and selectivity over other PIPs (including the more common phosphatidyl inositol (4,5)-trisphosphate, PI(4,5)P2 or more simply PIP2).6 While a number of experimental tools exist for the investigation of the membrane binding of peripheral proteins7, the exact molecular and energetic details of protein-membrane interactions, essential for understanding the function and regulation of these proteins, are difficult to elucidate using these methods.
Molecular Dynamics (MD) simulations have been established as a valuable tool for the study of protein-membrane interactions8, on time and length scales not readily accessible to experimental methods. In conjunction with free energy calculation methodologies, MD approaches can be used to quantify strengths of interaction of lipids with proteins. Given the temperature T, the binding free energy ΔG is related to the dissociation constant KD by:
| (1) |
where R is the ideal gas constant and c⊖ is the standard reference concentration 1M. Several approaches exist for the calculation of free energies. Construction of a potential of mean force (PMF) profile along a physical reaction coordinate using, for example, umbrella sampling9 may additionally provide insights into the interaction process. While the umbrella sampling approach is well established for protein interactions with relatively simple ligands10, the long simulation times required for convergence and sampling necessary for accurate PMF calculation have limited its application to larger systems such as those involving membranes. The use of coarse-grained (CG) models can allow an improvement in simulation timescales of 2-3 orders of magnitude11, and have recently been combined with an umbrella sampling approach to quantify the free energy of dimerization of transmembrane helices (e.g. 12, 13) and the free energy of interaction of cardiolipin with cytochrome c oxidase14. Here, we extend the application of CG simulations in an umbrella sampling approach to peripheral-protein/membrane systems, quantifying the interaction of the GRP1 PH domain with model membranes containing PIP lipids. The GRP1 PH domain has been the focus of a number of computational studies, using a range of simulation methodologies15–17 and so provides a biologically important and well characterised test case for exploring the energetics of membrane binding.
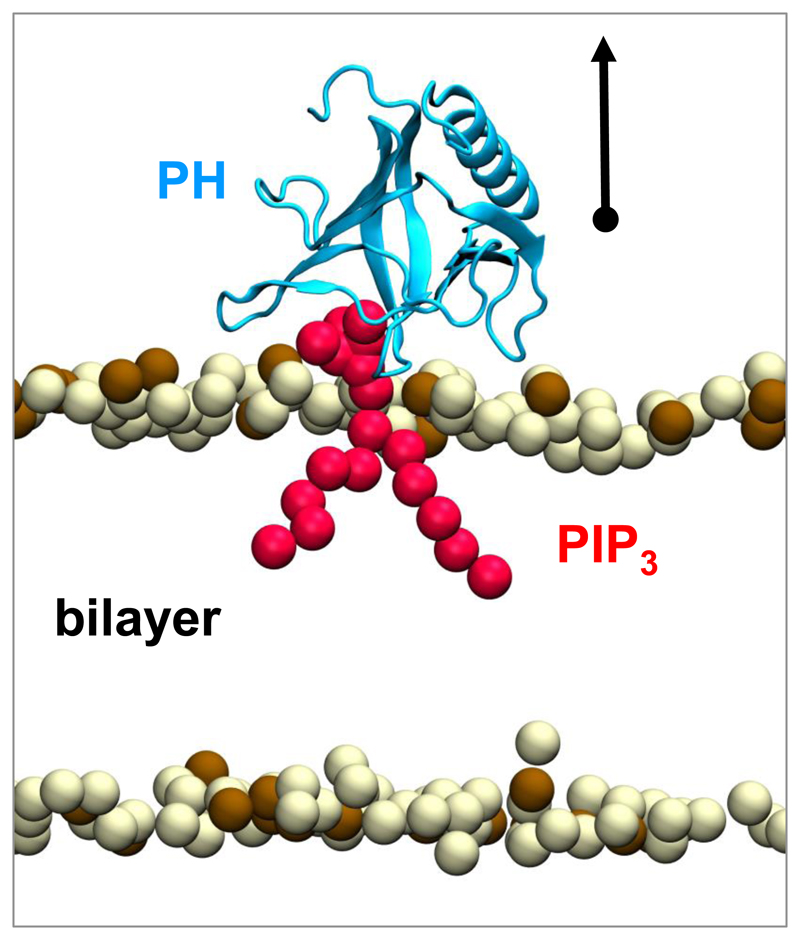
In order to calculate the free energy of binding of the GRP1 PH domain to PI(3,4,5)P3 and PI(4,5)P2 molecules, a GRP1-PH/bilayer complex was modelled using the crystal structure of the GRP1 PH domain bound to an Ins(1,3,4,5)P4 molecule (PDB: 1FGY). To this end we have aligned the bound Ins(1,3,4,5)P4 ligand with the head-group of a PIP3 molecule embedded in a preformed POPC:POPS (80:20) bilayer. The resultant complex was in agreement with experimental and computationally derived binding models.15,17,18 In particular, the tilt angle of GRP1-PH relative to the bilayer (given by the angle of the vector between residues C292 and F296 to the membrane surface; 47°), and penetration of GRP1-PH the membrane (given by depth of residue V278 compared to the average lipid phosphate plane; 0.18 nm), are within the range of models based on EPR site-directed spin labelling results18 (46 ± 7° and 0.24 ± 0.2 nm, respectively). To further confirm the initial orientation of GRP1-PH, we have also run coarse-grained simulations in which an initially displaced GRP1-PH molecule was allowed to freely diffuse and associate with a PIP3-containing membrane. A similar final binding orientation of GRP1-PH was reached in each of five repeat simulations (Supplementary Figure 1).
The atomistic GRP1-PH structure was converted to a coarse-grained representation using the MARTINI force field (version 2.1)11 to produce an initial PIP3-bound structure for umbrella sampling (Figure 1). A GRP1-PH/PIP2 complex was also generated by replacing the bound PIP3 molecule with a PIP2 molecule. Using a steered molecular dynamics simulation, the GRP1-PH domain was pulled away from the bound PIP2 or PIP3 lipid along the membrane normal, as indicated by the arrow in Figure 1 (note that the PIP phosphate bead was restrained relative to its initial position during this simulation). Snapshots at various protein-lipid separations (measured from the PH domain centre-of-mass to the backbone phosphate of the PIP molecule) were used as initial configurations to define a reaction pathway for a series of umbrella sampling simulations. Each window was simulated for 1000 ns. The restraint on the lipid phosphate-1 bead was maintained in these simulations. Comparison of PMFs generated without this restraint suggests that this allows a substantive reduction in the simulation time required for convergence without significantly altering the profile obtained (Supplementary Figure 3). In order to reduce the simulation time required for adequate sampling, restraints orthogonal to the membrane normal were introduced. The restraint constant used should allow reduction of the orthogonal space to be sampled at large protein-ligand separations while not significantly altering the behaviour of GRP1-PH when bound, as this may introduce errors. Restraint constants in the range of 50 to 500 kJ mol-1 nm-2 were tested and a value of 100 kJ mol-1 nm-1 selected as an optimal compromise based on histograms of orthogonal GRP1-PH centre-of-mass displacement (Supplementary Figure 4).
Figure 1.
Initial structure of the GRP1-PH domain bound to a PIP3 in a lipid bilayer membrane. GRP1-PH is shown in blue in a cartoon representation; PIP3 is shown in red. The lipid bilayer is represented by the headgroup phosphate particles of the POPC and POPS molecules, shown in light and dark brown respectively. The grey arrow indicates direction of pulling used generate the reaction pathway for the subsequent umbrella sampling windows.
To run the simulations we have used an automated pipeline, generalised for any protein-membrane system, which also simplified the process of setting up umbrella simulations and ensured consistency of the parameters used. Initial values for the total number of umbrella sampling windows, window spacing and simulation time of each window were chosen such that adequate sampling along the reaction coordinate (protein-lipid separation) and temporal and spatial convergence were achieved for the GRP1-PH/PIP3 system (Supplementary Figures 2, 5). These values were found to be suitable for all subsequent systems reported in this study.
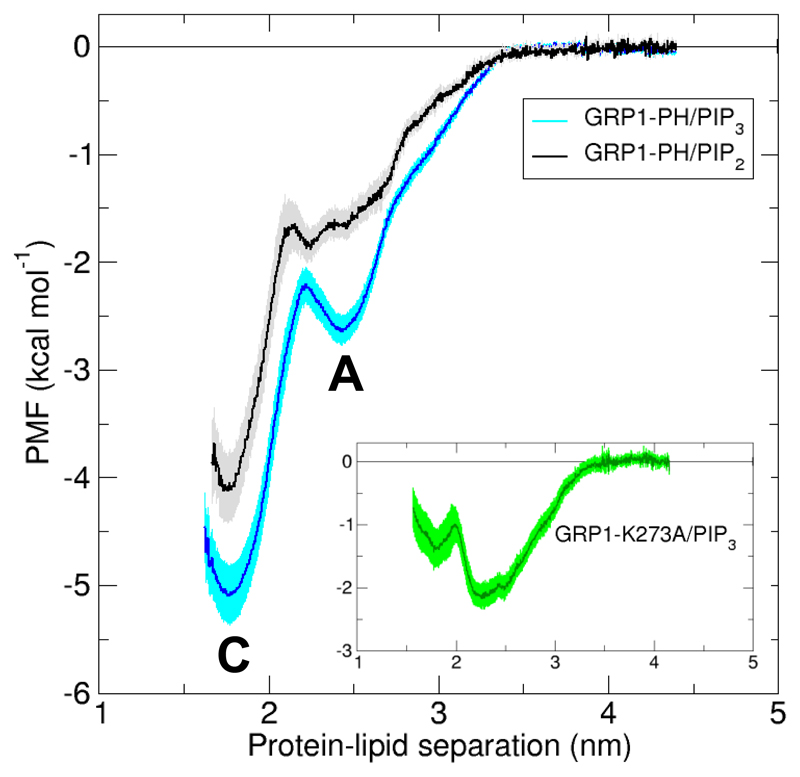
From the umbrella sampling simulations, 1D PMF profiles of GRP1-PH binding to PIP3 and PIP2 were generated (Figure 2). Both profiles have a global minimum at a protein-lipid separation of ~1.8 nm, with a well depth of -5.3 kcal mol-1 for PIP3 and -3.8 kcal mol-1 for PIP2, and a shallower well at a larger separation of ~2.2-2.6 nm. Note that the second minimum is deeper for PIP3 compared to PIP2. These profiles suggest favourable binding to both species with around a ~10-fold preference for PIP3 (calculated using Equation 1). A PMF profile generated from an initial structure of GRP1-PH bound to a PI(3,4)P2 molecule is the same as for PI(4,5)P2 within the errors indicated by bootstrap analysis (Supplementary Figure 6) (note that as a result of the low resolution of the CG model, the CG representations of PI(4,5)P2 and PI(3,4)P2 are essentially the same; only the initial alignment differs between the two systems). Experimental observations have reported selectivities of PIP3 over PI(3,4)P2 and PI(4,5)P2 of ~5-200 and ~50-200 fold, or greater, respectively.19–25
Figure 2.
PMF profiles for wild-type GRP1-PH bound to PIP3 or PIP2 (main Figure) and GRP1-PH-K273A mutant bound to PIP3 (inset). Protein-lipid separation is measured from the protein centre-of-mass to the lipid 1-phosphate along the membrane normal. Error estimates were obtained from bootstrap analysis.
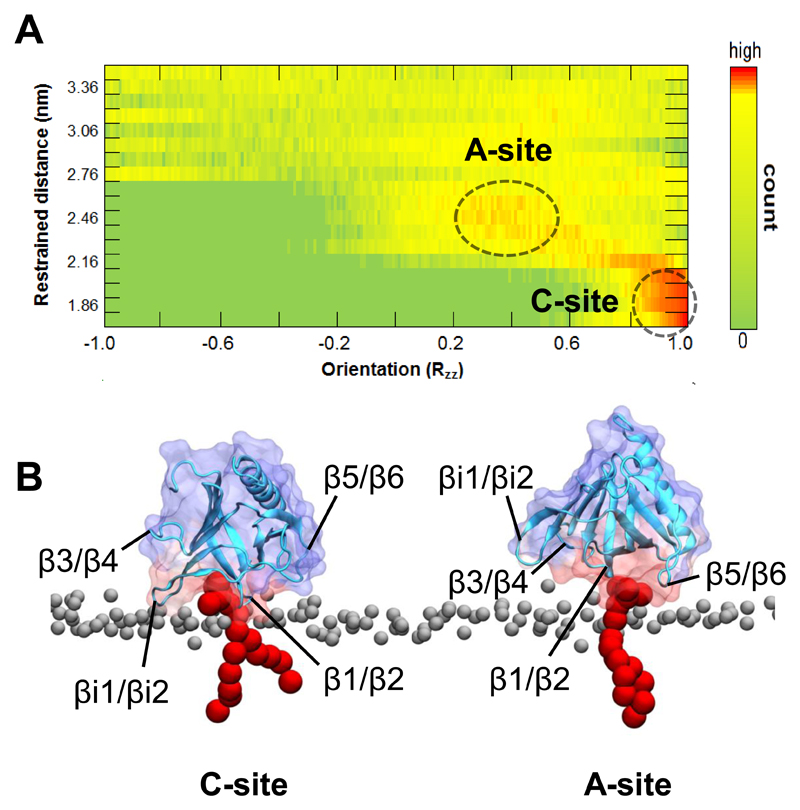
The orientation of the GRP1-PH domain relative to the membrane and the residues that contact lipids (using a cut-off distance of 0.5 nm to define a contact) were investigated in each umbrella window for both the GRP1-PH/PIP3 and GRP1-PH/PIP2 systems. In windows covering protein-lipid separations ~1.7 to 2.1 nm (corresponding to the first i.e. deeper well in Figure 2), GRP1-PH remained stably bound in the initial orientation (Figure 3A). The residues with the highest frequency of contacts with the POPC/POPS lipids (residues 277-283 and 322-323), defining the membrane-binding interface, are consistent with experimental results17,18. The residues interacting specifically with the bound PIP ligand (residues K273, G276, R277, V278, K279, T280, K282, R284, R305, K343, N354 and H355; Supplementary Figure 7) are, with the exception of G276 and V278, those observed to contact IP4 in the original crystal structure26 and are also in agreement with previous experimental and computational results15,17,18,27. This initial binding site (the C-site in Figure 3B) corresponds to the ‘canonical’ PIP-binding site common amongst many PH domains3.
Figure 3.
A. Three-dimensional histogram of the orientation of the GRP1-PH domain relative to the membrane throughout the first 18 umbrella sampling windows (which covers the distance until the PMF profile levels off), for the GRP1-PH/PIP3 system. RZZ is the component of the rotation matrix relative to the initial configuration (which corresponds to RZZ = 1); the 'restrained distance' indicates the protein centre-of-mass to lipid backbone phosphate separation at which each window was restrained. B. Representative structures of GRP1-PH bound to PIP3 in the ‘canonical’ (C) and ‘atypical’ (A) modes. GRP1-PH is shown in cartoon representation in blue; the surface is coloured by average number of lipid contacts in the windows where each mode is dominant. PIP3 is shown in red and phosphate groups of POPC/POPS in grey.
Disruption of the canonical binding site by mutation of four key basic residues in the binding pocket, K273, R284, R305 and K343, to alanine (GRP1-4A mutant) resulted in the disappearance of the first well from the PIP3-bound PMF profile. The second well was, however, retained, though with a reduced depth of -1 kcal mol-1 (Supplementary Figure 8). With the single mutation K273A, shown experimentally to effectively eliminate binding22, the first well was retained, however the deeper minimum occurs at the second well, with a value of -2.1 kcal mol-1 (Figure 2, inset). Using Equation 1, this ~3 kcal mol-1 change in minima compared to the wildtype corresponds to a ~100 fold reduction in affinity. Retention of the second well on mutation of the canonical binding site suggests that it may correspond to a secondary (i.e. non-canonical or atypical) binding mode, distinct from the canonical site.
Orientational analysis of windows covering the range ~2.2 to 2.6 nm (corresponding to the second well in Figure 2) in the wild-type GRP1-PH/PIP3 system reveals a second orientation becomes more favourable at these greater protein-lipid separations (Figure 3A). A pattern of residue contacts distinct from the initial windows, though with some overlap, is observed with both POPC/POPS (R322, Y298, K323, K279, and K302) and PIP3 (K282, R283, R284, R322, K323, W281 and K279; see Figure 3B; Supplementary Figure 5) molecules. This secondary PIP interaction site is located on the opposite side of the β1/β2 loop flanking the canonical site (the A-site in Figure 3B), corresponding to though slightly displaced from the ‘atypical’ (or non-canonical28) PIP-binding site identified from the crystal structures of several other PH domains29–31 (Supplementary Figure 9).
Though PIP interactions are usually observed at only one of the canonical and the atypical sites for a given PH domain, interaction with both has been recently reported for the ASAP1 PH domain29. As proposed for ASAP1, the secondary site in the GRP1 PH domain may act as a general anionic phospholipid interaction site to promote correct binding via cooperativity: a second anionic interaction site for GRP1-PH has previously been suggested based on experimental evidence32. Some interaction at the secondary site was also observed in windows covering protein-lipid separations in the range ~2.2 to 2.6 nm in the GRP1-PH/PIP2 system, though in general these were less stable than for PIP3, with e.g. fewer contacts per frame observed (Supplementary Figure 7). This explains the presence of the smaller well in the PMF at this protein-lipid separation, and may be due to the lower charge of PIP2.
Use of a polarizable MARTINI water model33 instead of the standard coarse-grained water model has been shown to e.g. improve estimation of the free energies of partitioning of charged amino acid sidechains into hydrophobic environments (e.g. reference 34). We therefore performed additional umbrella sampling simulations for the GRP1-PH/PIP3 system using polarizable MARTINI water. In addition to substantially increasing the time required for convergence, the use of the polarizable water model altered the depth of the PMF minimum corresponding to the canonical PIP3/PH interaction compared to the non-polarizable model (Supplementary Figure 10). The polarizable water model also appeared to increase the complexity of the underlying free energy landscape which, given our incomplete understanding of the relative influence of water polarizability on protein-water and PIP-water interactions, is perhaps not surprising.
We also calculated the PMF profile for GRP1-PH bound to a PIP3 molecule with a reduced net charge of -5, compared to the original -7 net charge (Supplementary Figure 11). The depth of the first well was reduced by around 2 kcal mol-1 and the second by around 1 kcal mol-1 for the -5e charged PIP3. PIP2 also has a net charge of -5e: however, the -5 PIP3 profile is shallower and in shape more closely resembles that of -7 PIP3, suggesting that the interactions of PIP2 and PIP3 can be distinguished in the coarse-grained model used.
PMF profiles were also generated from initial structures in which the PIP molecule was replaced by either the zwitterion lipid POPC or the anionic lipid POPS. In contrast to the well-shape of the profiles above, the wild-type GRP1-PH/POPC and GRP1-PH/POPS profiles exhibit an energy barrier, with PMF changes upon interaction of +0.7 kcal mol-1 and 0.0 kcal mol-1 respectively (Supplementary Figure 12). This suggests that binding of the PH domain to POPC/POPS alone is unfavourable, consistent with the lack of experimentally-observed binding to membranes in the absence of PIP lipids in many cases.17,23,32
We note that in our estimation of PMFs we have estimated errors from bootstrap analysis. Although widely employed, this approach does not adequately identify all sources of error. Therefore the additional ‘control’ PMFS as discussed provide a better overall measure of the level of interpretation which our results can sustain.
A number of experimentally determined dissociation constants of GRP1 from PIP-containing membranes have been published, ranging between ~5 nM and 1 μM (in the range typical for PH domains2) depending on the experimental conditions used (e.g. binding affinity increases at acidic pH, at higher temperatures, and upon the inclusion of background anionic lipids).17,23–25,27,32 From Equation 1, using the appropriate temperature reported in each study, these dissociation constants correspond to binding free energies between ~ -8 and -11 kcal mol-1. The well depth read from the PMF does not directly correspond to the binding free energy35: only one dimension is considered, the orthogonal restraints used will bias the effective concentration of GRP1-PH in the system, and despite the restraint constant being selected to minimise the effect on the bound state, some perturbation is still likely to have occurred. Additional energy terms are required to account for these. Following the work of Doudou et al.35, the contribution of the restraints in the bound state for the GRP1-wt/PIP3 system was found to be relatively small (-0.3 kcal mol-1), however the effect on relative unbound volume explored is significant, leading to a final standard binding free energy of -3.2 kcal mol-1. This appears to underestimate the experimental values. Many of the experimental studies, however, used conditions different from those used in our simulations. In particular, many experimental methods commonly employ membrane compositions with 2-3% PIP lipids, while in the current study there is only one PIP molecule available to associate with the PH domain. It is likely, particularly given the presence of the atypical binding site, that GRP1-PH is able to simultaneously bind two or more PIP molecules when PIP is present in sufficient quantity. The presence of background anionic lipids has been show experimentally to increase the binding affinity of GRP1-PH for PIP-containing membranes by around 10 fold17,23,24,27,32; cooperativity between multiple bound PIPs may be expected to have an even greater effect. If a second or third PIP3 molecule bound, for example, in the atypical site has a similar contribution to the binding energy to the PIP3 molecule in the canonical site (as is suggested by preliminary PMF estimations), the well depth of the PMF would be expected to up to double or triple while the additional correction terms would remain relatively unchanged, yielding a binding free energy in the region of that seen experimentally.
Overall, our simulations with the GRP1-PH system are able to correctly identify the binding preference of PIP3 over PIP2 and the absence of favourable binding to membranes lacking PIP. A pronounced effect of the single K273A mutation on the PMF profile was clearly observable, in line with experimental observations. The shape of the PMF profiles and further analysis also revealed a non-canonical lipid interaction site known amongst PH domains but not previously reported for GRP1-PH. This is notable in particular due to increasing understanding of the importance of ‘coincidence detection’, the requirement for multiple lipids or proteins to be present for the targeted recruitment of specific membrane binding proteins1. A recent study36 highlighted the key importance of lipid cooperativity in membrane recruitment of PH domains.
The GRP1-PH/PIP system investigated here provides a paradigm for how umbrella sampling coupled with coarse-grained simulations can be used to investigate peripheral membrane protein-lipid interactions. We have shown that this type of methodology is sensitive enough to capture differences in the binding of proteins to different lipid species and the effects of mutations seen experimentally, and therefore it could be used to predict these preferences where no experimental data is available. Having established an automated pipeline for setup and parameters to achieve adequate sampling and convergence for the GRP1-PH system, expansion of this methodology to other classes of peripheral proteins and membrane compositions is feasible, and shows promise as a fast and automated way in which to quantify protein-lipid interactions.
Supporting Information
Simulation and calculation details; evolution of binding simulations; evolution of PMF profiles; effects of removing transverse restraints; transverse distance dependence on restrain constant; comparison of C and A sites in PH domains; GRP1-PH/PIP contacts; effect of using polarizable water; effect of reducing charge on PIP3; additional PMF profiles for GRP1-PH bound to PI(3,4)P2, POPC, POPS and GRP1-4A-PH bound to PIP3.
Acknowledgements
A.C.K. and M.S.P.S. were funded by the Wellcome Trust. F.B.N. was supported by the Clarendon Fund and Lincoln College, University of Oxford. We acknowledge the EPSRC and the UK High-End Computing Consortium for Biomolecular Simulation for time on the ARCHER supercomputer.
References
- (1).Moravcevic K, Oxley CL, Lemmon MA. Conditional Peripheral Membrane Proteins: Facing up to Limited Specificity. Structure. 2012;20(1):15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).DiNitto JP, Lambright DG. Membrane and Juxtamembrane Targeting by PH and PTB Domains. Biochim Biophys Acta. 2006;1761(8):850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- (3).Stahelin RV, Scott JL, Frick CT. Cellular and Molecular Interactions of Phosphoinositides and Peripheral Proteins. Chem Phys Lipids. 2014;182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Di Paolo G, De Camilli P. Phosphoinositides in Cell Regulation and Membrane Dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- (5).Clodi M, Vollenweider P, Klarlund J, Nakashima N, Martin S, Czech MP, Olefsky JM. Effects of General Receptor for Phosphoinositides 1 on Insulin and Insulin-like Growth Factor I-Induced Cytoskeletal Rearrangement, Glucose Transporter-4 Translocation, and Deoxyribonucleic Acid Synthesis. Endocrinology. 1998;139(12):4984–4990. doi: 10.1210/endo.139.12.6351. [DOI] [PubMed] [Google Scholar]
- (6).Klarlund JK. Signaling by Phosphoinositide-3,4,5-Trisphosphate Through Proteins Containing Pleckstrin and Sec7 Homology Domains. Science (80-. ) 1997;275(5308):1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- (7).Scott JL, Musselman CA, Adu-Gyamfi E, Kutateladze TG, Stahelin RV. Emerging Methodologies to Investigate Lipid-Protein Interactions. Integr Biol (Camb) 2012;4(3):247–258. doi: 10.1039/c2ib00143h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stansfeld PJ, Sansom MSP. Molecular Simulation Approaches to Membrane Proteins. Structure. 2011;19(11):1562–1572. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- (9).Roux B. The Calculation of the Potential of Mean Force Using Computer Simulations. Comput Phys Commun. 1995;91(1–3):275–282. [Google Scholar]
- (10).de Ruiter A, Oostenbrink C. Free Energy Calculations of Protein–ligand Interactions. Curr Opin Chem Biol. 2011;15(4):547–552. doi: 10.1016/j.cbpa.2011.05.021. [DOI] [PubMed] [Google Scholar]
- (11).Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J Chem Theory Comput. 2008;4(5):819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- (12).Sengupta D, Marrink SJ. Lipid-Mediated Interactions Tune the Association of Glycophorin A Helix and Its Disruptive Mutants in Membranes. Phys Chem Chem Phys. 2010;12(40):12987–12996. doi: 10.1039/c0cp00101e. [DOI] [PubMed] [Google Scholar]
- (13).Aci-Sèche S, Sawma P, Hubert P, Sturgis JN, Bagnard D, Jacob L, Genest M, Garnier N. Transmembrane Recognition of the Semaphorin Co-Receptors Neuropilin 1 and Plexin A1: Coarse-Grained Simulations. PLoS One. 2014;9(5):e97779. doi: 10.1371/journal.pone.0097779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Arnarez C, Marrink SJ, Periole X. Identification of Cardiolipin Binding Sites on Cytochrome c Oxidase at the Entrance of Proton Channels. Sci Rep. 2013;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MSP. Biophysical and Computational Studies of Membrane Penetration by the GRP1 Pleckstrin Homology Domain. Structure. 2011;19(9):1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lumb CN, Sansom MSP. Finding a Needle in a Haystack: The Role of Electrostatics in Target Lipid Recognition by PH Domains. PLoS Comput Biol. 2012;8(7):e1002617. doi: 10.1371/journal.pcbi.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lai C-L, Srivastava A, Pilling C, Chase AR, Falke JJ, Voth GA. Molecular Mechanism of Membrane Binding of the GRP1 PH Domain. J Mol Biol. 2013;425(17):3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chen H-C, Ziemba BP, Landgraf KE, Corbin JA, Falke JJ. Membrane Docking Geometry of GRP1 PH Domain Bound to a Target Lipid Bilayer: An EPR Site-Directed Spin-Labeling and Relaxation Study. PLoS One. 2012;7(3):e33640. doi: 10.1371/journal.pone.0033640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Klarlund JK, Rameh LE, Cantley LC, Buxton JM, Holik JJ, Sakelis C, Patki V, Corvera S, Czech MP. Regulation of GRP1-Catalyzed ADP Ribosylation Factor Guanine Nucleotide Exchange by Phosphatidylinositol 3,4,5-Trisphosphate. J Biol Chem. 1998;273(4):1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- (20).Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and Promiscuity in Phosphoinositide Binding by Pleckstrin Homology Domains. J Biol Chem. 1998;273(46):30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- (21).Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural Basis for Discrimination of 3-Phosphoinositides by Pleckstrin Homology Domains. Mol Cell. 2000;6(2):373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- (22).Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. Distinct Polyphosphoinositide Binding Selectivities for Pleckstrin Homology Domains of GRP1-like Proteins Based on Diglycine versus Triglycine Motifs. J Biol Chem. 2000;275(42):32816–32821. doi: 10.1074/jbc.M002435200. [DOI] [PubMed] [Google Scholar]
- (23).Corbin JA, Dirkx RA, Falke JJ. GRP1 Pleckstrin Homology Domain: Activation Parameters and Novel Search Mechanism for Rare Target Lipid. Biochemistry. 2004;43(51):16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Manna D, Albanese A, Park WS, Cho W. Mechanistic Basis of Differential Cellular Responses of Phosphatidylinositol 3,4-Bisphosphate- and Phosphatidylinositol 3,4,5-Trisphosphate-Binding Pleckstrin Homology Domains. J Biol Chem. 2007;282(44):32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- (25).Pilling C, Landgraf KE, Falke JJ. The GRP1 PH Domain, like the AKT1 PH Domain, Possesses a Sentry Glutamate Residue Essential for Specific Targeting to Plasma Membrane PI(3,4,5)P(3) Biochemistry. 2011;50(45):9845–9856. doi: 10.1021/bi2011306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural Basis of 3-Phosphoinositide Recognition by Pleckstrin Homology Domains. Mol Cell. 2000;6(2):385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- (27).He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. Molecular Mechanism of Membrane Targeting by the GRP1 PH Domain. J Lipid Res. 2008;49(8):1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lemmon MA. Membrane Recognition by Phospholipid-Binding Domains. Nat Rev Mol Cell Biol. 2008;9(2):99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- (29).Jian X, Tang W, Zhai P, Roy N, Luo R, Gruschus J, Yohe M, Chen P, Li Y, Byrd R, et al. Molecular Basis for Cooperative Binding of Anionic Phospholipids to the PH Domain of the Arf GAP ASAP1. Structure. 2015 doi: 10.1016/j.str.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ceccarelli DFJ, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. Non-Canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9. J Biol Chem. 2007;282(18):13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- (31).Hyvönen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M. Structure of the Binding Site for Inositol Phosphates in a PH Domain. EMBO J. 1995;14(19):4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Knight JD, Falke JJ. Single-Molecule Fluorescence Studies of a PH Domain: New Insights into the Membrane Docking Reaction. Biophys J. 2009;96(2):566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yesylevskyy SO, Schäfer LV, Sengupta D, Marrink SJ. Polarizable Water Model for the Coarse-Grained MARTINI Force Field. PLoS Comput Biol. 2010;6(6):e1000810. doi: 10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Singh G, Tieleman DP. Using the Wimley-White Hydrophobicity Scale as a Direct Quantitative Test of Force Fields: The MARTINI Coarse-Grained Model. J Chem Theory Comput. 2011;7(7):2316–2324. doi: 10.1021/ct2002623. [DOI] [PubMed] [Google Scholar]
- (35).Doudou S, Burton NA, Henchman RH. Standard Free Energy of Binding from a One-Dimensional Potential of Mean Force. J Chem Theory Comput. 2009;5(4):909–918. doi: 10.1021/ct8002354. [DOI] [PubMed] [Google Scholar]
- (36).Vonkova I, Saliba A-E, Deghou S, Anand K, Ceschia S, Doerks T, Galih A, Kugler KG, Maeda K, Rybin V, et al. Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains. Cell Rep. 2015;12(9):1519–1530. doi: 10.1016/j.celrep.2015.07.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.