Abstract
HIV+ patients have an increased risk for tuberculosis disease despite clinical management with ARTs. We established a culture model of Mtb-infection in PBMCs from HIV+ PPD+ donors on suppressive ART (median 6.4 years) with negligible viral loads (median <50 copies/mL) and stable CD4+ T cell counts (517 cells/mm^3). We observed that HIV+ patient lymphocytes harbored a recruitment defect to Mtb-infected macrophages. To investigate these immune defects on a per cell basis, purified CD4+ T cells from HIV patients were assessed by label-free quantification protein mass spectrometry. CD4+ T cells from HIV patients displayed diminished nucleoprotein levels – notably of histone variant H2a.Z and ribonucleoprotein A1. Only within healthy donors, transcriptional regulatory histone variant H2a.Z expression was correlated to the extent of IFN-γ induction upon Mtb-infection. Our findings may explain why HIV patients exhibit prolonged immune cell dysfunction despite suppressive ART, and implicate a per cell defect of CD4+ T cells.
Graphical abstract
1. INTRODUCTION
Globally, tuberculosis (TB) disease remains a highly prevalent and life threatening disease to humans- where it is estimated that about a third of the world’s population is infected with TB (WHO fact sheet 2015). Once an individual is infected with the species Mycobacterium tuberculosis (Mtb), a small subset (5–10%) of the infected population will go on to develop primary active tuberculosis (ATB) disease, while over 90% carry the infection in a latent or subclinical stage (1). This observation highlights the inter-individual variability of host immune responses in the containment of Mtb infection.
Host adaptive immune responses are critical in the effective containment of TB disease, demonstrated by the dramatic increase in the reactivation risk of TB during HIV disease (2). Though successful clinical treatment with antiretroviral therapy (ART) reduces this risk and can restore many aspects of HIV-induced immunological dysfunction (3), incomplete restoration is common, and undermines effector immune responses in HIV+ patients (4). One hallmark of chronic HIV disease is the persistent immunological inflammation observed in HIV patients despite suppressive ART (reviewed in (5)). Immune phenotyping studies have reported that peripheral CD4+ T cells from a subset of HIV+ patients on suppressive ART regimens continue to display features of increased cellular exhaustion (PD-1+) (6), turnover (Ki67+) (7), and senescence (CD28-) (8). Despite these findings, the exact cause of immune dysfunction in HIV patients on fully suppressive ART is unclear. While the inflammatory effects from the local tissue environment and cytokine milieu have been implicated (7), enhanced models of disease at the cellular immunological level are lacking.
Studies modeling ex vivo Mtb infection of HIV+/ART patient peripheral immune cells can provide physiological insights to this dysfunction by recapitulating the dynamic response to tuberculosis disease. These mechanistic immunological studies could explain why CD4+ T cells from HIV patients on suppressive ART exhibit lingering signs of dysfunction- particularly during co-infections with Mtb. The aim of this study was to establish an ex vivo infection model using a live reporter Mtb strain in primary human immune cells. Other groups that have performed similar studies report the formation of granuloma-like immune cell complexes in healthy donor PBMCs (peripheral blood mononuclear cells) infected with Mtb (9,10). To build upon insights from these prior studies, we established a system to assess recall immune responses in patients with prior exposure to TB. We hypothesized that the quality of CD4+ T cell responses predict the likelihood of patients with latent TB to progress to disease, in which the latter is much more likely in patients who are HIV+ and PPD+ (2,4). We collected PBMCs from HIV+ and healthy PPD+ (Purified Protein Derivative) donors, and performed infections with an auxotrophic, gfp (green fluorescent protein)- expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD).
In accordance with our hypothesis that HIV+/ART PPD+ donors would display features of immune anergy, we report that HIV+/ART patients with latent TB (PPD+) indeed display an impaired ability to recruit activated leukocytes to the Mtb-infected core. Furthermore, Mtb-infection cultures from HIV+/ART PPD+ patients failed to produce chemoattractant proteins, pro-inflammatory macrophage cytokines, and Th1 effector cytokines. To understand the cause of this impaired effector mechanism within discrete CD4+ T cells from HIV+/ART PPD+ donors, we assessed the enrichment of proteins from purified primary CD4+ T cells from HIV+/ART patients via LFQ (label-free quantification) protein mass spectrometry. This unbiased screening enables the elucidation of endogenous expression levels of key proteins critical for CD4+ T cells to engage in downstream effector mechanisms, such as those against Mtb-infection. We report a dysregulated transcription factor network (e.g., histone variant H2a.Z, ribonucleoproteins, and c-myc) in CD4+ T cells from HIV patients with limited effector capabilities against Mtb-infected cells. These findings may explain in part the nature of partial CD4+ T cell anergy in HIV+/ART patients, and elucidates potential therapeutic strategies against tuberculosis disease.
2. MATERIALS AND METHODS
2.1 Human subjects
Written informed consent was obtained from all healthy adults and HIV+ patients prior to obtaining the biological specimens and clinical data used in this study (Table 1). Each donor attested to his or her medical fitness and willingness and ability to donate blood. Donors were assessed for their PPD status based on medical history (healthy donors) or clinical chart review (HIV donors). For this latter cohort, tuberculosis disease status was ascertained based on clinical diagnostic chart reviews of HIV patients seen at the outpatient 1917 clinic at the University of Alabama at Birmingham.
Table 1.
Cohort description of PPD+/TB+ donors
| Healthy PPD+ | HIV+ PPD+ | |
|---|---|---|
| n | 20 | 13 |
| Male % | 55.0% | 46.2% |
| Black/African American | 55.0% | 77.0% |
| Median Age (I.Q.R.) | 44 (27.8 – 55.9) | 41.1 (36.1–48.1) |
| Median CD4+ T cell count (I.Q.R.) | \ | 517 (393 – 683) |
| Median CD4+ T cell nadir (I.Q.R.) | \ | 301 (161 – 407) |
| Median Viral Load (I.Q.R.) | \ | 49 (47 – 49) |
| Median years of ART (I.Q.R.) | \ | 6.4 (1.6 – 11.9) |
2.2 Isolation of plasma and primary peripheral bloods cells
PBMCs were isolated from peripheral blood drawn into EDTA (ethylenediaminetetraacetic acid) tubes by density gradient centrifugation using Ficoll–Paque Plus (GE Healthcare Life Sciences, Uppsala, SE). PBMCs washed with PBS (phosphate buffered saline) were frozen in aliquots of fetal bovine serum (FBS) with 10% dimethyl sulfoxide (Sigma, St Louis, MO), and stored in a liquid nitrogen cryofreezer until use. Cryopreserved PBMCs were rapidly thawed and washed in RPMI 1640 (HyClone GE Healthcare Life Sciences, Logan, UT) supplemented with 10% FBS (HyClone), 1% penicillin/streptomycin and 2 mM L–glutamine (Corning Cellgro, Manassass, UA) (hereafter referred to as R10++ medium). PBMCs were counted and assessed for viability with a Guava EasyCyte flow cytometer (EMD Millipore, Billerica, MA), pelleted, and then re–suspended at an adequate cell number for subsequent experiments. Cellular viability was further defined by positivity with an amine reactive dead cell dye, and the median lymphocyte viability ranged from 85–95%.
2.3 Ex vivo Mtb primary infection cultures and quantification of surface area using ICY software
PBMCs were infected with Mtb at an multiplicity of infection (MOI) of 1:1 (bacillus:cell) in the antibiotic-free RPMI + 10% FBS media in 24 well polystyrene cell culture plates at 1 million cells per 1 mL of culture media. Tuberculosis infections were performed with an auxotrophic, gfp - expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD) chosen to allow experiments under BSL2 conditions (11,12). For infection culture studies of monocyte, PBMCs from each donor were plated onto 24-well plates as indicated above, and non-adherent cells were removed after 2 hours of culture. The remaining adherent monocytes were subsequently infected with Mtb at an MOI of 1:1 (bacillus:cell) after estimating a 10% composition of monocytes within donor PBMCs. The Mtb H37Rv derived auxotroph strain was grown as previously described, and the growth media was supplemented with 24 µg/ml pantothenate and 50 µg/ml L-leucine (12). Culture images over the course of 1 – 4 days were captured on the EVOS Cell Imaging Systems (Thermo Fisher Scientific) in overlapping bright field and green fluorescence field, and an average of 20 separate captures in 10× fields per sample were evaluated and collected in triplicates to quantify mean surface areas for each infection culture (µm^2). Measurement of leukocyte aggregate surface area was captured for each Mtb (gfp)-containing lymphocyte aggregate with the freehand ellipse tool in the Icy bioimage informatics platform http://icy.bioimageanalysis.org/ (13).
2.4 Bioplex multi-analyte assay
Multi-analyte assays were performed using the MILLIPLEX® MAP Human Cytokine/Chemokine 41-plex Magnetic Bead Panel Immunology Multiplex Assay (Merck Milliplore, Billerica, MA) designed to measure a panel of human cytokines and chemokines in multiplex. 24-hour supernatant cultures were collected and assayed according to the protocol provided by the manufacturer (Merck Milliplore), and samples were analyzed using a Bio-Plex 200 instrument. The output median fluorescence intensity (MFI) data was analyzed using weighted 5-parameter logistic for calculating the cytokine and chemokine concentrations in the samples based on the standard curves. Data analysis was performed using Bio-Plex manager 6.0 software (Bio-Rad, Hercules, CA). Cytokine responses in the Mtb infection culture were calculated as the difference in the mean measured cytokine of the background expression levels to the experimental condition.
2.5 Mass Spectrometry
Sample Preparation
Cryopreserved PBMCs were thawed, and proteins were extracted from untouched CD4+ T cells (CD4+ T cell enrichment kit from Stemcell Technologies, Cambridge, MA) using the mammalian protein extraction reagent (M-PER) (Thermo Fisher Scientific, Waltham, MA). Proteins were then quantified using Pierce Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Fisher Scientific). Following the separation of 10 µg of protein by 1D polyacrylamide gel electrophoresis, each lane was cut into equal molecular weight fractions and prepared for analysis via nano-HPLC electrospray ionization multistage tandem mass spectrometry according to the methods published by our group previously (14).
Peptide Filtering, Grouping, and Quantification
The list of peptide identities generated based on SEQUEST search results were filtered using Scaffold (Proteome Software, Portland Oregon). The Scaffold program was used to filter and group all peptides to generate and retain high confidence identities, while also generating normalized spectral counts (N-SC’s) across all samples for relative quantification. The filter cut-off values were set with minimum peptide length of >5 amino acids, with no 1+ charge states, peptide probabilities of >80% confidence intervals, and with the number of peptides per protein set at ≥2. In addition, upon setting protein probabilities to a >99.0% confidence interval, and with a false discovery rate (FDR) of <1.0, 1267 high confidence proteins were identified. Relative quantification across experiments was performed via spectral counting (15,16), and spectral count abundances were subsequently normalized between samples.
Pairwise Statistical & Systems Analysis
For the proteomic data generated, two separate non-parametric statistical analyses were performed between each pair-wise comparison. These non-parametric analyses included (1) the calculation of weight values by significance analysis of microarray (SAM; cut off >|0.6| (17)) and (2) t-test (single tail, unequal variance, cut off of p < 0.05), which were subsequently sorted according to the highest statistical relevance in each comparison. For protein abundance ratios determined with N-SC’s, we set a ≥1.5 fold change as the threshold for significance, determined empirically by analyzing the inner-quartile data from the control experiment indicated above using natural logarithmic plots, where the Pearson’s correlation coefficient (R) was 0.98, and >99% of the normalized intensities fell between +/−1.5 fold. For these analyses, a passing score was defined as any two of the three tests (SAM, t-test, or fold change) achieving their defined respective threshold.
Gene ontology assignments and pathway analysis were carried out using MetaCore using non-supervised protein interaction networks (GeneGO Inc., St. Joseph, MI). Interactions identified within MetaCore were manually correlated using curated full text articles within the software’s algorithms (18,19).
2.6 Statistical analysis
All analyses with more than 2 groups were performed with 2-way ANOVA, with a Sidak’s multiple comparisons post-test correction. Analyses comparing 2 groups were performed with Mann-Whitney test for non-normally distributed values. All correlation analyses were performed with Pearson correlation coefficient assessment of normally distributed data. Within the manuscript text, all clinical experimental values are reported as median ± interquartile range. The statistical software used was GraphPad Prism v5.0d (Graphpad Software, La Jolla, CA, USA). P values for all statistical analyses are depicted as follows: *p<0.05, **p<0.01, ***p<0.001.
3. RESULTS
3.1 HIV patients display a leukocyte recruitment defect in an ex vivo culture model of Mtb-infected donor monocytes
Infection with HIV is a significant risk factor that predisposes an individual for Mtb infection and subsequent progression to active disease, and can be reduced with the initiation with ART (3). T cells from HIV patients display a diminished ability to respond to Mtb protein/peptide antigen, partially explaining why host immunity is compromised within this cohort (20). In order to understand host immune mediators of Mtb infection during the course of HIV disease during successful ART, PPD+ donors were recruited for the study. HIV+/ART PPD+ donors matched by age and gender to healthy PPD+ donors were assessed for their current CD4+ T cell counts, CD4+ T cell nadir, viral load, and years of successful ART (Table 1).
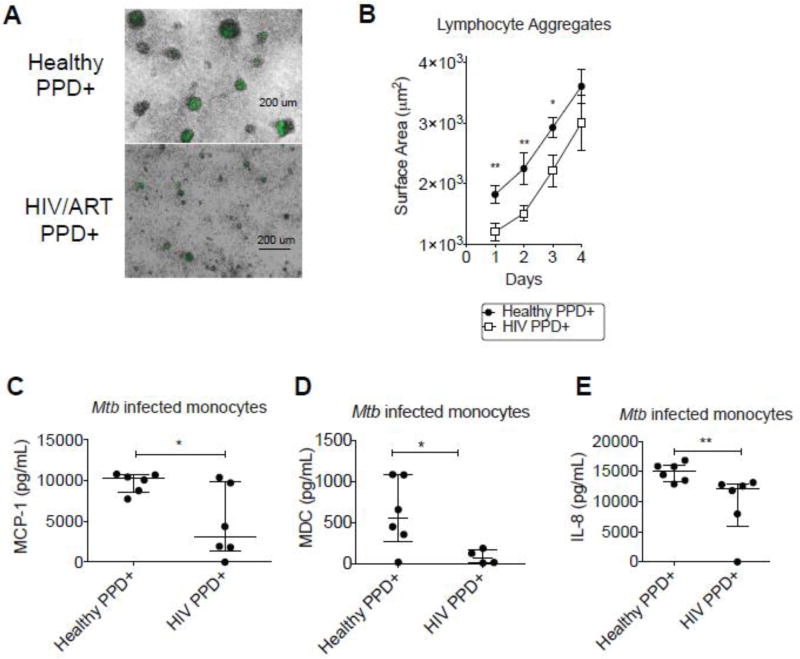
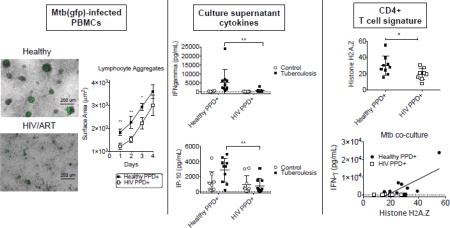
Ex vivo Mtb infection models have been established before, with some reporting the formation of large granuloma structures in PBMCs infected with Mtb (9). A recent comprehensive study established an in vitro granuloma model in PBMCs and established differences between healthy individuals and those with latent TB infection (10). We assessed immune cells from HIV+/ART patients with prior history of TB to determine if there was an impaired immune response to Mtb infection. We isolated PBMCs from these patients and set up an ex vivo model of infection with a reporter gfp expressing Mtb organism (11). We observed that leukocytes from HIV+PPD+ donors formed smaller and more diffuse aggregates around gfp+ Mtb infected host monocytes (Figure 1A). To determine a more quantitative assessment of leukocyte aggregation to gfp+ Mtb infected monocytes, images were captured using fluorescence microscopy. Upon 24 hours of culture at an MOI (multiplicity of infection) of 1:1, the leukocyte aggregates surrounding Mtb-infected monocytes were quantified. The surface area of leukocyte aggregates was diminished in HIV+/ART+ PPD+ donors (1205 ± 512 µm^2) compared to those of healthy PPD+ donor PBMCs (1820 ± 641 µm^2) (Figure 1B). This difference persisted into Days 2–3 post-infection, with leukocyte aggregates approaching 2925 ± 723 µm^2 in healthy PPD+ donor samples, and 2215 ± 940.7 µm^2 in HIV+/ART PPD+ samples (Figure 1B).
Figure 1. HIV patients display a leukocyte recruitment defect in an ex vivo culture model of Mtb-infected donor monocytes.
(A) Cryopreserved PBMCs from healthy PPD+ donors (n=20) and HIV+ PPD+ donors on ART (n=13) were infected with an auxotrophic, gfp -expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD) at an MOI of 1:1, and representative 24-hour culture images were captured and shown here as merged bright and GFP fields. Represented donors are a subset of individuals represented in Table 1.
(B) Mean surface areas (um^2) for leukocyte aggregates clustering around Mtb infected monocytes were quantified with the freehand ellipse tool in the Icy bioimage informatics platform http://icy.bioimageanalysis.org/. An average of 20 separate captures in 10× fields per sample were evaluated and collected in triplicates to quantify mean surface areas for each infection culture (µm^2), and statistical analysis was performed with a Mann-Whitney test for non-normally distributed values (P values indicated as follows: *p<0.05, **p<0.01, ***p<0.001). Data represent the mean +/− standard error of the mean (SEM) for 10 donors in each group.
(C) MCP-1 (D) MDC and (E) IL-8 were assessed by multiplex cytokine analysis from 24-hour culture supernatants from each donor. Cryopreserved PBMCs were infected with an auxotrophic, gfp -expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD) at an MOI of 1:1 24-hour cultures). Horizontal lines indicate median of each value, with positive and negative error bars depicting interquartile ranges. Data were analyzed using Mann-Whitney test for non-normally distributed values (P values indicated as follows: *p<0.05, **p<0.01, ***p<0.001).
To determine the cause of this leukocyte recruitment defect in HIV+ donors, we assessed a panel of human cytokines and chemokines on 24-hour supernatants from each infection culture. Out of the various soluble factors assessed, MCP-1 and MDC were most potently diminished in isolated Mtb-infected monocytes, suggesting that the induction of this class of chemoattractant proteins was inherently impaired in monocytes from HIV patients (Figure 1C and 1D). These monocyte chemoattractant proteins function to recruit leukocytes, and may be recruiting Mtb-reactive leukocytes to infected monocytes (21). Additionally, the neutrophil chemoattractant IL-8 was also diminished in monocytes from HIV patients (Figure 1E).
In summary, HIV donor leukocytes are diminished in their ability to recruit leukocytes to Mtb-infected monocytes, and may reflect a defect in the induction of leukocyte chemoattractant proteins.
3.2 Culture supernatants from HIV donor ex vivo model of Mtb- infection have diminished induction of inflammatory IL-1 family cytokines
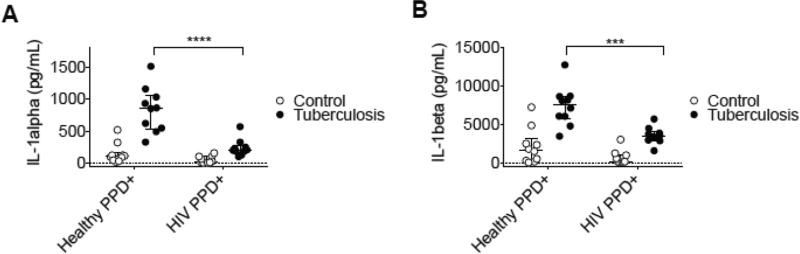
Based on the observation that HIV donor ex vivo cultures had a diminished induction of leukocyte recruitment cytokines, we determined whether additional effector cytokines were altered in HIV donor Mtb infection cultures. It has been recently reported that IL-1α is an upstream regulator of other leukocyte recruitment cytokine networks (e.g., IL-6/IL-8) (22). Additionally, there are multiple roles of human recombinant interleukin 1 during homeostasis and disease (23), and via a macrophage-mediated mechanism (24). Due to their potential role in containment of Mtb-infection and recruitment of activated leukocytes, we assessed IL-1 family cytokines in the culture supernatants from the primary Mtb infection cultures.
Assessment of culture supernatants revealed that donor PBMCs from HIV+/ART patients had diminished levels of IL-1 family cytokines (Figure 2A). This result strongly corroborates a recent study reporting that IL-1α was the strongest predictor of Mtb infection within HIV patients (25), underscoring its critical effector role in response to infection. Similarly, the same result was seen with IL-1β (Figure 2B), and is significant given its role in host resistance to M. tuberculosis (26). These findings indicate that the leukocyte recruitment deficit observed in Mtb-infection cultures from HIV+/ART donors is concurrent with an impaired induction of the IL-1 family of effector cytokines.
Figure 2. Culture supernatants from HIV donor ex vivo model of Mtb- infection have diminished induction of inflammatory IL-1 family cytokines.
(A) IL-1α and (B) IL-1β were assessed by multiplex cytokine analysis from 24-hour culture supernatants from each donor. Cryopreserved PBMCs were infected with an auxotrophic, gfp - expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD) at an MOI of 1:1 24-hour cultures). Scatter plots depicting each of these values are represented as open circles indicating values from uninfected PBMCs in control wells or closed circles indicating values from Mtb infected wells. H Horizontal lines indicate median of each value, with positive and negative error bars depicting interquartile ranges. Data were analyzed using 2-way ANOVA statistical analysis with a Sidak post-test (P values indicated as follows: *p<0.05, **p<0.01, ***p<0.001).
3.3 Th1 signaling pathways are dysfunctional from an ex vivo Mtb culture infection model from HIV donors
Based on our finding that Mtb-infection cultures from HIV donors have an impairment of both leukocyte recruitment capabilities and the induction of the inflammatory IL-1 family of cytokines, we asked whether T helper immune responses known to be protective against Mtb were also attenuated. Prior studies have implicated the role of IL-1 in the activation of T lymphocytes (27) and in priming their proliferation and antigen-specific functions (28,29). Additionally, studies in clinical tuberculosis patients and in animal models of disease have shown that efficient control of TB requires a robust “Type 1” immune response, elucidated in mouse models of disease (30), and in human genetic studies of IL-12 receptor deficiency (31).
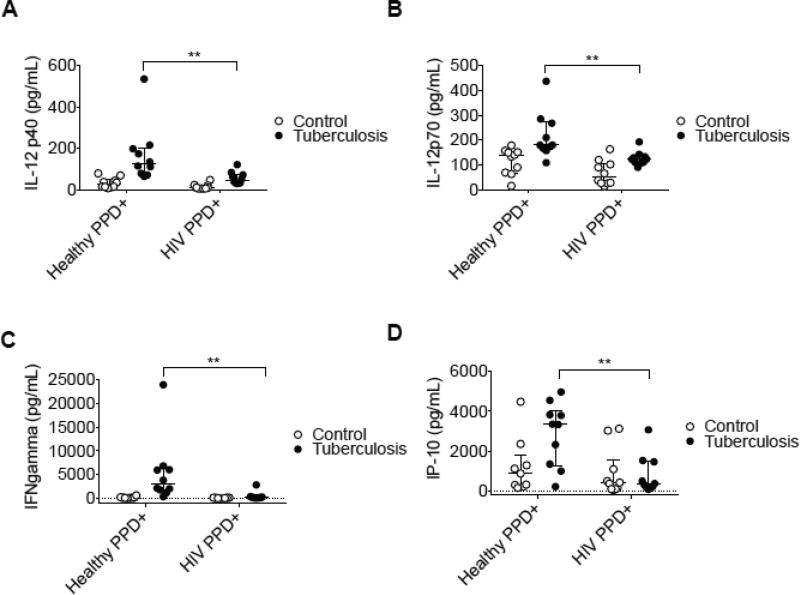
We assessed Th1 polarizing cytokines (IL-12) as well as Th1-defining cytokines (IFN-γ, and IP-10) in the culture supernatants from HIV donor PBMC Mtb infection cultures. IL-12p70 is a Th1 priming cytokine secreted by activated macrophages, and has been shown in in vitro (32) and in vivo models of Mtb infection (33). We show here that levels of IL-12p40 and IL-12p70 are impaired in culture infection supernatants in HIV patients, in accord with the fact that there may be diminished priming of monocytes during chronic HIV disease (Figure 3A and 3B).
Figure 3. Th1 signaling pathways are dysfunctional from an ex vivo Mtb culture infection model from HIV donors.
(A) IL12p40 (B) IL12p70 (C) IFN-γ and (D) IP-10 were assessed by multiplex cytokine analysis from 24-hour culture supernatants from each donor. Cryopreserved PBMCs were infected with an auxotrophic, gfp -expressing Mtb strain (H37Rv derivative, ΔpanCD, ΔleuD) at an MOI of 1:1 24-hour cultures). Scatter plots depicting each of these values are represented as open circles indicating values from uninfected PBMCs in control wells or closed circles indicating values from Mtb infected wells. Horizontal lines indicate median of each value, with positive and negative error bars depicting interquartile ranges. Data were analyzed using 2-way ANOVA statistical analysis with a Sidak post-test (P values indicated as follows: *p<0.05, **p<0.01).
IFN-γ is a major product of Th1 cells and influences naïve CD4+ T cell differentiation toward a Th1 phenotype (34). In addition to the reduction of Th1 polarizing cytokines in HIV culture supernatants, the primary Th1-associated cytokine IFN-γ was also diminished (Figure 3C). IP-10 (IFN-γ - inducible protein) is induced in cells upon stimulation with IFN-γ (35). We report that IP-10, a Th1 associated cytokine, is also reduced in HIV culture supernatants (Figure 3D). This finding was significant in consideration of the critical role that IP-10 has in effector T cell generation and trafficking (36). Additionally, we report that Mtb - infected leukocytes potently induce these Th1 effector molecules from day 1 – 4 of infection in healthy PPD+ donors, whereas the effect was not observed in HIV+/ART PPD+ donor cultures (Supplemental Figure 1A and 1B).
These findings indicate that HIV donors not only have a deficit in their leukocyte recruitment capacity and induction of IL-1 family of effector cytokines, there is also a profound defect in their ability to mount a protective Th1 response against Mtb.
3.4 The Protein Interaction Network (PIN) of CD4+ T cells from healthy vs. HIV+ donors reveal diminished expression of transcriptional nucleoproteins
To determine whether baseline T cell phenotypes were altered in our HIV+/ART patient cohort, we performed a cell surface expression analysis of markers of T cell exhaustion (PD-1) (37), senescence (CD28) (8), and activation (CD38) (38) (Supplemental Table 1). Within our cohort of ART suppressed HIV patients, there were no significant differences in T cell phenotype - likely due to the prolonged length of successful ART suppression (Supplemental Figure 2).
Due to the lack of obvious surface phenotype that may define the Th1-polarized immune cell defect, we decided to perform an unbiased screen of CD4+ T cells from a subset of HIV patients (Supplemental Table 1). We have previously shown that cellular anergy contributes to cellular dysfunctionality of discrete CD4+ T cells from HIV patients (39). Cellular anergy results in the inability to mount a complete response against target - oftentimes induced by the chronic nonspecific stimulation during viral infections (40,41). Within T cells, this influences cell proliferation, cytokine induction, cell signaling, and cell metabolism (42). In our prior work, we demonstrated that latently HIV infected T cells display hallmarks of cellular anergy – exhibited by attenuated NF-κB induction and altered levels of T cell anergy-related proteins (e.g., GRAIL, ICER, and Dryk1a) (39).
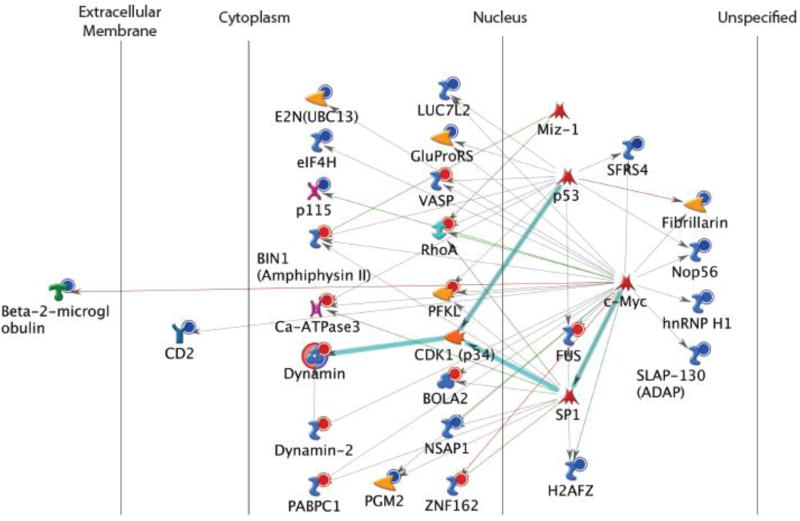
Upon performing LFQ protein mass spectrometry on purified CD4+ T cells from HIV+/ART patients, we observed several enriched PIN (protein interaction network) signatures (Figure 4). The most enriched PIN signature was of the key nucleoproteins- notably of the histone protein variant (H2AFZ) and ribonucleoprotein A1 (hnRNP H1). Both of these nucleoproteins were diminished in discrete CD4+ T cells from HIV+/ART patients. Upon assessment of protein enrichment by normalized spectral count (N-SC) values, beta-2 microglobulin, and CD2 (LFA-2) were also shown to be potently diminished in CD4+ T cells from HIV+ donors. These results indicated that a network of immune priming and signaling molecules were defective during chronic HIV disease (Supplemental Figure 3). Taken together, these findings highlight the coordination of several key endogenous nucleoproteins within CD4+ T cells that are poised to initiate immune cell responses upon TCR engagement (i.e., cognate antigen recognition). The full GO (gene ontology) enrichment analysis report of the top 5 networks is listed in Supplemental Table 2.
Figure 4. The Protein Interaction Network (PIN) of CD4+ T cells from healthy PPD+ vs. HIV+ PPD+ donors reveal diminished expression of transcriptional nucleoproteins.
Purified CD4+ T cells from donors (S. Table 1) were harvested, and protein lysates were purified and loaded equally (10 µg) onto a nano-HPLC electrospray ionization multistage tandem mass spectrometer. Gene ontology assignments and pathway analysis were carried out using MetaCore using non-supervised protein interaction networks. The Scaffold program was used to filter and group all peptides to generate and retain high confidence identities, while also generating normalized spectral counts (N-SC’s) across all samples for relative quantification. Of the 1267 proteins identified upon setting protein probabilities to a >99.0% confidence interval, non-parametric analyses included (1) the calculation of weight values by significance analysis of microarray (SAM; cut off >|0.6| (17)) and (2) t-test (single tail, unequal variance, cut off of p < 0.05), which were subsequently sorted according to the highest statistical relevance in each comparison. The pathway legend can be found on the Metacore website: https://ftp.genego.com/files/MC_legend.pdf
Our results from this unbiased screening approach to determine whether “anergy-like” defects could be observed in CD4+ T cells from our HIV+/ART patient cohort reveal the profound attenuation of a complex of proteins that initiate transcriptional regulation- notably of key nucleoproteins involved in transcriptional regulation (e.g., H2a.Z and hnRNP h1 proteins).
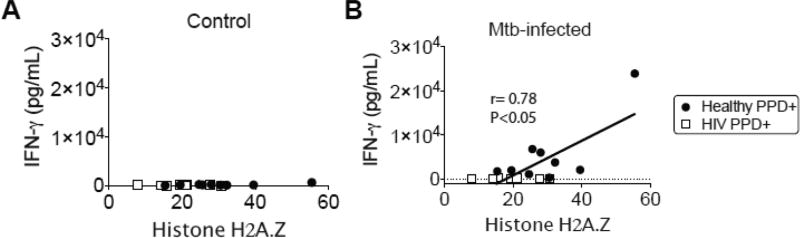
3.5 CD4+ T cell levels of endogenous histone isoform (H2a.Z) is correlated with IFN-γ induction in healthy PPD+ donors
We next explored whether the endogenous nucleoprotein attenuation in CD4+ T cells from HIV patients may correlate with the inter-individual variability observed in Mtb-induced Th1 responses. Based on our prior data showing a robust induction of several Th1-priming and – associated cytokines (Figure 3), we performed correlation assessment to these cytokine levels. We assessed the normalized spectral count (N-SC) mass spectrometry data for relative quantification of H2a.Z and correlated this value to the ex vivo Mtb infection culture supernatant levels from each donor. We show that the levels of endogenous H2a.Z in PPD+ donors were correlated to the extent of IFN-γ induction from healthy donors (r= 0.78, P<0.05) but not in HIV+ PPD+ donors (r= 0.03, P<n.s.) (Figure 5A and 5B).
Figure 5. CD4+ T cell levels of endogenous histone isoform (H2a.Z) is correlated with IFN-γ induction in healthy PPD+ donors.
The normalized spectral count (N-SC) mass spectrometry data for relative quantification of H2a.Z was correlated this value to the ex vivo Mtb infection culture supernatant levels from each donor. H2a.Z relative quantification correlation analyses were performed for IFN-γ cytokine concentrations in (A) control and (B) Mtb-infected cultures (r= 0.78, P<0.05) from healthy PPD+ (r= 0.78, P<0.05) and HIV+ PPD+ (r= 0.03, P<n.s.) donors. Statistical analyses were performed with Pearson’s correlation coefficient.
Our results implicate that the inability of patient CD4+ T cells to mount a recall response to Mtb antigen may be tightly correlated with the diminished levels of endogenous nucleoproteins at the individual cellular level. These transcription factor proteins are likely poised to mobilize the cellular transcriptional machinery upon host recognition of cognate antigen. This finding is significant as it suggests a potential mechanism for the immune hypo-responsiveness to Mtb recall antigen within HIV patients (43–45). These findings may elucidate why chronic HIV patients have such poor clinical outcomes related to TB disease management. Studies aimed at improving the transcriptional induction of critical Th1 effector molecules, through specific activation of key transcription factors and their associated regulatory proteins, may be one approach to improve immune responses in this cohort.
In conclusion, we report that there is a significant association between the attenuated levels of the transcriptional regulatory nucleoproteins, and the subsequent ability of the CD4+ T cells from HIV+/ART patients to induce Th1 effector cytokines upon exposure to Mtb antigen.
4. DISCUSSION
Antiretroviral therapy (ART) has been successful at extending years of life in HIV patients (46), suppressing viral replication (47), and increasing CD4+ T cell numbers (48). However, immunological complications persist in the T cell compartment from HIV patients on fully suppressive ARTs (6–8), perhaps caused by ineffective containment of HIV in localized tissue inflammation sites. This may lead to long-term consequences on immune cells during suppressive ART, thus influencing the containment of co-pathogens such as Mtb.
To address these questions, we performed an immunological study assessing Mtb recall responses from HIV+/ART patients. We report that Mtb-infected monocytes from HIV+/ART patients display diminished lymphocyte recruitment and an overall defect in recruiting leukocytes to infected cells (Figure 1A and 1B). We also show that Mtb-infected monocytes from HIV+/ART patients display diminished induction of the chemoattractant proteins MCP-1/ and MDC (Figure 1C and 1D). Concurrently, we observed a lack of IL-1α/β (Figure 2) and Th1 priming cytokines in the Mtb infection cultures from HIV patient immune cells (Figure 3). To determine whether these downstream effector functions were influenced by basal CD4+ T cell defects, we performed a label-free quantification (LFQ) high throughput mass spectrometry screen of purified CD4+ T cells from HIV patients. Indeed, we discovered that CD4+ T cells from HIV+/ART patients possessed diminished levels of endogenous regulatory transcriptional elements (Figure 4 and 5).
These findings illustrate the paradigm of discretely dysfunctional CD4+ T cells from chronically infected HIV patients – despite effective clinical management with ART. This novel finding is mutually exclusive to the clinical reports showing that diminished CD4+ T cell count strata are robustly predictive for the risk for Mtb infection and subsequent progression to active disease (3,49). Our study adds an important dimension to this well-documented clinical observation, and reveals that there is a dysfunctional signature on a “per cell” basis within CD4+ T cells from HIV patients on chronic ART. These findings were elucidated by utilizing an ex vivo model of Mtb infection- enabling a better understanding of functional recall responses to live Mtb organism. Additionally, they may address some of the root causes for the increased false negative tuberculin skin test (TST) results within HIV+TB+ due to immune cell anergy (50,51). Addressing this “per cell” defect can give us a better sense of the role of how both the innate and adaptive immune response work together to contain and eradicate TB – so that enhanced diagnostics and vaccines can be developed.
Mass spectrometry (MS)- based proteomic analyses is a powerful tool for the discovery of novel targets that are altered in host immune cells, particularly when linked to their downstream effector responses. The ability to perform label-free quantification (LFQ) methods for an unbiased readout of novel targets reveals a better insight into the global cellular defects inherent in primary immune cells during disease states. These MS methods for the global quantification of proteins have allowed for the discovery of hundreds of candidate biomarkers of disease, demonstrated by a recent study of host protein biomarkers identifying active tuberculosis in HIV uninfected and co-infected individuals (52). In our study, we utilized this as a powerful discovery tool to assess the “per cell” defect of purified CD4+ T cells from HIV patients (S. Table 1). The use of label-free quantification (LFQ) high throughput mass spectrometry, as well as the equal loading of protein concentrations of purified CD4+ T cells from all donors, facilitates the discovery of endogenous cellular defect of discrete cells in a highly unbiased fashion. Utilizing this next-generation technology, we discovered that CD4+ T cells from HIV+/ART patients possessed diminished levels of endogenous regulatory transcriptional elements that have been previously reported to exert important CD4+ T cell effector functions (Figure 4 and 5).
One of these elements, the nucleoprotein histone 2a.Z isoform (H2a.Z), has been shown to control the extent of nucleosome positioning at the -1 nucleosome of the transcription start sites (TSS) of several key genes upon TCR activation in CD4+ T cells (53). In this study, genomewide maps of nucleosome positions in both resting and activated human CD4+ T cells were generated, and H2a.Z was elevated upon TCR engagement and signaling. This in turn contributed to the extensive nucleosome reorganization in promoters/enhancers to allow transcriptional activation in activated CD4+ T cells (53). A later study confirmed that this histone isoform promotes transcriptional competence (54). Concordant with these finding, our observation that the histone variant H2a.Z was diminished in CD4+ T cells from HIV+/ART patients may allude to a defect in the repositioning of critical transcription factors necessary to activate effector immune molecules upon cellular activation. Additionally, another regulatory protein elucidated from our screen was the heterogeneous nuclear ribonucleoprotein h1 isoform (hnRNP h1), an RNA binding proteins that forms a complex with heterogeneous nuclear RNA. Interestingly, it has been reported that this class of regulatory proteins are subverted by HIV-1 molecular mechanisms within the infected CD4+ T cell. Notably, this group reported that HIV-1 mediated the suppression of T cell activation through the subversion of complex transcriptional networks inside the infected cell (55).
These combined findings (enriched cellular processes listed in S. Table 2) highlights the paradigm of a dysregulated transcription factor network (e.g., histone variant H2a.Z, c-myc, and ribonucleoproteins) in CD4+ T cells from HIV patients with limited effector capabilities against Mtb-infected cells. These findings may explain in part the nature of CD4+ T cell anergy in HIV+/ART patients, and elucidates potential therapeutic strategies against tuberculosis disease.
5. CONCLUSIONS
In this study, we report the novel application of a lab-adapted reporter Mtb infection model for use in primary immune cell culture. On the basis of this model, we report that there is a leukocyte recruitment defect to Mtb-monocytes from HIV+/ART donors. Furthermore, we show that HIV patient cultures had reduced MCP and MDC chemoattractants, IL-1α/β, and Th1 cytokines. In terms of clinical application, these results may explain in part why many HIV+ TB+ patients display cutaneous anergy for the PPD tuberculin skin test. Additionally, these findings shed an important mechanistic pathway, combining aspects of both innate and adaptive immunity, as to why effector T cell responses to Mtb are limited in HIV patients even after suppressive ART. Additionally, an unbiased LFQ (label-free quantification) protein mass spectrometry analysis on purified CD4+ T cells from HIV patients reveals a defect of key regulatory proteins. We report a dysregulated transcription factor network (e.g., histone variant H2a.Z, and various ribonucleoproteins) in CD4+ T cells from HIV patients with limited effector capabilities against Mtb-infected cells. However, the small number of patient samples, as well as the use of an auxotrophic Mtb strain, limits the generalizability of this study. Validation within a larger cohort of HIV patients, as well as a broader cohort of immune suppressed individuals, can translate this finding into enhanced therapeutics and enhanced tuberculosis diagnostics.
Supplementary Material
Highlights.
There is a lymphocyte recruitment defect to Mtb-macrophages from HIV+/ART donors
HIV+/ART Mtb-infection cultures have diminished, IL-1a/b and Th1 cytokine induction
Mass spectrometry analysis on purified CD4+ T cells from HIV patients was performed
Label free quantification was performed to generate unbiased Protein Interaction Networks
There is a “per-CD4+ T cell” defect of nucleoproteins in patients with poor Mtb-responses
Acknowledgments
We thank our collaborators and participants at the Jefferson County department of Public Health, the UAB Vaccine Research Center, the UAB Comprehensive Cancer Center and Mass Spectrometry Shared Facility, and the NIH Fogarty International Center. We also thank Marion Spell at the UAB CFAR flow cytometry core, and Dr. Jim Sun for invaluable advice on establishing Mtb infection cultures.
FUNDING
This work was supported by the National Heart Lung and Blood Institute at the National Institutes of Health (NHLBI/NIH) (F32 HL121924 to L.S.). This work was also supported by a National Cancer Institute (NCI/NIH) grant (P30 CA013148) to the UAB Comprehensive Cancer Center and by the National Center for Advancing Translational Research (NCATS/NIH) (UL1TR001417).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST DISCLOSURE
The authors declare no conflict of interest.
References
- 1.Vynnycky E, Fine PE. Lifetime risks, incubation period, and serial interval of tuberculosis. Am J Epidemiol. 2000 Aug 1;152(3):247–63. doi: 10.1093/aje/152.3.247. [DOI] [PubMed] [Google Scholar]
- 2.Selwyn PA, Hartel D, Lewis VA, Schoenbaum EE, Vermund SH, Klein RS, et al. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N Engl J Med. 1989 Mar 2;320(9):545–50. doi: 10.1056/NEJM198903023200901. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland JS, Young JM, Peterson KL, Sanneh B, Whittle HC, Rowland-Jones SL, et al. Polyfunctional CD4(+) and CD8(+) T cell responses to tuberculosis antigens in hiv-1-infected patients before and after anti-retroviral treatment. J Immunol. 2010 Jun 1;184(11):6537–44. doi: 10.4049/jimmunol.1000399. [DOI] [PubMed] [Google Scholar]
- 4.Girardi E, Sabin CA, d'Arminio Monforte A, Hogg B, Phillips AN, Gill MJ, et al. Incidence of tuberculosis among hiv-infected patients receiving highly active antiretroviral therapy in europe and north america. Clin Infect Dis. 2005 Dec 15;41(12):1772–82. doi: 10.1086/498315. [DOI] [PubMed] [Google Scholar]
- 5.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–55. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabmeier-Pfistershammer K, Steinberger P, Rieger A, Leitner J, Kohrgruber N. Identification of PD-1 as a unique marker for failing immune reconstitution in hiv-1- infected patients on treatment. J Acquir Immune Defic Syndr. 2011 Feb 1;56(2):118–24. doi: 10.1097/QAI.0b013e3181fbab9f. [DOI] [PubMed] [Google Scholar]
- 7.Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis. 2011 Oct 15;204(8):1217–26. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vivar N, Ruffin N, Sammicheli S, Hejdeman B, Rethi B, Chiodi F. Survival and proliferation of CD28- T cells during HIV-1 infection relate to the amplitude of viral replication. J Infect Dis. 2011 Jun 1;203(11):1658–67. doi: 10.1093/infdis/jir156. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor N, Pawar S, Sirakova TD, Deb C, Warren WL, Kolattukudy PE. Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS One. 2013;8(1):e53657. doi: 10.1371/journal.pone.0053657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guirado E, Mbawuike U, Keiser TL, Arcos J, Azad AK, Wang SH, Schlesinger LS. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio. 2015 Feb 17;6(1):e02537–14. doi: 10.1128/mBio.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson SL, Dascher CC, Sambandamurthy VK, Russell RG, Jacobs WR, Bloom BR, Hondalus MK. Protection elicited by a double leucine and pantothenate auxotroph of mycobacterium tuberculosis in guinea pigs. Infect Immun. 2004 May;72(5):3031–7. doi: 10.1128/IAI.72.5.3031-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Schaaf K, Duverger A, Wolschendorf F, Speer A, Wagner F, et al. Protein phosphatase, mg2+/mn2+-dependent 1A controls the innate antiviral and antibacterial response of macrophages during HIV-1 and mycobacterium tuberculosis infection. Oncotarget. 2016 Mar 29;7(13):15394–409. doi: 10.18632/oncotarget.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat Methods. 2012 Jun 28;9(7):690–6. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 14.Meares GP, Liu Y, Rajbhandari R, Qin H, Nozell SE, Mobley JA, et al. PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol Cell Biol. 2014 Oct;34(20):3911–25. doi: 10.1128/MCB.00980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005 Oct;4(10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004 Jul 15;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 17.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999 Oct 15;286(5439):531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 18.Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: Open-source protein annotation software with data visualization. Anal Chem. 2009 Dec 1;81(23):9819–23. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekins S, Bugrim A, Brovold L, Kirillov E, Nikolsky Y, Rakhmatulin E, et al. Algorithms for network analysis in systems-adme/tox using the metacore and metadrug platforms. Xenobiotica. 2006;36(10–11):877–901. doi: 10.1080/00498250600861660. [DOI] [PubMed] [Google Scholar]
- 20.Geldmacher C, Schuetz A, Ngwenyama N, Casazza JP, Sanga E, Saathoff E, et al. Early depletion of mycobacterium tuberculosis-specific T helper 1 cell responses after HIV-1 infection. J Infect Dis. 2008 Dec 1;198(11):1590–8. doi: 10.1086/593017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007 Apr;117(4):902–9. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound il-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17031–6. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinarello CA, Cannon JG, Mier JW, Bernheim HA, LoPreste G, Lynn DL, et al. Multiple biological activities of human recombinant interleukin 1. J Clin Invest. 1986 Jun;77(6):1734–9. doi: 10.1172/JCI112495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juffermans NP, Florquin S, Camoglio L, Verbon A, Kolk AH, Speelman P, et al. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J Infect Dis. 2000 Sep;182(3):902–8. doi: 10.1086/315771. [DOI] [PubMed] [Google Scholar]
- 25.Waruk JL, Machuki Z, Mesa C, Juno JA, Anzala O, Sharma M, et al. Cytokine and chemokine expression profiles in response to mycobacterium tuberculosis stimulation are altered in hiv-infected compared to hiv-uninfected subjects with active tuberculosis. Tuberculosis (Edinb) 2015 Sep;95(5):555–61. doi: 10.1016/j.tube.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, et al. Caspase-1 independent il-1beta production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol. 2010 Apr 1;184(7):3326–30. doi: 10.4049/jimmunol.0904189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichtman AH, Chin J, Schmidt JA, Abbas AK. Role of interleukin 1 in the activation of T lymphocytes. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9699–703. doi: 10.1073/pnas.85.24.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci U S A. 2009 Apr 28;106(17):7119–24. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Sasson SZ, Hogg A, Hu-Li J, Wingfield P, Chen X, Crank M, et al. IL-1 enhances expansion, effector function, tissue localization, and memory response of antigen-specific CD8 T cells. J Exp Med. 2013 Mar 11;210(3):491–502. doi: 10.1084/jem.20122006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993 Dec 1;178(6):2243–7. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998 May 29;280(5368):1432–5. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 32.Skeen MJ, Miller MA, Shinnick TM, Ziegler HK. Regulation of murine macrophage IL-12 production. Activation of macrophages in vivo, restimulation in vitro, and modulation by other cytokines. J Immunol. 1996 Feb 1;156(3):1196–206. [PubMed] [Google Scholar]
- 33.Flesch IE, Hess JH, Huang S, Aguet M, Rothe J, Bluethmann H, Kaufmann SH. Early interleukin 12 production by macrophages in response to mycobacterial infection depends on interferon gamma and tumor necrosis factor alpha. J Exp Med. 1995 May 1;181(5):1615–21. doi: 10.1084/jem.181.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986 Apr 1;136(7):2348–57. [PubMed] [Google Scholar]
- 35.Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315(6021):672–6. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 36.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002 Apr 1;168(7):3195–204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 37.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 expression on hiv-specific T cells is associated with t-cell exhaustion and disease progression. Nature. 2006 Sep 21;443(7109):350–4. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 38.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999 Apr;179(4):859–70. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 39.Seu L, Sabbaj S, Duverger A, Wagner F, Anderson JC, Davies E, et al. Stable phenotypic changes of the host T cells are essential to the long-term stability of latent HIV-1 infection. J Virol. 2015 Jul;89(13):6656–72. doi: 10.1128/JVI.00571-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins MK, Pardoll DM, Mizuguchi J, Quill H, Schwartz RH. T-cell unresponsiveness in vivo and in vitro: Fine specificity of induction and molecular characterization of the unresponsive state. Immunol Rev. 1987 Feb;95:113–35. doi: 10.1111/j.1600-065x.1987.tb00502.x. [DOI] [PubMed] [Google Scholar]
- 41.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007 Oct;27(4):670–84. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–34. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 43.Purified protein derivative (PPD)-tuberculin anergy and HIV infection: Guidelines for anergy testing and management of anergic persons at risk of tuberculosis. MMWR Recomm Rep. 1991 Apr 26;40(RR-5):27–32. [PubMed] [Google Scholar]
- 44.Huebner RE, Schein MF, Hall CA, Barnes SA. Delayed-type hypersensitivity anergy in human immunodeficiency virus-infected persons screened for infection with mycobacterium tuberculosis. Clin Infect Dis. 1994 Jul;19(1):26–32. doi: 10.1093/clinids/19.1.26. [DOI] [PubMed] [Google Scholar]
- 45.Dolan MJ, Clerici M, Blatt SP, Hendrix CW, Melcher GP, Boswell RN, et al. In vitro T cell function, delayed-type hypersensitivity skin testing, and CD4+ T cell subset phenotyping independently predict survival time in patients infected with human immunodeficiency virus. J Infect Dis. 1995 Jul;172(1):79–87. doi: 10.1093/infdis/172.1.79. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa F, May M, Phillips A. Life expectancy living with HIV: Recent estimates and future implications. Curr Opin Infect Dis. 2013 Feb;26(1):17–25. doi: 10.1097/QCO.0b013e32835ba6b1. [DOI] [PubMed] [Google Scholar]
- 47.Egger M, May M, Chêne G, Phillips AN, Ledergerber B, Dabis F, et al. Prognosis of hiv-1-infected patients starting highly active antiretroviral therapy: A collaborative analysis of prospective studies. Lancet. 2002 Jul 13;360(9327):119–29. doi: 10.1016/s0140-6736(02)09411-4. [DOI] [PubMed] [Google Scholar]
- 48.Bucy RP, Hockett RD, Derdeyn CA, Saag MS, Squires K, Sillers M, et al. Initial increase in blood CD4(+) lymphocytes after HIV antiretroviral therapy reflects redistribution from lymphoid tissues. J Clin Invest. 1999 May 15;103(10):1391–8. doi: 10.1172/JCI5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henostroza G, Harris JB, Chitambi R, Siyambango M, Turnbull ER, Maggard KR, et al. High prevalence of tuberculosis in newly enrolled HIV patients in zambia: Need for enhanced screening approach. Int J Tuberc Lung Dis. 2016 Aug;20(8):1033–9. doi: 10.5588/ijtld.15.0651. [DOI] [PubMed] [Google Scholar]
- 50.Selwyn PA, Sckell BM, Alcabes P, Friedland GH, Klein RS, Schoenbaum EE. High risk of active tuberculosis in hiv-infected drug users with cutaneous anergy. JAMA. 1992;268(4):504–9. [PubMed] [Google Scholar]
- 51.Converse PJ, Jones SL, Astemborski J, Vlahov D, Graham NM. Comparison of a tuberculin interferon-gamma assay with the tuberculin skin test in high-risk adults: Effect of human immunodeficiency virus infection. J Infect Dis. 1997 Jul;176(1):144–50. doi: 10.1086/514016. [DOI] [PubMed] [Google Scholar]
- 52.Achkar JM, Cortes L, Croteau P, Yanofsky C, Mentinova M, Rajotte I, et al. Host protein biomarkers identify active tuberculosis in HIV uninfected and co-infected individuals. EBioMedicine. 2015 Sep;2(9):1160–8. doi: 10.1016/j.ebiom.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008 Mar 7;132(5):887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008 Nov 6;456(7218):125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang ST, Sova P, Peng X, Weiss J, Law GL, Palermo RE, Katze MG. Next-generation sequencing reveals hiv-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. MBio. 2011;2(5) doi: 10.1128/mBio.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.