Abstract
Exposure to petrogenic polycyclic aromatic hydrocarbons (PPAHs) in the food chain is the major human health hazard associated with the Deepwater Horizon oil spill. C4-Phenanthrenes are representative PPAHs present in the crude oil and could contaminate the seafood. We describe the metabolism of a C4-phenanthrene regioisomer retene (1-methyl-7-isopropyl-phenanthrene) in human HepG2 cells as a model for metabolism in human hepatocytes. Retene because of its sites of alkylation cannot be metabolized to a diol-epoxide. The structures of the metabolites were identified by HPLC-UV-fluorescence detection and LC–MS/MS. O-Monosulfonated-retene-catechols were discovered as signature metabolites of the ortho-quinone pathway of PAH activation catalyzed by aldo-keto reductases. We also found evidence for the formation of bis-ortho-quinones where the two dicarbonyl groups were present on different rings of retene. The identification of O-monosulfonated-retene-catechol and O-bismethyl-O-monoglucuronosyl-retene-bis-catechol supports metabolic activation of retene by P450 and aldo-keto reductase isozymes followed by metabolic detoxification of the ortho-quinone through interception of redox cycling by catechol-O-methyltransferase, uridine 5′-diphospho-glucuronosyltransferase, and sulfotransferase isozymes. We propose that catechol conjugates could be used as biomarkers of human exposure to retene resulting from oil spills.
Graphical abstract
INTRODUCTION
The Deepwater Horizon oil spill in the Gulf of Mexico in 2010 was the largest release of crude oil in U.S. history.1,2 Over 200 million gallons of crude oil were released from the Macondo well in the Gulf of Mexico.3–5 Polycyclic aromatic hydrocarbons (PAHs) are potentially the most toxic and persistent components of crude oil.6 This toxicity relates to their carcinogenicity where IARC has listed benzo[a]pyrene as a Group 1 known human carcinogen, other PAHs are listed as Group 2A (probable human carcinogens), and occupational exposures to PAH mixtures is regarded as a Group 1 carcinogen.7–9 Petrogenic PAHs (PPAHs) uniquely present in crude oil differ from pyrogenic PAH since they are either extensively alkylated or oxygenated, but there has been no assessment of PPAH toxicity. Examination of the composition of the Macondo Oil Spill shows that it consists predominately of naphthalenes [2 rings], phenanthrenes [3 rings], and chrysenes [4 rings]. No benzo[a]pyrene is present. Each of these ring systems can contain 1–4 alkyl groups. For example, C4-phenanthrenes could be alkylated so that four separate methyl groups could substitute any of the C10 ring carbons or one ring carbon could be substituted with a butyl or isobutyl group. Thus, the number of regioisomers and isomers possible for each ring system is large. Contamination of the food chain with alkylated PPAHs is a major hazard associated with human health.10 In our previous studies, we described metabolic activation of two representative alkylated PPAHs, that is, 5-methylchrysene (regioisomer of C1-chrysenes), 6-ethylchry-sene (regioisomer of C2-chrysenes), and a representative oxygenated PAH phenanthrene-9,10-quinone in human hepatoma (HepG2) cells as a model to predict metabolism in human hepatocytes.11–13 However, no studies have yet been performed on alkylated phenanthrenes.
In our previous studies on 5-methylchrysene and 6-ethylchrysene, we found evidence for the formation of diol-epoxides and ortho-quinones on the terminal benzo-rings. Importantly, we found evidence for bis-electrophiles, for example, bis-diol-epoxides, bis-ortho-quinones, and the formation of a monodiol-epoxide and mono-ortho-quinone within the same structure. These bis-electrophiles have the potential to be highly toxic due to their potential cross-linking properties with DNA and protein.
Retene (1-methyl-7-isopropyl-phenanthrene) is a representative regioisomer of C4-phenanthrenes found in the Macondo oil spill.14 Retene cannot be metabolically activated to diol-epoxide due to its ring substitution by alkyl groups. In our study of retene metabolism, we considered four possible routes of metabolic activation (Scheme 1). First, we predicted the ortho-quinone pathway, involving conversion of trans-dihydrodiols in the terminal ring to catechols followed by either formation of phase II conjugates, or their oxidation to ortho-quinones. These quinones could be reduced back to the catechols to establish redox cycling leading to oxidative DNA damage. Second, as retene contains two terminal rings within its structure, activation to bis-electrophiles, for example, bis-ortho-quinones, was also possible. Third, we predicted that hydroxylation on the alkyl side chain followed by formation of sulfate conjugates leading to DNA adducts could be another pathway of metabolic activation. Fourth, we predicted that retene could act as a co-reductant in a peroxidase reaction leading to the formation of stable carbonium ion on the isopropyl group side chain and subsequent DNA adduct formation.
Scheme 1.
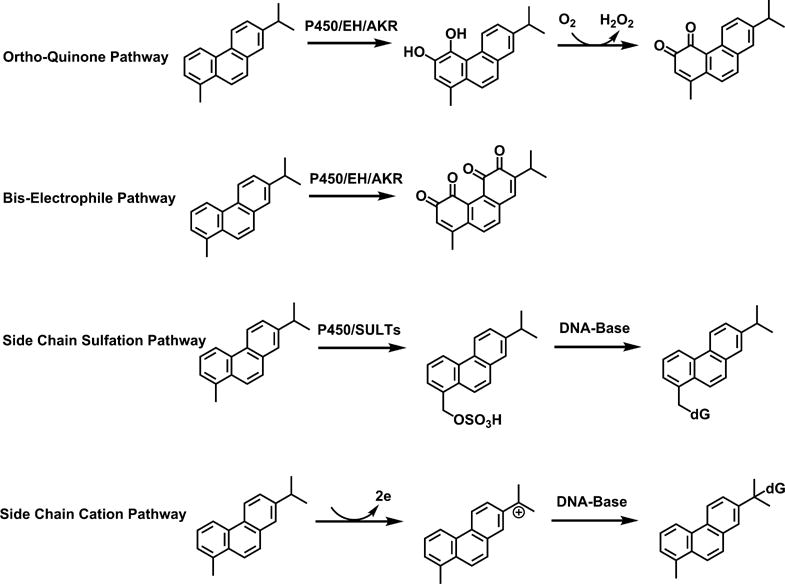
Possible Metabolic Activation Pathways of Retene
We find that the metabolism of retene involved the formation of mono-ortho-quinones and the formation of bis-ortho-quinones, which likely led to the metabolic activation of retene following ingestion. We propose that monocatechol and bis-catechol conjugates of retene represent potential human exposure biomarkers for retene that may result from consuming seafood contaminated by crude oil spills.
MATERIALS AND METHODS
Caution: These chemicals are dangerous
All PAHs are potentially hazardous and should be handled in accordance with the National Institutes of Health Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Reagents
Cell culture media and reagents were all obtained from Invitrogen Co. (Carlsbad, CA) except fetal bovine serum (FBS), which was purchased from Hyclone (Logan, UT). Retene was purchased from American Custom Chemicals Corporation (San Diego, CA). 3-Hydroxy-retene, 4-hydroxy-retene, and 6-hydroxy-retene were synthesized by the Synthetic Organic Chemistry Core at UTMB-Galveston as described in the Supporting Information. All other chemicals used were of the highest grade available, and all solvents were HPLC grade.
Cell Culture
HepG2 (human hepatocellular carcinoma) cells were obtained from American Type Culture Collection and maintained as previously described.11 Cultured cells with a passage number of 10–20 were used in the experiments to reduce variability due to long-term culture conditions. Cultured cells were authenticated by short-terminal repeat DNA analysis and were mycoplasma free (DNA Diagnostics Center Medical, Fairfield, OH).
Detection and Identification of Retene Metabolites in HepG2 Cells
The confluent HepG2 cells were plated in a six-well plate (~5 × 106). The cells were washed twice and then treated with MEM (without phenol red) containing 10 mM glucose as an energy source and 1 μM retene (DMSO, 0.2% v/v). The culture media were collected at 0 and 24 h, respectively, and subsequently acidified with 0.1% formic acid before extraction as previously described.11 The extract from the cell culture media was reconstituted in 150 μL of methanol.
For HPLC-UV-FLR analysis, a 10 μL aliquot of the reconstituted extract was analyzed on a tandem Waters Alliance 2695 chromatographic system with a Waters 2996 Photodiode Array (PDA) Detector and a Waters 2475 Multi λ Fluorescence (FLR) Detector (Waters Corporation, Milford, MA). Separations were accomplished on a Zorbax-ODS C18 analytical column (5 μm, 4.6 mm × 250 mm) with a Zorbax-ODS analytical guard column (5 μm, 4.6 mm × 12.5 mm) (DuPont Co., Wilmington, DE) at ambient temperature. The mobile phase consisted of 5 mM ammonium acetate and 0.1% trifluoroacetic acid (TFA) (v/v) in H2O (solvent A) and 5 mM ammonium acetate and 0.1% TFA in acetonitrile (solvent B) and was delivered at a flow rate of 0.5 mL/min. The linear gradient elution program was as follows: 5% to 95% B over 30 min, followed by an isocratic hold at 95% B for another 10 min. At 40 min, B was returned to 5% in 1 min, and the column was equilibrated for 19 min before the next injection. The total run time for each analysis was 60 min. Eluants from the column were introduced sequentially into the PDA detector and the FLR detector. Excitation wavelength (λex) and emission wavelength (λem) for FLR detector were set at 259 and 370 nm, respectively, based on the spectral properties of retene (Figure S1). The optimal pair of λex and λem of retene was employed to detect its metabolites based on the assumption that most retene metabolites show fluorescence signals at these wavelengths.
For ion trap LC–MS/MS analysis, a 10 μL aliquot of the reconstituted extract was analyzed on a Waters Alliance 2690 HPLC system (Waters Corporation, Milford, MA) coupled to a Finnigan LTQ linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA). The column, mobile phase, flow rate, and the linear gradient elution program were the same as described above. During LC–MS/MS analysis, up to 10 min of the initial flow were diverted to the waste before evaluation of eluants. The mass spectrometer was operated in both the positive and negative ion modes with an electrospray ionization (ESI) source. Eluants were monitored on the LTQ using product ion scan (MS2), subsequent MS/MS/MS (MS3), and pseudo selected reaction monitoring (SRM) modes. The mass spectrometry parameters included spray voltage (3 kV in positive ion mode, 5 kV in negative ion mode), sheath gas flow rate (40 arbitrary units in both ion modes), auxiliary gas flow rate (15 arbitrary units in both ion modes), capillary temperature (275 °C in both ion modes), capillary voltage (38 V in positive ion mode, −19 V in negative ion mode), and tube lens (20 V in positive ion mode, −22.05 V in negative ion mode). An isolation width of three bracketed around the m/z of interest, activation Q of 0.25, and activation time of 30 ms were used for data acquisition. Xcalibur software version 2.0 (Thermo Scientific, San Jose, CA) was used to control the LC–MS/MS system and to process data. The preliminary information on metabolite structures was obtained by interpreting the corresponding MS2 and MS3 spectra of retene metabolites from ion trap LC–MS/MS.
In some instances, another 5 μL aliquot of the reconstituted extract was analyzed on a nano-Acquity ultraperformance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA). Separations were accomplished on a nano-UPLC C18 column (1.7 μm BEH130, 150 μm × 100 mm) (Waters Corporation, Milford, MA) at 50 °C. The mobile phase consisted of 0.1% formic acid (v/v) in H2O (solvent A) and 0.1% formic acid (v/v) in acetonitrile (solvent B) and was delivered at a flow rate of 1.6 μL/min. The linear gradient elution program was as follows: an isocratic hold at 5% B for 5 min, 5% to 95% B over 30 min, followed by an isocratic hold at 95% B for another 10 min. At 46 min, B was returned to 5% in 2 min, and the column was equilibrated for 12 min before the next injection. The total run time for each analysis was 60 min. The mass spectrometer was operated in the positive and negative ion modes, respectively, with a nanoelectrospray ionization (nano-ESI) source after accurate calibration with the manufacturer’s calibration mixture. The ionization voltage was set to 1.5 kV, and the capillary temperature was set to 200 °C. Full scan spectra were acquired with a resolving power of 60 000 full-width half-maximum (fwhm) in a mass range from m/z 100–600. Xcalibur software version 2.0 (Thermo Scientific, San Jose, CA) was used to control the Orbitrap mass spectrometry and to process data.
RESULTS
Detection of Retene Metabolites in HepG2 Cells by HPLC-UV-FLR
Comparison of UV chromatograms at λmax 258 nm at 0 h (Figure 1A) and 24 h (Figure 1B) showed that 12 metabolites of retene were detected in the organic phase of the ethyl acetate-extracted acidified media from HepG2 cells. The media were acidified prior to extraction so that conjugates would be protonated and extracted into the organic phase. The peak attributed to retene at 0 h was almost completely absent at 24 h, which suggested that retene was rapidly metabolized by HepG2 cells over this time course. The corresponding UV spectra of the 12 metabolites were extracted from the PDA detector and are shown in Figure S2. The UV spectra of metabolites 4–12 were similar to that of retene showing that aromaticity has been retained, whereas metabolites 1–3 are predicted to have lost their aromaticity.
Figure 1.
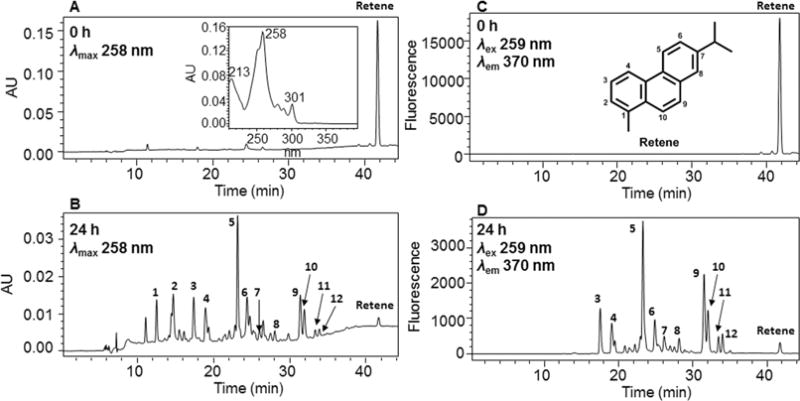
HPLC detection of retene metabolites in human HepG2 cells. (A) UV chromatogram at λmax 258 nm at 0 h. (B) UV chromatogram at λmax 258 nm at 24 h. (C) FLR chromatogram at λex 259 nm and λem 370 nm at 0 h. (D) FLR chromatogram at λex 259 nm and λem 370 nm at 24 h. Human HepG2 cells (~5 × 106) were treated with retene (1 μM, 0.2% (v/v) DMSO) in MEM (without phenol red) containing 10 mM glucose. The cell media were collected at 0 and 24 h, respectively, and subsequently acidified with 0.1% formic acid before extraction with ethyl acetate. The extracts were analyzed by HPLC-UV-FLR detection.
Comparison of FLR chromatograms at λex 259 nm and λem 370 nm at 0 h (Figure 1C) and 24 h (Figure 1D) showed that there were ten fluorescence peaks in Figure 1D corresponding to the metabolites 3–12 in Figure 1B, which thus validated the presence of the retene fluorophore. The peaks corresponding to metabolites 1 and 2 were only detected in the UV chromatogram (Figure 1B) but not detected in the FLR chromatogram (Figure 1D), which suggested the loss of the retene ring fluorophore.
Evidence for the Ortho-Quinone Pathway
Identification of O-monosulfonated-retene-dihydrodiol, O-monosulfonated-retene-catechol, O-monoglucuronosyl-retene-catechol, retene-dione, and monohydroxy-retene-dione indicated the occurrence of the ortho-quinone pathway for the metabolic activation of retene in HepG2 cells.
A single isomer of the O-monosulfonated-retene-dihydrodiol was detected at 22.80 min following its dehydration in the ion source to yield monodehydrated O-monosulfonated-retene-dihydrodiol and was assigned by comparing the MS3 chromatograms (m/z 329 → 249 →) at 0 h (Figure 2A) and 24 h (Figure 2B) in the negative ion mode (see Table 1 for mass transitions). The corresponding MS2 spectrum (m/z 329) of this metabolite showed the characteristic loss of the sulfate group (80 amu) from the deprotonated molecular ion (Figure 2C), and the MS3 spectrum (m/z 329 → 249 →) of this metabolite showed the subsequent loss of one CH3 group from the alkyl side chain (Figure 2D). This metabolite is not a monophenol-O-monosulfate. The O-monosulfonated retene dihydrdodiol (contains 1-OH and 1 -OSO3H group before dehydration) and the catechol-O-monosulfate (contains 1-OH and 1-OSO3H group) have retention times of 22.80 and 22.79 min, respectively, and would elute earlier than a monophenol-O-monosulfate, which has only one −OSO3H group and the same nominal mass as the monodehydrated O–monosulfo-nated retene dihydrodiol. The specific position of the O-monosulfonated-dihydrodiol could not be assigned using mass spectrometry.
Figure 2.
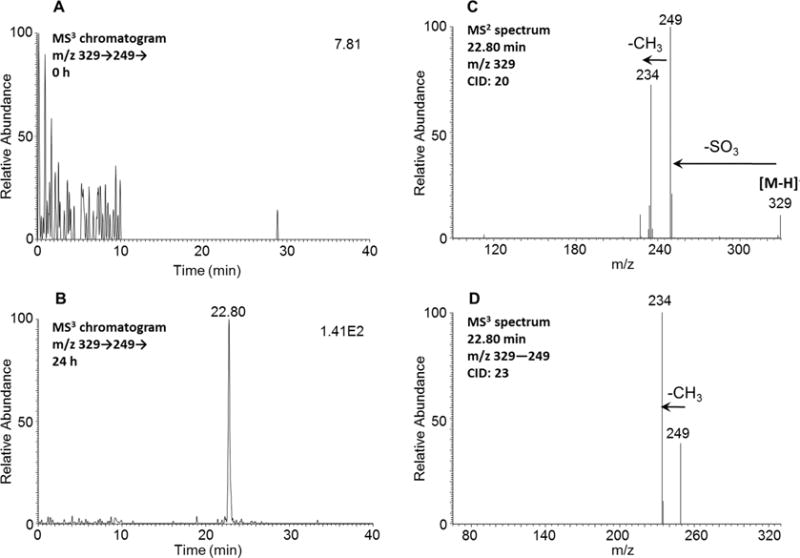
Detection of monodehydrated O-monosulfonated-retene-dihydrodiol in human HepG2 cells. (A) MS3 chromatogram at 0 h. (B) MS3 chromatogram at 24 h. (C) MS2 spectrum of the peak at 22.80 min. (D) MS3 spectrum of the peak at 22.80 min. The samples were prepared as described in the legend to Figure 1 and were subsequently analyzed on an ion trap LC–MS/MS in the negative ion mode.
Table 1.
Mass Transitions for Retene Metabolites in HepG2 Cells
| metabolite no. | retene metabolites | retention time (min) | mode | m/z |
|---|---|---|---|---|
| − | dehydrated O-sulfonated dihydrodiol | 22.80 | negative | 329 [M−H]−, 249 [M−H–SO3]−, 234 [M−H–SO3–CH3]− |
| 3 | O-sulfonated bis-phenol | 17.31, 17.67 | negative | 345 [M−H]−, 265 [M−H–SO3]−, 247 [M−H–SO3–H2O]− |
| 5 | O-sulfonated catechol | 20.85, 21.44, 22.79 | negative | 345 [M−H]−, 265 [M−H–SO3]−, 250 [M−H–SO3–CH3]−, 222 [M−H–SO3–CH3–CO]− |
| − | quinone | 32.33, 35.39 | positive | 265 [M + H]+, 247 [M+H–H2O]+, 237 [M+H–CO]+, 219 [M+H–H2O–CH3]+ |
| 2 | bis-dihydrodiol | 15.61 | positive | 303 [M + H]+, 285 [M+H–H2O]+, 256 [M+H–H2O–CO−H]+, 242 [M+H–H2O–CO–CH3]+ |
Five isomers at 17.31, 17.67, 20.85, 21.44, and 22.79 min corresponding to either O-monosulfonated-retene-catechols or O-monosulfonated-retene-bis-phenols were detected by monitoring the MS3 chromatograms (m/z 345 → 265 →) at 0 h (Figure 3A) and 24 h (Figure 3B) in the negative ion mode. The corresponding MS2 spectra (m/z 345) of these five metabolites showed the characteristic loss of the sulfate group (80 amu) from the deprotonated molecular ion. The MS3 spectra (m/z 345 → 265 →) of the metabolites at 17.31 and 17.67 min showed the subsequent loss of one H2O group, and a representative MS3 spectrum of the metabolite at 17.31 min is shown in Figure 3C. This fragmentation pattern is inconsistent with that of O-monosulfonated-phenols of 5-methylchrysene and 6-ethylchrysene,11,12 which indicated that the metabolites at 17.31 and 17.67 min were O-monosulfonated-retene-bis-phenols (see Table 1 for mass transitions). The MS3 spectra (m/z 345 → 265 →) of the remaining metabolites at 20.85, 21.44, and 22.79 min showed the subsequent loss of one CH3 group followed by the loss of one CO group but no loss of H2O, and a representative MS3 spectrum of the metabolite at 22.79 min is shown in Figure 3D. This fragmentation pattern is consistent with that of O-monosulfonated-catechols of 5-methylchrysene and 6-ethylchrysene,11,12 which strongly indicated that the metabolites at 20.85, 21.44, and 22.79 min were O-monosulfonated-retene-catechols (see Table 1 for mass transitions). In this assignment, O-monosulfonated bis-phenols showed the loss [M−H–SO3–H2O], and the O-monosulfo-nated catechols showed the loss of [M−H–SO3–CH3–CO]. Comparisons of the retention times of peaks at 17.31 and 22.79 min with metabolites 3 and 5 on HPLC-UV-FLR in Figure 1 were in agreement.
Figure 3.
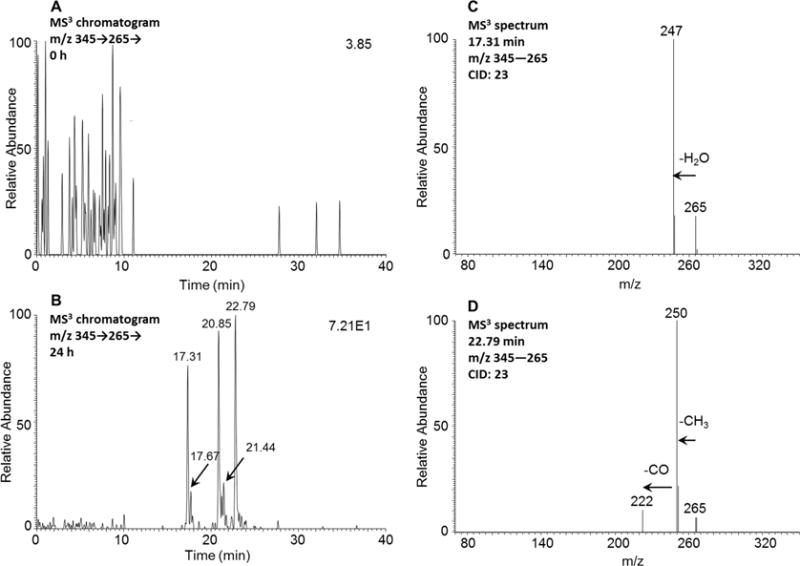
Detection of either an O-monosulfonated-retene-catechol or an O-monosulfonated-retene-bis-phenol in human HepG2 cells. (A) MS3 chromatogram at 0 h. (B) MS3 chromatogram at 24 h. (C) MS3 spectrum of the peak at 17.31 min. (D) MS3 spectrum of the peak at 22.79 min. The samples were prepared as described in the legend to Figure 1 and were subsequently analyzed on an ion trap LC–MS/MS in the negative ion mode.
Two isomers of either O-monoglucuronosyl-retene-catechols or O-monoglucuronosyl-retene-bis-phenols were detected in HepG2 cells by monitoring the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the negative ion mode with retention times of 14.92 and 17.92 min (Figure 4). MS spectra of these isomers provided the accurate masses and molecular formulas of O-monoglucuronosyl-retene-catechols or O-monoglucuronosyl-retene-bis-phenols to within 4 ppm (Figure 4). As both are present, the analyte at 14.72 min is assigned as the O-monoglucuronsyl retene bis-phenol, and the analyte at 17.92 min is assigned as the O-monoglucuronsyl retene catechol based on the observation that bis-phenol conjugates eluted before catechol conjugates as observed for the O-monosulfonated bis-phenols and the O-monsulfonated catechols (see above).
Figure 4.
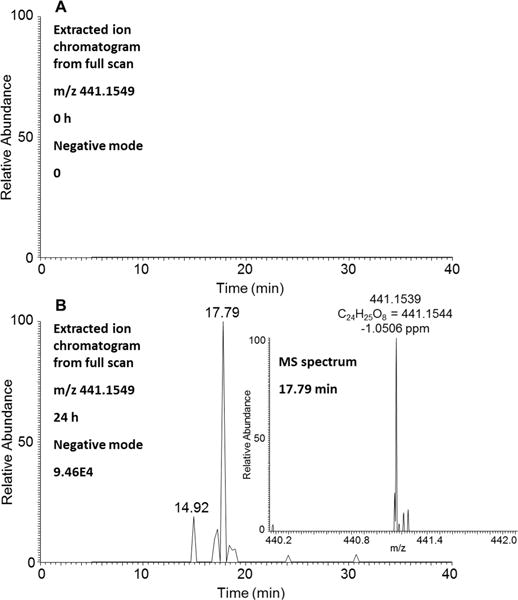
Detection of either O-monoglucuronosyl-retene-catechols or O-monoglucuronosyl-retene-bis-phenols in human HepG2 cells. (A) Extracted ion chromatogram of Orbitrap full scan at 0 h. (B) Extracted ion chromatogram of Orbitrap full scan at 24 h and MS spectrum of the peak at 17.79 min. The samples were prepared as described in the legend to Figure 1 and were subsequently analyzed on an Orbitrap LC–MS/MS in the negative ion mode.
Two isomers of retene-diones at 32.33 and 35.39 min were detected by monitoring the MS2 chromatograms (m/z 265) at 0 and 24 h in the positive ion mode (Table 1). The corresponding MS2 spectra (m/z 265) of these metabolites showed the sequential loss of H2O (18 amu) and CO (28 amu) from the protonated molecular ion (Table 1), which supported the presence of either an ortho-quinone or a remote quinone, which cannot be distinguished based on mass spectrometry. However, we favor formation of the retene-o-quinones since three different O-monosulfonated retene catechols were detected. These two diones could not be related to any of the metabolites observed in Figure 1 due to their low abundance and presumptive reactivity.
Four isomers of monohydroxy-retene-diones were detected in HepG2 cells by monitoring the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the positive ion mode (Figure S3). Two isomers of monohydroxy-retene-diones were detected with different retention times in the negative ion mode (Figure S3). Monohydroxy-retene-dione could be derived from either an ortho-quinone or a remote quinone, and these alternatives cannot be distinguished based on mass spectrometry.
Evidence for the Bis-Electrophile Pathway
Evidence for metabolic activation of retene in HepG2 cells to form a bis-electrophile containing two ortho-quinones within the same structure was obtained. Identification of retene-bis-dihydrodiol, retene-bis-dione, and O-bismethyl-O-monoglucuronosyl-retene-bis-catechol indicated the occurrence of the bis-electrophile pathway for the metabolic activation of retene in HepG2 cells.
A single isomer of retene-bis-dihydrodiol at 15.61 min was detected by monitoring the MS2 chromatograms (m/z 303) at 0 and 24 h in the positive ion mode (Table 1). Structural validation for the bis-dihydrodiol was obtained by the loss of the UV chromophore for retene, loss of the fluorophore for retene, and the corresponding MS2 spectra (m/z 303) for this metabolite, which showed the sequential loss of H2O (18 amu), CO (28 amu), and CH3 (15 amu) from the protonated molecular ion (Table 1). Comparison of the retention times of retene-bis-dihydrodiol with metabolite 2 on HPLC-UV-FLR in Figure 1 showed good agreement.
Two isomers of retene-bis-diones with retention times of 17.22 and 19.51 min were detected in HepG2 cells by monitoring the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the negative ion mode (Figure 5). Retene-bis-dione could be derived from either an ortho-quinone or a remote quinone precursor, and these alternatives cannot be distinguished based on mass spectrometry. However, the ability to detect the bis-dihydodiol precursor indicates that subsequent formation of the bis-o-quinone is likely. We did not relate the bis-dione peaks to those in Figure 1 because we used different LC conditions for the nano-UPLC orbitrap studies.
Figure 5.
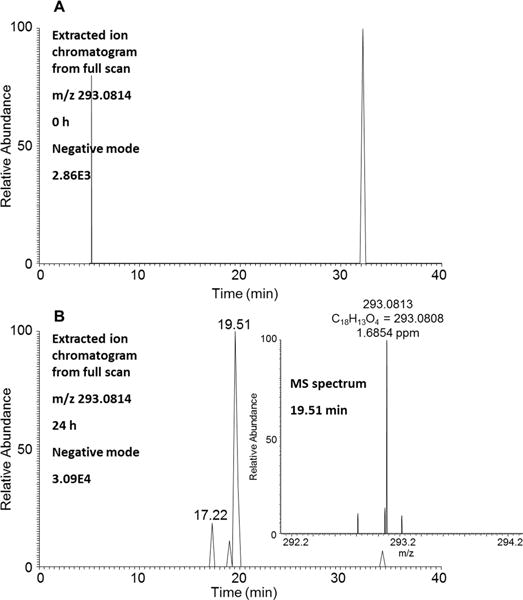
Detection of retene-bis-diones in human HepG2 cells. (A) Extracted ion chromatogram of Orbitrap full scan at 0 h. (B) Extracted ion chromatogram of Orbitrap full scan at 24 h and MS spectrum of the peak at 19.51 min. The samples were prepared as described in the legend to Figure 1 and were subsequently analyzed on an Orbitrap LC–MS/MS in the negative ion mode.
Three isomers of O-bismethyl-O-monoglucuronosyl-retene-bis-catechols were detected by comparing the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the positive ion mode (Figure 6). Five isomers of O-bismethyl-O-monoglucuronosyl-retene-bis-catechols were detected with different retention times in the negative ion mode (Figure 6). In particular, the unique biotransformation of O-bismethylation strongly indicated the formation of the bis-catechol, which thus confirmed the occurrence of bis-ortho-quinone pathway.
Figure 6.
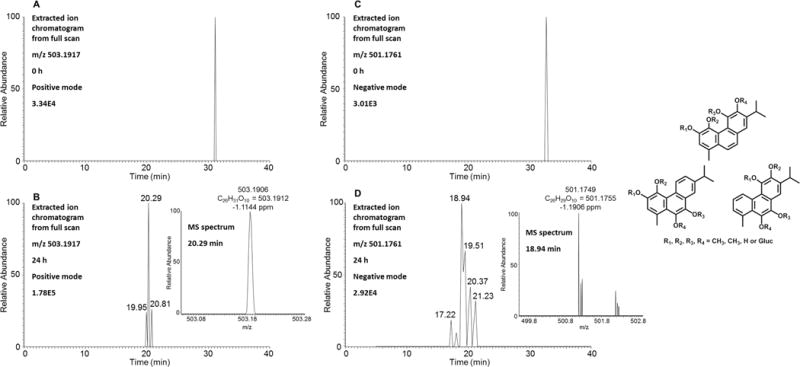
Detection of O-bismethyl-O-monoglucuronosyl-retene-bis-catechols in human HepG2 cells. (A) Extracted ion chromatogram of Orbitrap full scan in the positive mode at 0 h. (B) Extracted ion chromatogram of Orbitrap full scan in the positive mode at 24 h and MS spectrum of the peak at 20.29 min. (C) Extracted ion chromatogram of Orbitrap full scan in the negative mode at 0 h. (D) Extracted ion chromatogram of Orbitrap full scan in the negative mode at 24 h and MS spectrum of the peak at 18.94 min. The samples were prepared as described in the legend to Figure 1 and were subsequently analyzed on an Orbitrap LC-MS/MS.
Evidence for the Other Pathways
3-Hydroxy-retene, 4-hydroxy-retene, and 6-hydroxy-retene were synthesized as authentic standards (Figure S4 and Supporting Information) to determine whether any of the metabolites detected in human HepG2 cells supported hydroxylation on the retene ring. Comparison of the retention times and UV spectra of three synthetic standards with metabolites 9–12 on HPLC-UV-FLR in Figure 1 showed similarity, which suggested that these metabolites were monophenols. No evidence for hydroxylation on the side chain or side chain cation formation was obtained.
DISCUSSION
Retene is a representative regioisomer of C4-phenanthrene present in crude oil that may enter the food chain after oil spills. We examined its metabolism in human HepG2 cells knowing that human exposure pathways involve ingestion. Human HepG2 cells were selected as an alternative to human hepatocytes. Previous studies by us on the PPAH, 5-methylchrysene and 6-ethylchrysene were performed in these cells making direct comparison of the metabolic profiles possible. We also found that HepG2 cells have inducible P4501A1, the major P450 isoform involved in the metabolic activation of PAHs.15,16 We did not select HepaRG cells, which contain a larger compliment of P450 isozymes, since they have not been characterized with regards to their AKR expression, which plays a role in PPAH activation.
We proposed four possible pathways of retene metabolic activation: formation of ortho-quinones, formation of bis-electrophiles, formation of side chain sulfonates, and the formation of a side chain cation. We found evidence for the first two pathways but no evidence for activation on the side chain either by hydroxylation or by cation formation. Retene metabolites representative of the first two pathways were identified by HPLC-UV-FLR and LC-MS/MS (Table 1). Metabolite 2 in Figure 1 was identified as an bis-dihydrodiol. Metabolite 3 in Figure 1 was identified as an O-monosulfo-nated-bis-phenol. Metabolite 5 in Figure 1 was identified as an O-monosulfonated-catechol. Metabolites 9, 10, 11, and 12 in Figure 1 were identified as regio-isomers of retene monophenols, but no monophenol-O-monoconjugates were detected.
On the basis of the peak areas of the metabolites on UV and FLR chromatograms, the major metabolic activation pathways of retene in human HepG2 cells involved formation of ortho-quinones (Scheme 2). We also found evidence for the metabolic activation of retene on both terminal rings to form bis-electrophiles containing bis-ortho-quinones (Scheme 2). We suspect that the underdetection of these metabolites was probably due to the potential of bis-electrophiles to form protein and DNA cross-links.
Scheme 2. Proposed Metabolic Pathway of Retene in Human HepG2 Cellsa.
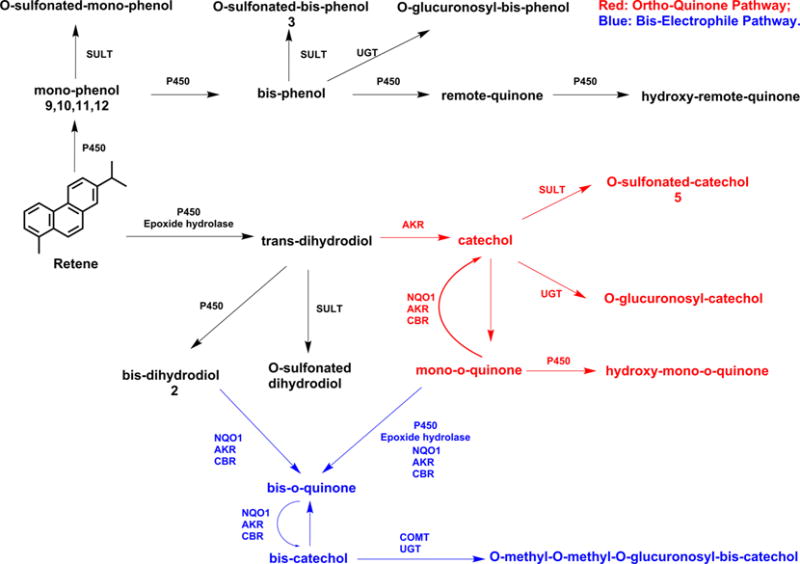
aNumbers for each metabolite correspond to the metabolites labeled in the UV and fluorescence chromatograms in Figure 1.
We did not find any evidence for hydroxylation on the retene side chain followed by formation of a sulfate in human HepG2 cells, which is consistent with the previous findings that hydroxymethylation of 5-methylchrysene, 6-methylchrysene, and 6-ethylchrysene is not an important metabolic activation pathway in humans.11,12,17 Monodehydrated O-monosulfonated-retene-dihydrodiol shown in Figure 2 had an identical nominal mass as the sulfate conjugate of side chain monohydroxy-retene, but the former metabolite was produced in the ion source from the O-monosulfonated retene dihydrodiol and could be distinguished from the phenol-O-monosulfate based on retention time. However, the fragmentation pattern in Figure 2 shows the loss of the sulfate group and the subsequent loss of CH3 group, which rules out the possibility of side chain hydroxylation on retene.
Several metabolites indicated that the ortho-quinone pathway was involved in the metabolic activation of retene. Interestingly, O-monosulfonated-retene-catechols and O-monosulfonated-retene-bis-phenols with identical accurate masses were distinguished by MS3 spectra for the first time, which confirmed the presence of both the ortho-quinone pathway and the formation of phenols.
The phase I and phase II isozymes responsible for the metabolic activation pathway of retene in human HepG2 cells remain to be identified. The cytochrome P450 1A1 and epoxide hydrolase present in human HepG2 cells are probably responsible for the formation of trans-dihydrodiols.15 Aldo-keto reductases (AKRs) such as AKR1C1, AKR1C2, and AKR1C3 present in human HepG2 cells could catalyze the oxidation of retene trans-dihydrodiols to catechols, which airoxidize to ortho-quinones.18–20 AKR1C4 although absent from HepG2 cells likely plays a similar role to the other AKR1C enzymes in hepatocytes where it is expressed.16 AKRs, NAD(P)H: quinone oxidoreductase 1 (NQO1), and carbonyl reductase (CBR) could catalyze the redox-cycling of ortho-quinones to reform the intermediate catechols.21 Sulfotransferases (SULTs), uridine 5′-diphospho-glucuronosyltransferases (UGTs), and catechol-O-methyltransferase (COMT) could be responsible for the formation of O-monosulfonated-catechols and O-bismethyl-O-monoglucuronosyl-retene-bis-catechols.22–24 Previous studies indicated that both UGT2B7 and SULT1A1 enzyme isoforms are expressed in human HepG2 cells and thus may be involved in the phase II conjugation of retene catechols.23,25
Elucidation of the metabolic pathways of retene in human liver cells can be used to identify exposure biomarkers of retene that could be used for biomonitoring human urine. In our ongoing studies with UTMB at Galveston, we have established a cohort of human subjects that may have had different exposures to contaminated seafood. As a next step, it would be interesting to determine whether retene bis-catechol conjugates can be detected in human urine and whether their levels vary by exposure group.
Supplementary Material
Acknowledgments
Funding
This publication was made possible by the Deepwater Horizon Research Consortia Grant Nos. U19 ES020676-05, P30 ES013508 (T.M.P.), and P30 ES006676 (R.P.H.) from the National Institute of Environmental Health Sciences (NIEHS), NIH, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NIH.
ABBREVIATIONS
- AKR
aldo-keto reductase
- CBR
carbonyl reductase
- COMT
catechol-O-methyltransferase
- P450
cytochrome P450
- ESI
electrospray ionization
- FBS
fetal bovine serum
- HPLC-UV-FLR
high performance liquid chromatography-ultraviolet-fluorescence
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- retene
1-methyl-7-isopropyl-phenanthrene
- NQO1, NAD(P)H
quinone oxidoreductase 1
- PPAH
petrogenic polycyclic aromatic hydrocarbon
- ROS
reactive oxygen species
- SULTs
sulfotransferases
- UGTs
uridine 5′-diphospho-glucuronosyltransferases
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.6b00457.
Excitation wavelength and emission wavelength spectra; UV spectra; extracted ion chromatograms; synthetic routes and details of synthesis (PDF)
ORCID
Trevor M. Penning: 0000-0002-3937-1066
Notes
The authors declare no competing financial interest.
References
- 1.Joye SB, MacDonald IR, Leifer I, Asper V. Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat Geosci. 2011;4:160–164. [Google Scholar]
- 2.Atlas RM, Hazen TC. Oil Biodegradation and Bioremediation: A Tale of the Two Worst Spills in US History. Environ Sci Technol. 2011;45:6709–6715. doi: 10.1021/es2013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, Carmichael CA, McIntyre CP, Fenwick J, Ventura GT, Van Mooy BA, Camilli R. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci U S A. 2012;109:20229–20234. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNutt MK, Camilli R, Crone TJ, Guthrie GD, Hsieh PA, Ryerson TB, Savas O, Shaffer F. Review of flow rate estimates of the Deepwater Horizon oil spill. Proc Natl Acad Sci U S A. 2012;109:20260–20267. doi: 10.1073/pnas.1112139108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryerson TB, Camilli R, Kessler JD, Kujawinski EB, Reddy CM, Valentine DL, Atlas E, Blake DR, de Gouw J, Meinardi S, Parrish DD, Peischl J, Seewald JS, Warneke C. Chemical data quantify Deepwater Horizon hydrocarbon flow rate and environmental distribution. Proc Natl Acad Sci U S A. 2012;109:20246–20253. doi: 10.1073/pnas.1110564109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein BD, Osofsky HJ, Lichtveld MY. The Gulf oil spill. N Engl J Med. 2011;364:1334–1348. doi: 10.1056/NEJMra1007197. [DOI] [PubMed] [Google Scholar]
- 7.IARC. IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans. Vol. 32. World Health Organization, International Agency for Research on Cancer; Lyon, France: 1983. Polynuclear Aromatic Compounds. Part 1: Chemical, Environmental and Experimental Data. [PubMed] [Google Scholar]
- 8.IARC. IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans. Vol. 92. World Health Organization, International Agency for Research on Cancer; Lyon, France: 2010. Some Non-heterocyclic Polycyclic Aromatic Hydrocarbons and Some Related Exposures. [Google Scholar]
- 9.IARC. IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Humans. Vol. 105. World Health Organization, International Agency for Research on Cancer; Lyon, France: 2011. Diesel and gasoline engine exhausts and some nitroarenes. [Google Scholar]
- 10.Ylitalo GM, Krahn MM, Dickhoff WW, Stein JE, Walker CC, Lassitter CL, Garrett ES, Desfosse LL, Mitchell KM, Noble BT, Wilson S, Beck NB, Benner RA, Koufopoulos PN, Dickey RW. Federal seafood safety response to the Deepwater Horizon oil spill. Proc Natl Acad Sci U S A. 2012;109:20274–20279. doi: 10.1073/pnas.1108886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Zhang L, Mesaros C, Hackfeld LC, Hodge RP, Blair IA, Penning TM. Metabolism of an Alkylated Polycyclic Aromatic Hydrocarbon 5-Methylchrysene in Human Hepatoma (HepG2) Cells. Chem Res Toxicol. 2015;28:2045–2058. doi: 10.1021/acs.chemrestox.5b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M, Mesaros C, Zhang S, Blair IA, Penning TM. Potential Metabolic Activation of a Representative C2-Alkylated Polycyclic Aromatic Hydrocarbon 6-Ethylchrysene Associated with the Deepwater Horizon Oil Spill in Human Hepatoma (HepG2) Cells. Chem Res Toxicol. 2016;29:991–1002. doi: 10.1021/acs.chemrestox.6b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, Zhang L, Mesaros C, Zhang S, Blaha MA, Blair IA, Penning TM. Metabolism of a representative oxygenated polycyclic aromatic hydrocarbon (PAH) phenanthrene-9,10-quinone in human hepatoma (HepG2) cells. Chem Res Toxicol. 2014;27:852–863. doi: 10.1021/tx500031p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NIST. Standard Reference Material (SRM) 2779, Gulf of Mexico Crude Oil. Natl Inst Stds & Technol Cert Analysis. 2012:1–7. [Google Scholar]
- 15.Burczynski ME, Penning TM. Genotoxic polycyclic aromatic hydrocarbon ortho-quinones generated by aldo-keto reductases induce CYP1A1 via nuclear translocation of the aryl hydrocarbon receptor. Cancer Res. 2000;60:908–915. [PubMed] [Google Scholar]
- 16.Burczynski ME, Lin HK, Penning TM. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs), electrophiles, and oxidative stress: implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res. 1999;59:607–614. [PubMed] [Google Scholar]
- 17.Koehl W, Amin S, Staretz ME, Ueng YF, Yamazaki H, Tateishi T, Guengerich FP, Hecht SS. Metabolism of 5-methylchrysene and 6-methylchrysene by human hepatic and pulmonary cytochrome P450 enzymes. Cancer Res. 1996;56:316–324. [PubMed] [Google Scholar]
- 18.Palackal NT, Burczynski ME, Harvey RG, Penning TM. The ubiquitous aldehyde reductase (AKR1A1) oxidizes proximate carcinogen trans-dihydrodiols to o-quinones: potential role in polycyclic aromatic hydrocarbon activation. Biochemistry. 2001;40:10901–10910. doi: 10.1021/bi010872t. [DOI] [PubMed] [Google Scholar]
- 19.Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J Biol Chem. 2002;277:24799–24808. doi: 10.1074/jbc.M112424200. [DOI] [PubMed] [Google Scholar]
- 20.Steckelbroeck S, Oyesanmi B, Jin Y, Lee SH, Kloosterboer HJ, Penning TM. Tibolone metabolism in human liver is catalyzed by 3alpha/3beta-hydroxysteroid dehydrogenase activities of the four isoforms of the aldo-keto reductase (AKR)1C subfamily. J Pharmacol Exp Ther. 2005;316:1300–1309. doi: 10.1124/jpet.105.091587. [DOI] [PubMed] [Google Scholar]
- 21.Shultz CA, Quinn AM, Park JH, Harvey RG, Bolton JL, Maser E, Penning TM. Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase, and carbonyl reductases to redox-cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-o-quinone. Chem Res Toxicol. 2011;24:2153–2166. doi: 10.1021/tx200294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Huang M, Blair IA, Penning TM. Detoxication of benzo[a]pyrene-7,8-dione by sulfotransferases (SULTs) in human lung cells. J Biol Chem. 2012;287:29909–29920. doi: 10.1074/jbc.M112.386052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Huang M, Blair IA, Penning TM. Interception of benzo[a]pyrene-7,8-dione by UDP glucuronosyltrans-ferases (UGTs) in human lung cells. Chem Res Toxicol. 2013;26:1570–1578. doi: 10.1021/tx400268q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Jin Y, Chen M, Huang M, Harvey RG, Blair IA, Penning TM. Detoxication of structurally diverse polycyclic aromatic hydrocarbon (PAH) o-quinones by human recombinant catechol-O-methyltransferase (COMT) via O-methylation of PAH catechols. J Biol Chem. 2011;286:25644–25654. doi: 10.1074/jbc.M111.240739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerink WM, Schoonen WG. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1592–1602. doi: 10.1016/j.tiv.2007.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


