Abstract
Background
Congenital nephrotic syndrome (CNS) is defined as nephrotic syndrome that manifests within the first 3 months of life. Mutations in the NPHS1 gene encoding nephrin, are a major cause for CNS. Currently, more than 173 different mutations of NPHS1 have been published as causing CNS, affecting most exons.
Methods
We performed mutation analysis of NPHS1 in a worldwide cohort of 20 families (23 children) with CNS. All 29 exons of the NPHS1 gene were examined using direct sequencing. New mutations were confirmed by demonstrating their absence in 96 healthy control individuals.
Results
We detected disease-causing mutations in 9 of 20 families (45%). Seven of the families showed a homozygous mutation, while two were compound heterozygous. In another 2 families, single heterozygous NPHS1 mutations were detected. Out of 10 different mutations discovered, 3 were novel, consisting of 1 splice site mutation and 2 missense mutations.
Conclusion
Our data demonstrate that the spectrum of NPHS1 mutations is still expanding, involving new exons, in patients from a diverse ethnic background.
Keywords: Mutation analysis, Congenital nephrotic syndrome, NPHS1
INTRODUCTION
Congenital Nephrotic Syndrome
Congenital nephrotic syndrome (CNS) is defined as nephrotic syndrome manifesting by the 90th day of life. CNS of the Finnish type (CNF; MIM#256300) is a recessively inherited disorder first described in highly inbred Finnish communities [1, 2]. CNF is characterized by massive proteinuria at birth, a large placenta and marked edema occurring within the first 3 months of life [1, 3–5]. Renal histology shows mesengial hypercellularity and matrix expansion that progresses with age towards complete mesangial sclerosis and capillary obliteration [2]. Irregular microcystic dilatation of the proximal tubules is the most typical histologic feature, but is not observed in all cases [6–8]. Ultrastructural analysis of the glomerular capillary loops show complete foot process effacement and swelling of endothelial cells [9]. The course of the disease is progressive, leading to end-stage renal disease by 2–3 years of age.
NPHS1
By positional cloning, CNF was shown to be caused by mutations in NPHS1 [10]. The Finmajor mutation (nt-121delCT, L41fsX91) and Finminor mutation (c.3325 C>T,R1109X) in the NPHS1 gene were the first mutations to be discovered and the most prevalent mutations of CNF in the Finnish population (98% of cases) [10]. However, these mutations are also found in other ethnic groups [11, 12]. Screening for NPHS1 mutations in patients of non-Finnish origin has shown that the frequency of NPHS1 mutations is lower than that in Finnish patients, accounting for 39–50% of non-Finnish cases with CNS [13, 14]. On the other hand, rare cases with a manifestation beyond the age of 90 days have also been published, indicating that different mutations in NPHS1 might cause a spectrum of clinical severity [15, 16]. To date, 173 different mutations in NPHS1 have been described (http://www.biobase-international.com). One striking finding among patients with CNS has been the detection of mutations in the NPHS2 gene, encoding podocin, which has been implicated in early-onset steroid-resistant nephrotic syndrome [17]. NPHS2 was shown to be mutated in almost 50% of cases with CNS who are of European origin [13]. In addition to the mutations in the NPHS1 and NPHS2 genes, further genetic heterogeneity has been demonstrated in CNS cases: PLCE1 and WT1 cause CNS and diffuse mesangial sclerosis (DMS) [18–21]. Mutations of LAMB2 cause Pierson syndrome, as a part of a syndromic entity [22] with nephrotic syndrome and microcoria, or as isolated nephrotic syndrome [23, 24].
Mitochondropathies in which the coenzyme Q10 biosynthesis pathway is disrupted may cause monogenic CNS along with neuromuscular symptoms as in mutations of the COQ2 [25], COQ6 [26] and PDSS2 genes [27].
Nephrin
NPHS1 codes for the nephrin protein, an essential component of the interpodocyte-spanning slit diaphragm [28]. Nephrin is a transmembrane protein of the Ig superfamily characterized by eight C2-type Ig-like domains and a fibronectin type III-like module in the extracellular region, a single transmembrane domain and a cytosolic C-terminal end [10]. Mutations in NPHS1 lead to disruption of the filtration barrier and cause massive protein loss. Nephrin plays a significant role in signaling between podocytes by interacting with CD2AP and podocin [29, 30].
METHODS
Patient and Data Ascertainment
Within a worldwide cohort of children with nephrotic syndrome referred to us since May 2008 for mutational analysis, we selected all the patients who had nephrotic syndrome onset within the first 90 days of life. These were a total of 25 patients from 22 families.
Patients with mutations in the other genes known to cause CNS were excluded from the study. The frequency of mutations, clinical signs, renal and extra-renal signs, and the results of the renal biopsy make up the basis for the choice of genes to be tested. First, we performed mutation analysis for NPHS2 and WT1 for all 25 patients since these are the most frequent monogenic causes of childhood NS [13]. One patient (A3318 II-1) was found to have a homozygous mutation in NPHS2 gene Ex2: c.353 C 1 T (H) (p.P118L) [31] and another patient (A3194 II-1) was revealed to have a novel heterozygous mutation in WT1 gene Ex8: c.1097 G>A (h) (p.R366H). These two patients were excluded from the cohort. Additionally, screening for all 31 exons of PLCE1 was performed in 2 patients (A3205 II-1 and A3360 II-1) with CNS because they revealed renal histology of DMS [18, 19]. However, none of these patients had a mutation in the PLCE1 gene. There were no additional signs for other CNS causing genes to be tested in our cohort; therefore, we performed mutation analysis for NPHS1 for the remaining 23 patients with CNS from 20 families.
Human subject research was approved by the University of Michigan Institutional Reviews Board and the Ethics Commission of the University of Freiburg, Germany. The diagnosis of CNS was made by pediatric nephrologists in specialized centers based on published criteria [32]. Following informed consent, detailed clinical and pedigree information was obtained by a standardized questionnaire available on www.renalgenes.org. Nephrotic range proteinuria was defined as proteinuria >40 mg/m2/h. When evaluating the frequency of mutations, we relate them to families rather than patients because siblings have identical mutations. When evaluating clinical data, we relate them to patients because siblings might differ in their clinical phenotypes. It was shown in one of our previous studies that out of the two siblings with the homozygous missense mutation in NPHS1 gene Ex14: c.1760 T 1 G (H) (p.L587R), only one developed nephrotic syndrome before the age of 90 days, while the other did not manifest until the age of 2 years [33].
Mutation Analysis
Genomic DNA was isolated from blood samples using the Puregene® DNA purification kit (Gentra, Minneapolis, Minn., USA) following the manufacturer’s guidelines. Mutation analysis by direct exon sequencing was performed using exon-flanking primers and by direct sequencing of all the exons for NPHS1, NPHS2, PLCE1 and PAX2. WT1 analysis was limited to exons 8 and 9 since mutations of this gene that account for isolated NS has almost exclusively been reported in these two exons [21, 34]. Exon primers for NPHS1, NPHS2, WT1 and PLCE1 have been published previously [14, 19, 21, 34, 35]. For sequence analysis the software SEQUENCHER 3.8 TM (Gene Codes, Ann Arbor, Mich., USA) was used. The published reference sequence of NPHS1 (NM_004646) was used as the relevant wild-type gene sequence. Sequencing of both DNA strands was performed for all detected mutations and other sequence variants. If parental samples were available, segregation of the variants was confirmed by direct sequencing of parental samples. For each novel mutation, its absence was demonstrated in 96 healthy control individuals of matched ethnic origin by direct sequencing. We here define ‘disease-causing mutations’ as the presence of both alleles of a recessive-disease gene (NPHS1 or NPHS2) and one allele of a dominant disease gene (WT1) that are absent from 96 healthy control individuals and from the ‘1,000 genomes’ database www.1000genomes.org).
Results
Patient Characteristics of the CNS Cohort
In this study, a worldwide cohort of 23 patients from 20 families with CNS was included. All patients were examined for NPHS1 mutations. Families were from the following ethnicities: 7 Caucasian, 2 Turkish, 4 Arabic, 3 Indian, 2 Pakistan, 1 Vietnamese and 1 Hispanic. Eight patients from 8 families were from consanguineous parents. Renal biopsy was performed in 7 of them and showed CNF (3 patients), DMS (2 patients), glomerular mesangial proliferation (1 patient) and mesangioprolifarative glomerulonephritis (1 patient). Because traditionally CNS is considered treatment refractory, 18 patients (78%) did not receive any therapy. In 3 patients (13%) steroid therapy was administered: 2 of them (A3319 II-1 and A3358 II-1) did not respond to the steroid therapy (steroid-resistant nephrotic syndrome). A3358 II-1 was then started on CPA and CsA therapy. The third patient (A3449 II-1) died 24 hours after the administration of steroid therapy so no data on the response was available. One patient (A3360 II-1) was partially responsive to cyclosporine A (CsA). One patient (A3325 II-1) was on antiproteinuric therapy with angiotensin-converting enzyme inhibitors.
NPHS1 Mutations
Mutation analysis by direct exon sequencing of all 29 exons of NPHS1 was performed for a total of 23 patients from 20 families. Both causative NPHS1 mutations were detected in 9 of the 20 families (45%; table 1); therefore, the CNS phenotype is fully explained. NPHS1 mutations represent a recessive single-gene cause of CNS. Recessive single-gene disease causes convey full penetrance of a disease. They are thus distinct from genetic variants that are found only to be associated with disease because associated variants usually explain only a low percentage of the phenotypic variance, as are the cases for instance in the MYH9/APOL1 [36] and HLA [37–42] variants that have been found in nephrotic syndrome.
Table 1.
Clinical and mutation information for 11 families with NPHS1 mutations detected
| Patient number |
Origin | Known Consan- guinity |
Age of onset |
Gender | Renal biopsy |
Treatment | Other clinical features | NPHS1 mutationa (Exon: nucleotide change; aminoacid change) |
Origin of mutation |
Initial phenotype | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous mutations | |||||||||||
| A3205 II-1 | Caucasian | No | 53 d | M | DMS | No treatment | none | EX18:c.2491C>T;p.R831C | Lenkkeri et al. 1999 | North America | CNS, Finnish type |
| A3235 II-3 | Arabic | Yes | 2 mo | F | Not done | No treatment | none | EX2: C.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type |
| A3236 II-1 | Indian Subcontinent | Yes | 1 mo | F | Not done | Conservative treatment | none | IVS 7+1 G>T; splice errorb | This study | ||
| A3325 II-1 | Pakistan | Yes | 2 mo | M | Not done | Albumin infusion, Lisinopril | Grand mal seizures; brother 4 y normal. | EX6:c.614-621delinsTT; p.T205,P206,R207>1205 | Lenkkeri et al. 1999 | Turkey | CNS, Finnish type |
| A3337 II-3 | Arabic | Yes | 1 mo | F | Not done | No treatment | Edema at birth, low set ears, depressed nasal bridge, high arched palatet; two deceased brothers (sample not available). | EX2: C.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type |
| A3416 II-1 | Indian Subcontinent | Yes | 13 d | M | Not done | No treatment | Premature (34 weeks). | EX2: c.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type |
| A3442 II-2 | Indian Subcontinent | No | 1 mo | M | CNF | ND | Microcephaly, aminoaciduria, 3+ glycosuria and acidosis suggesting proximal tubular defect. Died at 6 mo of age, his older sister died at age 3.5 y. Mother had oligohydramnios during pregnancy. | Ex9:c.1099 C>T; p.R367C | Lenkkeri et al. 1999 | France | CNS, Finnish type |
| Compound heterozygous mutations | |||||||||||
| A3322 II-2 | Caucasian | No | no data | M | Not done | No treatment | Degrees of proteinuria, not frank NS, hypothyroidism, hypertension, acidosis. | Ex22: c.2930A>G; p.Y977Cc | This study | ||
| EX27: c.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type | ||||||||
| A3322 II-3 | Caucasian | No | no data | F | Not done | No treatment | Degrees of proteinuria, not frank NS. | Ex22: c.2930 A>G; p.Y977Cc | This study | ||
| EX27: c.3478 C>T; p R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type | ||||||||
| A3322 II-4 | Caucasian | No | no data | F | Not done | No treatment | Degrees of proteinuria, not frank NS. | Ex22: c.2930 A>G; p.Y977Cc | This study | ||
| EX27: c.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type | ||||||||
| A3322 II-5 | Caucasian | No | no data | M | Not done | No treatment | Degrees of proteinuria, not frank NS. | Ex22: c.2930 A>G; p.Y977Cc | This study | ||
| EX27: c.3478 C>T; p.R1160X | Lenkkeri et al. 1999 | Italy | CNS, Finnish type | ||||||||
| A3326 II-1 | Hispanic | No | 1 mo | F | CNF | No treatment | Unilateral nephrectomy (3/2009) with normal renal function afterwards, left inguinal hernia. | Ex2: c.139delG; p.E46fsX127 | Heeringa et al. 2008 | Hispanic | Nephrotic syndrome |
| Ex13: c.1701 C>A; p.C567X | Beltcheva et al. 2001 | Non-Finnish | CNS, Finnish type | ||||||||
| Sinqle Heterozygous mutations | |||||||||||
| A3237 II-1 | Caucasian | No | 3 d | M | Not done | ND | CNS, born preterm (33+3), unexplained cardiorespiratory arrest day 5. | EX6; c. 644 T>G; p. L215R | This study | ||
| A3319 II-1d | Turkish | No | 42 d | F | Glomerular mesangial proliferation | SRNS | none | Ex 15: c.2014 G>A; p.A672T | Machuca et al. 2010 | France | CNS, Finnish type |
CNS = congenital nephrotic syndrome; CSA = cyclosporin-A; d = days; DMS = diffuse mesangial sclerosis; Ex = exon; F = female; FSGS = focal segmental glomerulosclerosis; M = male; mo = months; MPGN = mesangial proliferative glomerulonephritis; ND = No data; NS = nephrotic syndrome; SRNS = steroid resistant nephrotic syndrome; SSNS = steroid sensitive nephrotic syndrome; y = years.
All novel mutations were absent from 96 Turkish control individuals and from the 1,000 genomes project (http://www.1000genomes.org). Novel mutations are printed in bold. Novel missense mutations were conserved through evolution at least down to Danio rerio. RefSeq NM_004646 was used as relevant wild type gene sequence for human NPHS1.
The novel mutation is shown to be segregating from mother and father.
The novel mutation is shown to be segregating from father and the known mutation from mother.
A3319 II-1 also has a single heterozygous mutation in NPHS2 Ex5: c.729G>C; p.E273Q.
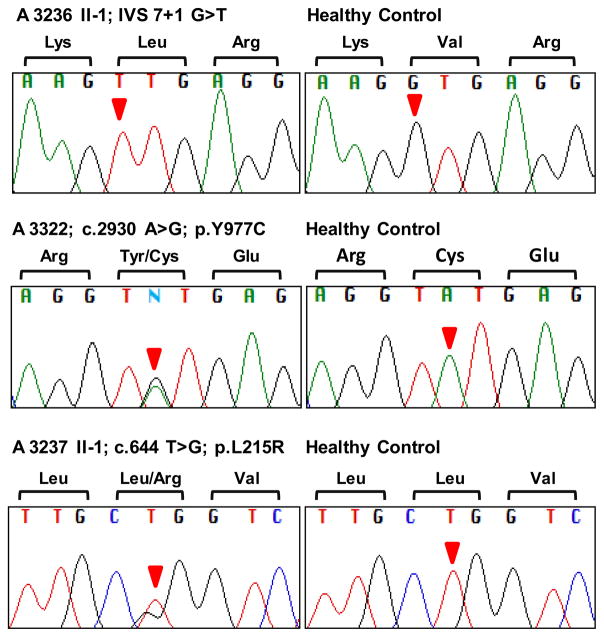
Nine families had disease-causing mutations in NPHS1. The affected individuals of 7 families were harboring the mutation homozygously (table 1). In these, we discovered one novel homozygous splice site mutation (IVS 7 + 1 G>T) in family A3236 (fig. 1). The affected individuals of 2 families (A3322 and A3326) were found to have compound heterozygous mutations (table 1). Family A3322 is compound heterozygous for a novel mutation in Ex22: c.2930 A>G (p.Y977C) along with the known mutation Ex27: c.3478 C>T (p.R1160X) [11] (fig. 1). p.Y977C is conserved down to Danio rerio with Poly-Phen1 [43] and PolyPhen2 [44] scores that are classified to be ‘probably damaging’ (online suppl. table 1, www.karger.com/doi/10.1159/000337379).
Fig. 1. Novel NPHS1 mutations found in this study.
One novel homozygous splice site mutation was found in NPHS1 (IVS 7+1 G>T) in patient A3236 II-1. Additionally, two novel heterozygous mutations, Ex22: c.2930 A>G; p.Y977C and Ex6: c.644 T>G; p.L215R, were detected in family A3322 (all affected) and A3237 II-1, respectively. For each mutation, sequence from the patient and a healthy control individual is shown.
Two patients from 2 families were found to have a single heterozygous mutation only (A3237 and A3319). One of these families (A3237) was carrying a novel single heterozygous mutation in Ex6: c.644 T>G (p.L215R) (fig. 1). p.L215R is conserved down to Caenorhabditis elegans with PolyPhen1 [43] and PolyPhen2 [44] scores that are classified to be ‘probably damaging’ (online suppl. table 1). Therefore, we speculate that our exon sequencing may have missed the second recessive mutation, e.g. a deletion or duplication or intronic mutations or mutations in the promotor region.
Overall, we discovered 10 different mutations, 3 of them novel, consisting of 1 splice site mutation (IVS 7 + 1 G>T) and 2 missense mutations (p.Y977C and p.L215R). We thereby extended the current NPHS1 mutation spectrum of 173 mutations (http://www.biobase-international.com) by 3 novel mutations.
DISCUSSION
In this study, we were able to define the disease-causing mutation in both alleles of NPHS1 in 9 of 20 families and in one allele only in 2 families. Three mutations were novel.
Biopsies were performed in 4 cases out of 11 CNS families in whom we detected a mutation in the NPHS1 gene. The results were CNF (2 patients), DMS (1 patient) and glomerular mesangial proliferation (1 patient). These data confirm the previous findings that NPHS1 mutations can cause a somewhat broader variety of histological phenotypes other than CNF [13, 14, 33].
Although CNF is classically known to be steroid resistant, several cases of steroid-sensitive patients with NPHS1 mutations have been reported previously [14, 45]. In our CNS cohort of patients, A3319 II-I, who carries the NPHS1 mutation in one allele only, was given steroid therapy. This individual had no response to therapy. Another finding was that patient A3325 II-1, who had a homozygous insertion-deletion in NPHS1, was clinically stable on lisinopril only. Previously, 1 patient compound heterozygous for Fin minor and a missense mutation was shown to respond to enalapril [46]. In another study, patients with homozygous missense mutations or patients with compound heterozygosity for a missense mutation and a frameshift mutation (or a small homozygous deletion causing a nonframeshift mutation) were shown to have a partial response to antiproteinuric therapy rarely [33].
We previously reported a CNS case of Hispanic origin (A1893) explained by the mutations in NPHS1 gene Ex2: c.139delG (h) (p.E46fsX127) and c.3482–2 A>G (h) (splice site) [14]. In the current study, we found the same mutation (p.E46fsX127), but this time in compound heterozygosity with Ex13: c.1701 C>A (h) (p.C567X) [47] in patient A3326 II-1, who was also of Hispanic origin. A3326 II-1 was biopsied and proven as CNF and underwent unilateral nephrectomy. The patient had normal renal function of his unilateral kidney afterwards. A previous study showed that CNS management with captopril and indomethacin therapy in combination with unilateral nephrectomy achieves significant improvements in plasma albumin and reduces the need for albumin infusions and time in hospital; therefore, second nephrectomy, dialysis and transplantation can be delayed until the 3rd year of life or longer [48].
Another finding in this study was that family A3416 from the Indian subcontinent was found to have the mutation p.R1160X. In a previous study, p.R1160X was shown to be suggestive of a founder effect and therefore commonly known as the ‘Maltese mutation’ [11]. This mutation was also detected in CNF cases of Indian/Bangladesh origin, but associated with a different allele [11]. In the same study, p.R1160X resulted in an unexpectedly mild CNF phenotype in about half of the cases [11]. In our study at least a part of the patients with this mutation have a very early onset of CNS (13 days, 1 month and 2 months, respectively).
The classical notion that NPHS1 mutations are seen in nephrotic syndrome cases with age of onset in the first 90 days of life was changed by the recent discovery that NPHS1 mutations may cause onset beyond the first 3 months [15]. Previously, it was demonstrated that homozygosity mapping is a useful tool for screening for homozygous disease causing mutations in NPHS1 [33, 49]. In our study, we also screened 9 families for NPHS1 mutations (A3113, A3191, A3310, A3317, A3321, A3323 (2 sibs), A3327, A3329, A3377) that had the onset of NS symptoms beyond 90 days of life with single nucleotide polymorphism arrays (Gene Chip®) from Affymetrix, Inc. with a resolution of 250K (Human Mapping 250K Styl Array). The method has been described in detail previously [33, 49]. However, none of these families were found to have a disease-causing mutation in the NPHS1 gene by direct sequencing (data not shown).
In previous studies, it was shown that approximately one half of CNS cases are caused by recessive mutations in NPHS1 [13, 14, 33]. The NPHS1 mutation rate in our cohort was 45%, accordingly.
Of the mutations described in this study, R1160X was the most frequent. This mutation was found homozygously in 3 families and was found in a compound heterozygous state with the novel mutation Y977C in one family with 4 affected siblings (A3322), accounting for 7 of 40 alleles (17.5%).
Regarding the families in which we did not detect disease-causing mutations in NPHS1, NPHS2, PLCE1 or WT1, we cannot exclude mutations in regulatory elements or introns or heterozygous whole exon deletion as the missing allele since we used an exon sequencing approach that might not detect these mutations. We speculate that mutations affecting other essential slit diaphragm proteins or interaction partners of nephrin may cause the disease in these patients.
Supplementary Material
Acknowledgments
We thank the patients and their physicians for contribution of blood samples and clinical data. This work was supported by grants to F.H. from the National Institutes of Health (DK076683, RC1-DK086542), from the NephCure Foundation and from the Thrasher Research Fund. F.H. is a Doris Duke Distinguished Clinical Scientist, the Frederick G.L. Huetwell Professor for the Cure and Prevention of Birth Defects and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.Norio R. Heredity in the congenital nephrotic syndrome. A genetic study of 57 Finnish families with a review of reported cases. Ann Paediatr Fenn. 1966;27(suppl):21–94. [PubMed] [Google Scholar]
- 2.Huttunen NP. Congenital nephrotic syndrome of Finnish type. Study of 75 patients. Arch Dis Child. 1976;51:344–348. doi: 10.1136/adc.51.5.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahvenainen EK, Hallman N, Hjelt L. Nephrotic syndrome in newborn and young infants. Ann Paediatr Fenn. 1956;2:227–241. [PubMed] [Google Scholar]
- 4.Hallman N, Hjelt L. Congenital nephrotic syndrome. J Pediatr. 1959;55:152–162. doi: 10.1016/s0022-3476(59)80083-4. [DOI] [PubMed] [Google Scholar]
- 5.Worthen HG, Vernier RL, Good RA. Infantile nephrosis; clinical, biochemical, and morphologic studies of the syndrome. AMA J Dis Child. 1959;98:731–748. [PubMed] [Google Scholar]
- 6.Huttunen NP, Rapola J, Vilska J, Hallman N. Renal pathology in congenital nephrotic syndrome of Finnish type: a quantitative light microscopic study on 50 patients. Int J Pediatr Nephrol. 1980;1:10–16. [PubMed] [Google Scholar]
- 7.Rapola J. Renal pathology of fetal congenital nephrosis. Acta Pathol Microbiol Scand A. 1981;89:63–64. doi: 10.1111/j.1699-0463.1981.tb00188.x. [DOI] [PubMed] [Google Scholar]
- 8.Rapola J, Sariola H, Ekblom P. Pathology of fetal congenital nephrosis: immunohistochemical and ultrastructural studies. Kidney Int. 1984;25:701–707. doi: 10.1038/ki.1984.77. [DOI] [PubMed] [Google Scholar]
- 9.Kaukinen A, Kuusniemi AM, Lautenschlager I, Jalanko H. Glomerular endothelium in kidneys with congenital nephrotic syndrome of the Finnish type (NPHS1) Nephrol Dial Transplant. 2008;23:1224–1232. doi: 10.1093/ndt/gfm799. [DOI] [PubMed] [Google Scholar]
- 10.Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, Kashtan CE, Peltonen L, Holmberg C, Olsen A, Trygg vason K. Positionally cloned gene for a novel glomerular protein – nephrin – is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 11.Lenkkeri U, Männikkö M, McCready P, Lamerdin J, Gribouval O, Niaudet PM, Antignac CK, Kashtan CE, Homberg C, Olsen A, Kestilä M, Tryggvason K. Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am J Hum Genet. 1999;64:51–61. doi: 10.1086/302182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchshuber A, Niaudet P, Gribouval O, Jean G, Gubler MC, Broyer M, Antignac C. Congenital nephrotic syndrome of the Finnish type: linkage to the locus in a non-Finnish population. Pediatr Nephrol. 1996;10:135–138. doi: 10.1007/BF00862052. [DOI] [PubMed] [Google Scholar]
- 13.Hinkes BG, Mucha B, Vlangos CN, Gbadegesin R, Liu J, Hasselbacher K, Hangan D, Ozaltin F, Zenker M, Hildebrandt F Arbeitsgemeinschaft für Paediatrische Nephrologie Study Group. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2) Pediatrics. 2007;119:e907–e919. doi: 10.1542/peds.2006-2164. [DOI] [PubMed] [Google Scholar]
- 14.Heeringa SF, Vlangos CN, Chernin G, Hinkes B, Gbadegesin R, Liu J, Hoskins BE, Ozaltin F, Hildebrandt F Members of the APN Study Group. Thirteen novel NPHS1 mutations in a large cohort of children with congenital nephrotic syndrome. Nephrol Dial Transplant. 2008;23:3527–3533. doi: 10.1093/ndt/gfn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippe A, Nevo F, Esquivel EL, Reklaityte D, Gribouval O, Tête MJ, Loirat C, Dantal J, Fischbach M, Pouteil-Noble C, Decramer S, Hoehne M, Benzing T, Charbit M, Niaudet P, Antignac C. Nephrin mutations can cause childhood-onset steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2008;19:1871–1878. doi: 10.1681/ASN.2008010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Doné SC, Khoshnoodi J, Bertorello A, Wartiovaara J, Berggren PO, Tryggvason K. Defective nephrin trafficking caused by missense mutations in the NPHS1 gene: insight into the mechanisms of congenital nephrotic syndrome. Hum Mol Genet. 2001;10:2637–2644. doi: 10.1093/hmg/10.23.2637. [DOI] [PubMed] [Google Scholar]
- 17.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 18.Gbadegesin R, Hinkes BG, Hoskins BE, Vlangos CN, Heeringa SF, Liu J, Loirat C, Ozaltin F, Hashmi S, Ulmer F, Cleper R, Ettenger R, Antignac C, Wiggins RC, Zenker M, Hildebrandt F. Mutations in PLCE1 are a major cause of isolated diffuse mesangial sclerosis (IDMS) Nephrol Dial Transplant. 2008;23:1291–1297. doi: 10.1093/ndt/gfm759. [DOI] [PubMed] [Google Scholar]
- 19.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nürnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C, Hennies HC, Goyal M, Wharram BL, Schachter AD, Mudumana S, Drummond I, Kerjaschki D, Waldherr R, Dietrich A, Ozaltin F, Bakkaloglu A, Cleper R, Basel-Vanagaite L, Pohl M, Griebel M, Tsygin AN, Soylu A, Müller D, Sorli CS, Bunney TD, Katan M, Liu J, Attanasio M, O’toole JF, Hasselbacher K, Mucha B, Otto EA, Airik R, Kispert A, Kelley GG, Smrcka AV, Gudermann T, Holzman LB, Nürnberg P, Hildebrandt F. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 20.Jeanpierre C, Denamur E, Henry I, Cabanis MO, Luce S, Cécille A, Elion J, Peuchmaur M, Loirat C, Niaudet P, Gubler MC, Junien C. Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. Am J Hum Genet. 1998;62:824–833. doi: 10.1086/301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mucha B, Ozaltin F, Hinkes BG, Hasselbacher K, Ruf RG, Schultheiss M, Hangan D, Hoskins BE, Everding AS, Bogdanovic R, Seeman T, Hoppe B, Hildebrandt F Members of the APN Study Group. Mutations in the Wilms’ tumor 1 gene cause isolated steroid resistant nephrotic syndrome and occur in exons 8 and 9. Pediatr Res. 2006;59:325–331. doi: 10.1203/01.pdr.0000196717.94518.f0. [DOI] [PubMed] [Google Scholar]
- 22.Zenker M, Aigner T, Wendler O, Tralau T, Müntefering H, Fenski R, Pitz S, Schumacher V, Royer-Pokora B, Wühl E, Cochat P, Bouvier R, Kraus C, Mark K, Madlon H, Dötsch J, Rascher W, Maruniak-Chudek I, Lennert T, Neumann LM, Reis A. Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet. 2004;13:2625–2632. doi: 10.1093/hmg/ddh284. [DOI] [PubMed] [Google Scholar]
- 23.Hasselbacher K, Wiggins RC, Matejas V, Hinkes BG, Mucha B, Hoskins BE, Ozaltin F, Nürnberg G, Becker C, Hangan D, Pohl M, Kuwertz-Bröking E, Griebel M, Schumacher V, Royer-Pokora B, Bakkaloglu A, Nürnberg P, Zenker M, Hildebrandt F. Recessive missense mutations in LAMB2 expand the clinical spectrum of LAMB2-associated disorders. Kidney Int. 2006;70:1008–1012. doi: 10.1038/sj.ki.5001679. [DOI] [PubMed] [Google Scholar]
- 24.Choi HJ, Lee BH, Kang JH, Jeong HJ, Moon KC, Ha IS, Yu YS, Matejas V, Zenker M, Choi Y, Cheong HI. Variable phenotype of Pierson syndrome. Pediatr Nephrol. 2008;23:995–1000. doi: 10.1007/s00467-008-0748-7. [DOI] [PubMed] [Google Scholar]
- 25.Diomedi-Camassei F, Di Giandomenico S, Santorelli FM, Caridi G, Piemonte F, Montini G, Ghiggeri GM, Murer L, Barisoni L, Pastore A, Muda AO, Valente ML, Bertini E, Emma F. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol. 2007;18:2773–2780. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- 26.Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rötig A, Nürnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Müller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocaña C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nürnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruotsalainen V, Ljungberg P, Wartiovaara J, Lenkkeri U, Kestilä M, Jalanko H, Holmberg C, Tryggvason K. Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc Natl Acad Sci USA. 1999;96:7962–7967. doi: 10.1073/pnas.96.14.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih NY, Li J, Karpitskii V, Nguyen A, Dustin ML, Kanagawa O, Miner JH, Shaw AS. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;286:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 30.Khoshnoodi J, Sigmundsson K, Ofverstedt LG, Skoglund U, Obrink B, Wartiovaara J, Tryggvason K. Nephrin promotes cell-cell adhesion through homophilic interactions. Am J Pathol. 2003;163:2337–2346. doi: 10.1016/S0002-9440(10)63590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber S, Gribouval O, Esquivel EL, Morinière V, Tête MJ, Legendre C, Niaudet P, Antignac C. NPHS2 mutation analysis shows genetic heterogeneity of steroid resistant nephrotic syndrome and low posttransplant recurrence. Kidney Int. 2004;66:571–579. doi: 10.1111/j.1523-1755.2004.00776.x. [DOI] [PubMed] [Google Scholar]
- 32.Arbeitsgemeinschaft für Pädiatrische Nephrologie. Short versus standard prednisone therapy for initial treatment of idiopathic nephrotic syndrome in children. Lancet. 1988;1:380–383. [PubMed] [Google Scholar]
- 33.Schoeb DS, Chernin G, Heeringa SF, Matejas V, Held S, Vega-Warner V, Bockenhauer D, Vlangos CN, Moorani KN, Neuhaus TJ, Kari JA, Macdonald J, Saisawat P, Ashraf S, Ovunc B, Zenker M, Hildebrandt F Members of the Gesellschaft für Paediatrische Nephrologie (GPN) Study Group. Nineteen novel NPHS1 mutations in a worldwide cohort of patients with congenital nephrotic syndrome (CNS) Nephrol Dial Transplant. 2010;25:2970–2976. doi: 10.1093/ndt/gfq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruf RG, Schultheiss M, Lichtenberger A, Karle SM, Zalewski I, Mucha B, Everding AS, Neuhaus T, Patzer L, Plank C, Haas JP, Ozaltin F, Imm A, Fuchshuber A, Bakkaloglu A, Hildebrandt F APN Study Group. Prevalence of WT1 mutations in a large cohort of patients with steroid-resistant and steroid sensitive nephrotic syndrome. Kidney Int. 2004;66:564–570. doi: 10.1111/j.1523-1755.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 35.Ruf RG, Lichtenberger A, Karle SM, Haas JP, Anacleto FE, Schultheiss M, Zalewski I, Imm A, Ruf EM, Mucha B, Bagga A, Neuhaus T, Fuchshuber A, Bakkaloglu A, Hildebrandt F Arbeitsgemeinschaft für Pädiatrische Nephrologie Study Group. Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol. 2004;15:722–732. doi: 10.1097/01.asn.0000113552.59155.72. [DOI] [PubMed] [Google Scholar]
- 36.Genovese G, Tonna SJ, Knob AU, Appel GB, Katz A, Bernhardy AJ, Needham AW, Lazarus R, Pollak MR. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int. 2010;78:698–704. doi: 10.1038/ki.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T, Ogawa A, Takahashi K, Uchiyama M. HLA-DQB1 allele associates with idiopathic nephrotic syndrome in Japanese children. Acta Paediatr Jpn. 1995;37:293–296. doi: 10.1111/j.1442-200x.1995.tb03317.x. [DOI] [PubMed] [Google Scholar]
- 38.Haeffner A, Abbal M, Mytilineos J, Konrad M, Krammer I, Bouissou F, Opelz G, Schärer K, Cambon-Thomsen A. Oligotyping for HLA-DQA, -DQB, and -DPB in idiopathic nephrotic syndrome. Pediatr Nephrol. 1997;11:291–295. doi: 10.1007/s004670050279. [DOI] [PubMed] [Google Scholar]
- 39.Bakr AM, El-Chenawy F. HLA-DQB1 and DRB1 alleles in Egyptian children with steroid-sensitive nephrotic syndrome. Pediatr Nephrol. 1998;12:234–237. doi: 10.1007/s004670050445. [DOI] [PubMed] [Google Scholar]
- 40.Bakr AM, El-Chenawi F, Al-Husseni F. HLA alleles in frequently relapsing steroid-dependent and -resistant nephrotic syndrome in Egyptian children. Pediatr Nephrol. 2005;20:159–162. doi: 10.1007/s00467-004-1730-7. [DOI] [PubMed] [Google Scholar]
- 41.Krasowska-Kwiecień A, Sancewicz-Pach K, Moczulska A. Idiopathic nephrotic syndrome in Polish children – its variants and associations with HLA. Pediatr Nephrol. 2006;21:1837–1846. doi: 10.1007/s00467-006-0271-7. [DOI] [PubMed] [Google Scholar]
- 42.Huang YY, Lin FJ, Fu LS, Lan JL. HLA-DR,-DQB typing of steroid-sensitive idiopathic nephrotic syndrome children in Taiwan. Nephron Clin Pract. 2009;112:c57–c64. doi: 10.1159/000213082. [DOI] [PubMed] [Google Scholar]
- 43.PolyPhen1. Prediction of functional effect of human nsSNPs. http://genetics.bwh.harvard.edu/pph/
- 44.PolyPhen2. Prediction of functional effect of human nsSNPs. http://genetics.bwh.harvard.edu/pph2/
- 45.Kitamura A, Tsukaguchi H, Hiramoto R, Shono A, Doi T, Kagami S, Iijima K. A familial childhood-onset relapsing nephritic syndrome. Kidney Int. 2007;71:946–951. doi: 10.1038/sj.ki.5002110. [DOI] [PubMed] [Google Scholar]
- 46.Patrakka J, Kestilä M, Wartiovaara J, Ruotsalainen V, Tissari P, Lenkkeri U, Männikkö M, Visapää I, Holmberg C, Rapola J, Tryggvason K, Jalanko H. Congenital nephrotic syndrome (NPHS1): features resulting from different mutations in Finnish patients. Kidney Int. 2000;58:972–980. doi: 10.1046/j.1523-1755.2000.00254.x. [DOI] [PubMed] [Google Scholar]
- 47.Beltcheva O, Martin P, Lenkkeri U, Tryggvason K. Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum Mutat. 2001;17:368–373. doi: 10.1002/humu.1111. [DOI] [PubMed] [Google Scholar]
- 48.Kovacevic L, Reid CJ, Rigden SP. Management of congenital nephrotic syndrome. Pediatr Nephrol. 2003;5:426–430. doi: 10.1007/s00467-003-1131-3. [DOI] [PubMed] [Google Scholar]
- 49.Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MT, Hinkes BG, Chaib H, Ashraf S, Schoeb DS, Ovunc B, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, Macdonald J, Nürnberg P, Otto EA. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machuca E, Benoit G, Nevo F, Tête MJ, Gribouval O, Pawtowski A, Brandström P, Loirat C, Niaudet P, Gubler MC, Antignac C. Genotype-phenotype correlations in non-Finnish congenital nephrotic syndrome. J Am Soc Nephrol. 2010;21:1209–121. doi: 10.1681/ASN.2009121309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.