Abstract
Defects in genes involved in DNA damage repair (DDR) pathway are emerging as novel biomarkers and targets for new prostate cancer drug therapies. A previous report revealed an association between an exceptional response to cisplatin treatment and a somatic loss of heterozygosity (LOH) of FANCA in a patient with metastatic prostate cancer who also harbored a germline FANCA variant (S1088F). Although germline FANCA mutations are the most frequent alterations in patients with Fanconi anemia, germline alterations are less common in prostate cancer. We hypothesized that the germline S1088F FANCA variant in combination with FANCA LOH was deleterious for FANCA function and contributed to the patient's exceptional response to cisplatin. We show that although it properly localizes to the nucleus, the S1088F FANCA mutant protein disrupts the FANC protein complex resulting in increased sensitivity to DNA damaging agents. Because molecular stratification is emerging as a strategy for treating men with metastatic, castrate-resistant prostate cancer harboring specific DDR gene defects, our findings suggest that more biomarker studies are needed to better define clinically relevant germline and somatic alterations.
Keywords: prostate cancer
INTRODUCTION
DNA damage repair (DDR) genes are frequently altered in metastatic prostate cancer and have been associated with sensitivity to poly (adenosine diphosphate [ADP]–ribose) polymerase (PARP) inhibitor therapy. Data from a Phase 2 study of olaparib for patients with metastatic castration-resistant prostate cancer (mCRPC) revealed that of the 16 out of 49 (33%) patients that had a response to olaparib, a significant number (14 of the 16 [88%]) of patients harbored one or more germline or somatic deleterious alterations in 12 DNA repair genes including a homozygous deletion of FANCA (Mateo et al. 2015). The full scope of biomarkers for molecular stratification for DDR targeted therapy or platinum needs characterization.
PARP1 is a key protein in the DNA single-strand break (SSB) repair pathway of base excision repair but also plays role in double-strand break (DSB) repair pathways (Schultz et al. 2003; Schreiber et al. 2006; Krishnakumar and Kraus 2010), which is why PARP1 inhibition, which leads to persistent SSBs that are converted to DSBs, and BRCA1/2 loss of function results in synthetic lethality in breast, ovarian, and prostate cancer (Fong et al. 2009). Therefore, deficiencies in proteins that are essential for homologous recombination (HR) and which afford a “BRCAness” phenotype (e.g., FANC proteins; Taniguchi et al. 2003; McCabe et al. 2006) sensitize cells to PARP1 inhibition. Several possible mechanisms for this have been suggested (De Lorenzo et al. 2013), but recent studies suggest that PARP1 interacts with the Fanconi anemia (FA) pathway to inhibit excessive nonhomologous end joining (NHEJ) during DNA damage and inhibition of PARP in FANC-deficient cells have hyperactivation of NHEJ and increased DNA damage producing a synthetic lethality phenotype (Du et al. 2016).
Fanconi anemia is a rare, genetically heterogeneous syndrome with increased predisposition to a broad range of cancers and bone marrow failure (Brosh et al. 2016). Mutations in 20 genes encoding the Fanconi complementary group of proteins (FANCA-FANCU) have been observed in FA patients (Dong et al. 2015; Park et al. 2016). FANC proteins are involved in chromosomal stability and cellular resistance to DNA interstrand cross-linkers (ICLs) such as mitomycin C (MMC) (Gurtan and D'Andrea 2006) or cisplatin. In cells without FANC gene alterations, the FANC proteins FANCA, B, C, E, F and G, and L form a complex (Garcia-Higuera et al. 2001; Meetei et al. 2003, 2004). During the S phase of the cell cycle, FANCL monoubiquitinates and activates FANCD2, triggering FANCD2's association with chromatin and its accumulation in nuclear foci. These foci mark the sites in which DSB repair occurs. Activated FANCD2 colocalizes with factors such as BRCA1, BRCA2/FANCD1, and RAD51, which are involved in HR-mediated DSB repair (Taniguchi et al. 2002). We previously reported a prostate cancer patient (PM12) with small-cell neuroendocrine prostate cancer, a relatively uncommon, aggressive prostate cancer phenotype with limited available treatment options and poor overall survival (Wang et al. 2014), and who showed a complete and durable remission after systemic cisplatin-based chemotherapy. Subsequent analysis identified a germline variant in the FANCA gene (S1088F) (Table 1) with the tumor bearing a loss of the wild-type allele (Beltran et al. 2015).
Table 1.
FANCA variant summary
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP | Genotype | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| FANCA | 16 | NM_000135.2:c.3263C>T | NP_000126.2:p.Ser1088Phe | Substitution | Ser>Phe | 17,233,497 | Heterozygous | SCV000494044 |
HGVS, Human Genome Variation Society; dbSNP, Database for Short Genetic Variations.
Somatic mutations in FANC genes occur with varying frequencies in prostate cancer with ∼6% of tumors harboring a homozygous FANCA deletion in localized TCGA (The Cancer Genome Atlas Research 2015) and CRPC (SU2C; Robinson et al. 2015), which is notable as other DNA repair defects are enriched in CRPC. Because FANCA is located at the telomere of Chromosome 16, deletion calls were scored manually in these data sets. Germline FANCA mutations in prostate cancer patients in the same cohorts are observed with minor allele frequency of 0.065. Using preclinical prostate cancer models including isogenic cell lines and patient-derived xenografts (PDXs) generated from the exceptional responder patient, we found that prostate cancer cells with FANCA deletion resulted in a higher sensitivity to cisplatin compared with cells with wild-type FANCA (Beltran et al. 2015). The impact of the germline FANCA (S1088F) variant on FANC complex function and cisplatin sensitivity remains uncharacterized and is the focus of this current study.
RESULTS
FANCA S1088F Variant Enhances Sensitivity to DNA Damaging Agents
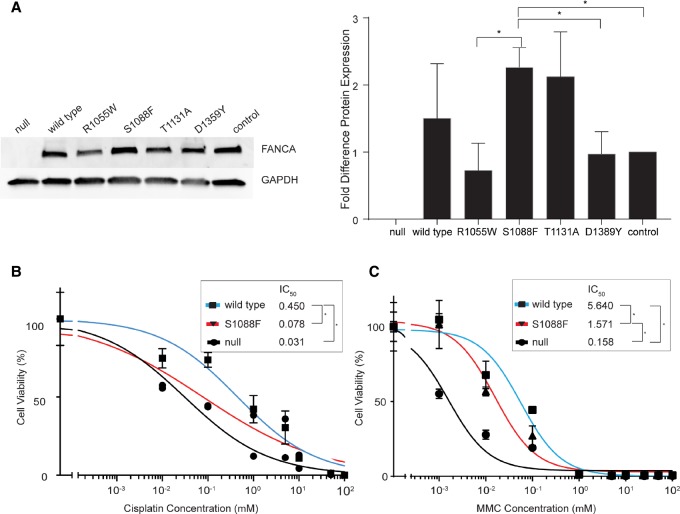
To investigate the consequence of the FANCA S1088F variant to drug sensitivity and DDR, we included a comparison to three mutations from the Fanconi Anemia Mutation Database (http://www.rockefeller.edu/fanconi/mutate/) that have been shown to result in strong (R1055W; seven reports), mild (T1131A; 19 reports), and weak (D1359Y; two reports) impact on MMC drug sensitivity and FANCD2 monoubiquitination (Adachi et al. 2002). We generated isogenic cell lines that express each of these FANCA mutant proteins, R1055W, T1131A, D1359Y, or S1088F, as well as the wild-type FANCA in the FANCA null cell line RA3087 (Zhou et al. 2012). Although moderate differences in protein levels were observed between specific mutant proteins (e.g., between R1055W and S1088F [P = 0.0374]; S1088F and D1359Y [P = 0.0439], and S1088F and control [P = 0.0126]), no significant difference was observed between cells expressing S1088F and wild-type FANCA (Fig. 1A). However, the S1088F FANCA cell line showed an enhanced sensitivity to cisplatin (IC50 = 0.078 µM) compared with cells expressing wild-type FANCA (IC50 = 0.45 µM, P = 0.004) but not as sensitive as FANCA null cells (IC50 = 0.031 µM, P < 0.0001, Fig. 1B). Similarly, the S1088F FANCA cells also showed an enhanced sensitivity to MMC (IC50 = 1.57 µM) compared with wild-type FANCA (IC50 = 5.64 µM, P = 0.0006, Fig. 1C) but are less sensitive than FANCA null cells (IC50 = 0.158 µM, P < 0.0001).
Figure 1.
(A) Western blot of FANCA expression levels in isogenic cell lines. The graph indicates fold difference in the level of FANCA protein normalized to the FANCA endogenous control (* specifies P < 0.0439). (B) Drug sensitivity to cisplatin representing IC50 of null, wild-type, and S1088F cell lines (* specifies significant difference P < 0.004). (C) Drug sensitivity to mitomycin C (MMC) illustrating the IC50 (* indicates significant difference P < 0.006).
FANCA S1088F Variant Alters FANC Complex Function and Results in Loss of Monoubiquination of FANCI
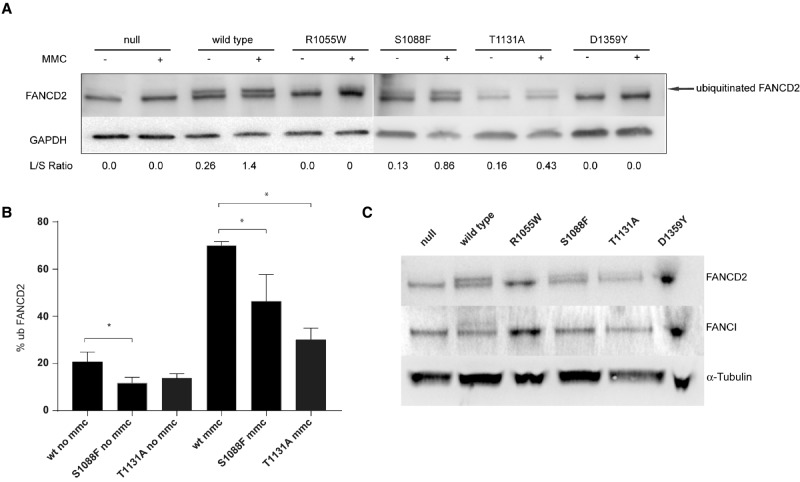
Intact FANC complex formation is essential for the monoubiquitination of FANCD2 (Garcia-Higuera et al. 2001). We therefore queried if FANCA S1088F variant impacted FANCD2/FANCI monoubiquitination. Cells were then treated with either dimethyl sulfoxide (DMSO) (vehicle) or 1 µM MMC for 24 h. Cells expressing the FANCA mutations show changes in the ratio of nonubiquitinated and monoubiquitinated FANCD2 following incubation in 1 µM MMC (Fig. 2A). As expected, cells expressing wild-type FANCA show both nonubiquitinated and monoubiquitinated FANCD2 isoforms with the monoubiquitination levels increasing from 20% to 70% following MMC treatment. No monoubiquitinated FANCD2 was observed either in vehicle or MMC-treated cell lines with no FANCA protein or expressing the deleterious R1055W and D1359Y FANCA mutants (Fig. 2B). Compared with wild-type FANCA, we observed S1088F and T1131A FANCA variants having lower levels of monoubiquitinated FANCD2 when untreated (11% and 13.5%, respectively) with a significant difference seen between wild-type FANCA and S1088F FANCA cell lines (P = 0.0328). Following MMC treatment we observe an increase in monoubiquitination of FANCD2 in S1088F and T1131A FANCA expressing cells to 46% and 30%, respectively. Both of these levels are significantly lower (P = 0.0235 and P = 0.0003, respectively) than monoubiquitinated FANCD2 levels in wild-type FANCA cells. None of the cell lines expressing the different FANCA mutants yielded detectable FANCI monoubiquitination levels, including cells expressing the S1088F FANCA mutant (Fig. 2C; Supplemental Fig. 1) following MMC treatment.
Figure 2.
(A) Western blot expression of FANCD2 with 1 µM mitomycin C (MMC) treatment or vehicle. Arrow indicates monoubiquitinated isoform and lower band is the nonubiquitinated FANCD2 protein. Longer/shorter (L/S) ratios are also included. (B) Bar graph specifies the percentage of monoubiquitinated FANCD2 with and without MMC treatment (* specifies significant difference P < 0.328). (C) Western blot expression of FANCD2 and FANCI with 1 µM MMC treatment.
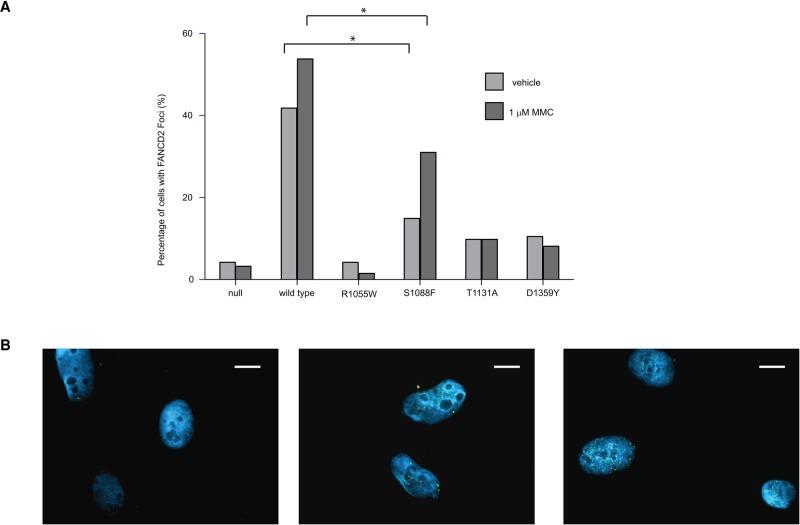
FANCD2/FANCI protein complex foci formation is observed during DNA repair (Smogorzewska et al. 2007) and is absent when there is a deleterious mutation in any of the FANC core complex genes. We scored each cell as FANCD2-foci positive (five or more fluorescent foci/nuclei) or negative (fewer than five fluorescent foci/nuclei) for the different cell lines expressing each of the FANCA mutations at baseline or after MMC treatment (results are summarized in Fig. 3A,B). At baseline, we observed little or no increase in the number of FANCD2 foci-positive nuclei in cells expressing FANCA-null (4.2%), T1131A (9.8%), and D1359Y (10.5%), all of which were significantly less than cells expressing wild-type FANCA (41.8%). As expected, MMC treatment increased the level of FANCD2 foci-positive cells that expressed wild-type FANCA (53.8%) but had no effect on FANCA-null cells (3.1%) or cells expressing deleterious FANCA mutations, R1055W (1.5%), T1131A (9.8%), or D1359Y (8.8%). Cells expressing the S1088F FANCA variant also displayed a fewer number of FANCD2 foci-positive nuclei (14.9%) compared with cells expressing wild-type FANCA expressing cells at baseline, but, unlike the deleterious mutations, MMC treatment resulted in a twofold increase (31%) in the number of FANCD2 foci-positive nuclei, suggesting that the S1088F variant results in a partial loss of function on FANCA function.
Figure 3.
(A) The graph shows the percentage of cells whose nuclei are identified as having five or greater FANCD2 foci with either vehicle or 1 µM mitomycin C (MMC) treatment. Fisher exact tests between wild-type FANCA and S1088F were performed and a significant difference was marked (*, P < 0.0211). (B) Typical FANCD2 foci formation in 1 µM MMC-treated cell lines: (left to right) B1 FANCA null, B2 FANCA wild type, and B3 FANCA S1088F cell lines. Blue, DAPI; green, FANCD2. Scale bar, ∼10 µm.
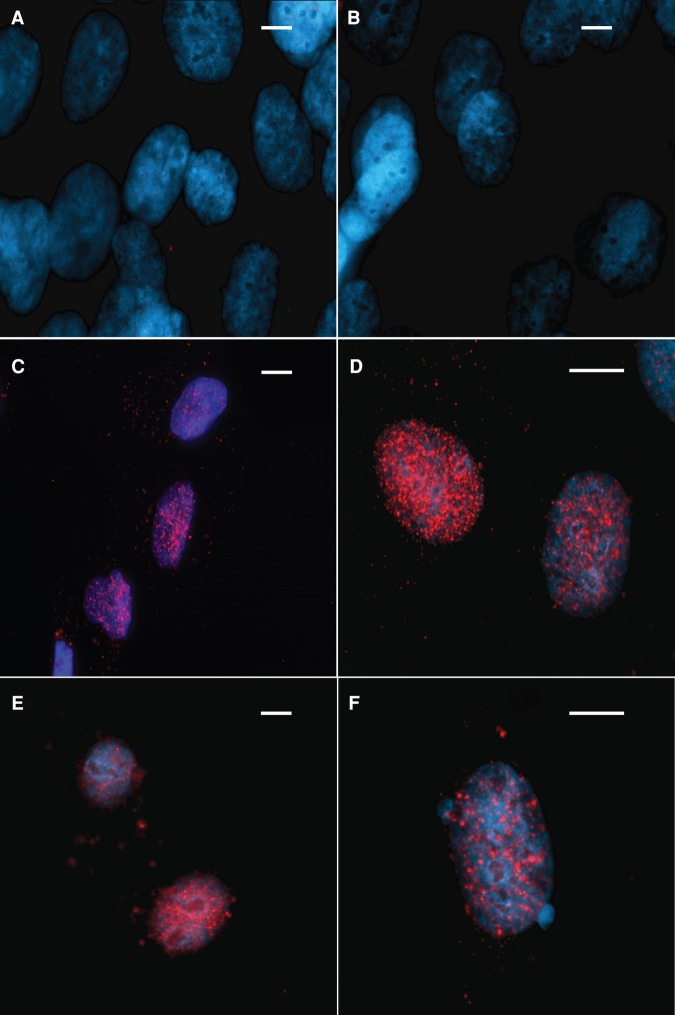
Nuclear localization of FANCA has been shown to be coordinated through a nuclear localization signal (NLS) in the FANCA protein, disruption of which leads to MMC sensitivity (Garcia-Higuera et al. 1999; Lightfoot et al. 1999). We assessed the localization of the S1088F FANCA variant and as a control, wild-type FANCA at baseline and after MMC treatment. Both the wild-type FANCA and the S1088F variant showed predominantly nuclear localization in the absence of MMC treatment suggesting that S1088F FANCA variant does not alter nuclear localization of the protein (Fig. 4).
Figure 4.
Cellular localization of FANCA protein. Red, FANCA; blue, DAPI. Nuclear localization of null FANCA control showing no FANCA signal with (A) no mitomycin C (MMC) and (B) 1 mM MMC. Nuclear localization of wild-type FANCA with (C) no MMC and (D) 1 mM MMC. Nuclear locatization of FANCA in the S1088F FANCA cell line with (E) no MMC and (F) 1 mM MMC. Scale bar, ∼10 µm.
FANCA S1088F Variant Sensitizes Cells to PARP Inhibitors In Vitro and In Vivo
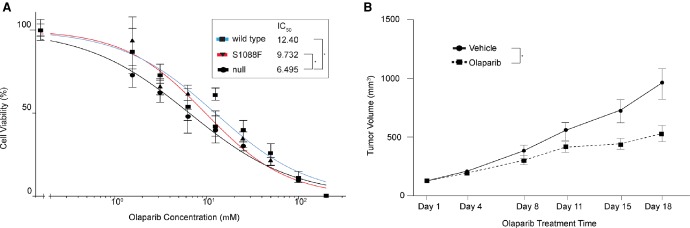
We queried if the S1088F variant sensitizes cells to olaparib. We found that the isogenic cell line expressing the S1088F variant showed an increase in sensitivity to olaparib; however, this change was not significant (S1088F FANCA IC50 = 9.7 µM vs. wild-type FANCA IC50 = 12.4 µM, P = 0.269) (Fig. 5A). As the in vitro models do not fully recapitulate the FANCA status observed in the patient PM12 tumor (germline variant S1088F; Beltran et al. 2015), we treated mice carrying a patient-derived xenograft (PDX) derived from PM12 tumor (LTL545) with olaparib for 18 d. The LTL545 tumors showed significantly enhanced sensitivity (P < 0.0001) to olaparib compared with the vehicle-treated group (Fig. 5B).
Figure 5.
(A) Dose–response to olaparib indicating the IC50 of null, wild-type, and S1088F (* specifies significant difference P < 0.0373). (B) Growth of the patient-derived xenograft (PDX) from patient PM12 (LTL545) with and without olaparib treatment over 18 d (*, P < 0.0001).
DISCUSSION
In the Phase 2 clinical trial of olaparib reported by Mateo et al. 15 out of the 16 mCRPC patients that showed favorable response to the PARP1 inhibitor olaparib harbored alterations in at least one of 12 DDR pathway genes assayed, 13 of which had BRCA1/2 or ataxia telangiectasia mutated (ATM) alterations (Mateo et al. 2015). This suggests that there are potentially other DDR gene alterations that may associate with olaparib response. DDR alterations have also been associated with platinum sensitivity (Cheng et al. 2016). In a recent SU2C-PCF study evaluating metastatic biopsies from patients with mCRPC, 19% of the 150 patients harbored BRCA1, BRCA2, and ATM aberrations, whereas 3.7% of patients harbored other DDR gene aberrations (Robinson et al. 2015). In an extended study to evaluate germline alterations in DDR genes across 692 patients (Prichard et al. 2016), 11.8% of metastatic prostate cancer patients were found to harbor deleterious germline DDR alterations, 64.5% of which were BRCA1/2 or ATM. We have previously published data showing an association between favorable response to cisplatin and FANCA loss (Beltran et al. 2015). Here, we show that although FANCA S1088F protein properly localizes to the nucleus, it alters FANC complex function, enhances sensitivity to DNA damaging agents, and sensitizes cells to PARP inhibitors in vitro and in vivo. This is consistent with previous reports that showed mutations in FANCA were associated with differing sensitivity to DNA cross-linking agents (Adachi et al. 2002; Taniguchi et al. 2003; Kachnic et al. 2010) or nuclear localization (Ferrer et al. 2005). All mutant FANCA expressing cell lines we analyzed including S1088F show no monoubiquitination of FANCI even after MMC treatment. The pathway by which FANCI and FANCD2 are monoubiquitinated is becoming better understood (Smogorzewska et al. 2007; Castella et al. 2015; Liang et al. 2016; Swuec et al. 2016; van Twest et al. 2016). A previous study has shown that a mutation in FANCI results in a protein that is unable to be monoubiquitinated and reduced FANCD2 monoubiquitination (Smogorzewska et al. 2007). Somatic mutations in FANC genes occur with varying frequencies in prostate cancer with ∼6% of tumors harboring a homozygous FANCA deletion, TCGA (The Cancer Genome Atlas Research 2015) or SU2C (Robinson et al. 2015). Although less common than BRCA and ATM alterations (19% combined mutation rate in mCRPC patients; Robinson et al. 2015), alterations in FANCA may also be informative in terms of inclusion criteria for PARP inhibitor or platinum trials.
METHODS
Cell Culture
The fibroblast cell line RA3087, which lacks the FANCA protein, was used as the parental cell line for the overexpression of the FANCA constructs. RA3087 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 15% fetal bovine serum (FBS) with penicillin and streptomycin. 293T cells were cultured in DMEM with 10% FBS with penicillin and streptomycin.
Generation of Mutant FANCA Constructs and Lentiviral Transduction into RA3087 Cells
Mutant FANCA expression plasmids were obtained by site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit, Agilent) from a wild-type open reading frame (ORF) construct (the primers used for generating FANCA mutations are listed in Table 2). Mutant and wild-type FANCA ORFs were then polymerase chain reaction (PCR)-amplified using high-fidelity Taq polymerase (Herculase II fusion DNA polymerases, Agilent) and primers that generated a FANCA ORF with a carboxy-terminal Flag tag. PCR fragments were cloned into pcr8/gw/topo entry vector (Invitrogen) and Gateway-cloned into a lentiviral expression vector (pHAGE_CMV_IP_HAFLAG; kindly provided by A. Smogorzewska) and Sanger sequence–verified. Each expression plasmid was transfected together with the packaging plasmids (SV40 Gag/Pol, SV40 VSVG, SV40 Rev, and SV40 Tat) described previously (Mostoslavsky et al. 2005). Supernatant containing lentiviral particles were collected, filtered, and placed on RA3087 cells with polybrene at a final concentration of 8 µg/ml. Following 24 h incubation, the media containing the virus is removed, and fresh complete media is added. Seventy-two hours after infection, the media is replaced with fresh media containing puromycin at 1 µg/ml. Following 7 d of puromycin selection, cells were collected to check for FANCA protein expression.
Table 2.
List of primers used for site-directed mutagenesis of FANCA
| FANCA-S1088F-F | 5′-CCGCCTGCCTTCGTTTGTCCTCTGCGGCA-3′ |
| FANCA-S1088F-R | 5′-CTGCCGCAGAGGACAAACGAAGGCACC-3′ |
| FANCA-R1055W-F | 5′-GATTTTCCGCAGATGGCTCCAGGCTC-3′ |
| FANCA-R1055W-R | 5′-GAGCCTGGCGCCATCTGCGGCAAATC-3′ |
| FANCA-T1131A-F | 5′-CTGACACAGGACATCGCCGCCCACTTCTTCAG-3′ |
| FANCA-T1131A-R | 5′-CTGAAGAAGTGGGCGGCGATGTCCTGTGTCAG-3′ |
| FANCA-D1359Y-F | 5′-CTGCATGTTGCTGTGTACATGTACTTGAAGC-3′ |
| FANCA-D1359Y-R | 5′-GCTTCAAGTACATGTACACAGCAACATGCAG-3′ |
Drug Treatment
Cisplatin drug assays were performed as before (Beltran et al. 2015). Briefly, cells were seeded on six-well tissue culture plates. Cell lines were treated with either vehicle (0.1% DMSO) or increasing doses of cisplatin. Following 4 d incubation, cells were trypsinized, reseeded in 96-well plates at 1000 cells per well, and cultured for a further 4 d. MMC and olaparib drug sensitivity assays were performed in a 96-well plate format where cells were seeded at 1000 cells per well 24 h before drug treatment. Media was removed and replaced with media containing the appropriate drug concentration. The cells were cultured for 4 d; the media was replaced with fresh media containing the appropriate drug dose and cultured for an additional 4 d. Cell viability was measured using Cell Titer-Glo luminescent assay (Promega Corporation), as we have previously described (Beltran et al. 2013) and according to the manufacturer's instructions. Cell viability dose response data were first normalized to data using the vehicle treated control and then analyzed using nonlinear regression in which the log (inhibitor) concentration versus normalized response curves were generated and the IC50 doses were calculated (GraphPad Software; www.graphpad.com).
Immunoblot Analysis
Protein lysates were prepared in RIPA buffer (radioimmunoprecipitation assay lysis buffer) supplemented with protease inhibitor cocktail and phosphatase inhibitors (Thermo Scientific). The total protein concentration of the soluble extract was determined using the BCA protein assay kit (Thermo Scientific). For FANCA each protein sample (20 µg) was resolved on a 4%–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), transferred onto a PDF (polyvinylidene difluoride) membrane (Bio-Rad), and incubated overnight at 4°C with primary antibodies. The antibodies used were anti-FANCA 1/5000 (A301-980A, Bethyl Laboratories). Following three washes with Tris-buffered saline with Tween 20 (TBST), the blot was incubated with horseradish peroxidase-conjugated secondary antibody and immune complexes were visualized by enhanced chemiluminescence detection (Luminata Forte WBLUF0500, Millipore). The blot was also probed with monoclonal antibody against anti-GAPDH 1/10000 (AB2302 Millipore). We performed three technical replicates and plotted the mean and SEM. To resolve monoubiquitinated FANCD2 from unubiquitinated FANCD2, protein samples were run on a 3%–8% Tris-acetate gel at 100 V for 90 min. Transfer to PDF membrane overnight 25 v at 4°C followed by incubation with anti-FANCD2 antibody 1/2000 (NB100-182, Novus Biologicals), horseradish peroxidase–conjugated secondary antibody, and chemiluminescence detection. Each of the isogenic cell lines has three biological replicates except T1131A which have two replicates. To demonstrate monoubiquitinated FANCI, previously FANCD2 blotted western blot membranes were stripped which with Restore PLUS western blot stripping buffer (Thermo Scientific) for 5 min then blocked for 30 min and incubated overnight at 4°C with anti-FANCI antibody A7 (Santa Cruz) diluted 1/500, followed by horseradish peroxidase–conjugated secondary antibody and chemiluminescence detection.
Immunofluorescent Labeling of Cells
Five sterile 12-mm circular coverslips were placed in each of the wells of a six-well dish. FANCA positive or negative cells were plated at a density of 1 × 105 to 2.5 × 105 cells per well. The next day, cells were treated with vehicle or 1 µM MMC (Sigma-Aldrich) for 24 h. Cells were washed twice in PBS. Following incubation in 0.5% Triton X-100 (Sigma-Aldrich) for 5 min, followed by fixation in 3.7% formaldehyde, permeabilized in 0.5% NP-40 in phosphate-buffered saline (PBS) for 10 min and washed once in PBS. Coverslips with cells were incubated in PBG blocking solution (0.2% w/v cold water fish gelatin [Sigma-Aldrich], 0.5% w/v BSA [Sigma-Aldrich] in PBS) for 20 min, and then were incubated with FANCD2 (Novus Biologicals) primary antibody (1/1000) in PBG at room temperature for 2 h. Cells were washed three times for 5 min each in PBG and incubated in secondary antibody (Alexa Fluor 488 Goat anti Rabbit; Life Technologies) (1/1000) dilution in PBG for 1 h at room temperature. Cells were then washed three times for 5 min and coverslips were mounted on slides in ProLong Gold Antifade Mountant with DAPI (Life Technologies). Foci formation was analyzed on an Olympus BX51 under 100× oil objective. Fisher exact tests were performed between groups. FANCA localization was performed as above with the Flag M2 antibody (Sigma-Aldrich) (1/500) incubated for 1 h and secondary antibody (Alexa Fluor 548 Goat anti Mouse; Life Technologies) (1/500) and incubated for 1 h.
Xenografts
Patient-derived xenografts were developed in collaboration with the Living Tumor Laboratory (www.livingtumorlab.com), University of British Columbia, as previously described (Lin et al. 2014; Beltran et al. 2015). For drug treatment studies, tumors were allowed to grow for 18 d (n = 16 total tumors per model) and were treated with vehicle or olaparib (intraperitoneal dosing of vehicle or olaparib, 50 mg/kg per day). Tumor volume based on caliper measurements and final tumor weight were assessed. The tumor weights at the final day were compared using a nonparametric method—the Wilcoxon rank-sum test. All tests are two-sided with a 0.05 level of significance.
ADDITIONAL INFORMATION
Data Deposition and Access
Genetic variant data were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) under accession number SCV000494044.
Ethics Statement
Patient PM12 was enrolled prospectively on an Institutional Review Board (IRB)-approved protocol at Weill Cornell Medicine (IRB #1305013903) and provided written informed consent that allowed for fresh tissue and blood collection, tumor and germline sequencing (results described in Beltran et al. 2015), and the development of a PDX model as part of the current study. The patient consent states that de-identified information may be used for research and participation may benefit society through an increased understanding of how to treat advanced cancer. The patient consented to publication of this article.
Acknowledgments
We thank Agata Smogorzewska for her technical assistance, cell lines (RA fibroblast isogenic cell lines), helpful comments, and insights on data presented in the manuscript. We thank the following WCM core Facilities: The Translational Research Program at WCM Pathology and Laboratory Medicine (Bing He, Yifang Liu, Leticia Dizon). This study was supported by the Englander Institute for Precision Medicine, Weill Cornell Medicine and New York-Presbyterian Hospital (D.C.W., V.S., H.B., M.A.R., D.S.R.), Department of Defense PC121341 (H.B.), Prostate Cancer Foundation, the Starr Cancer Consortium I7-A771 (H.B., M.A.R.), and the Terry Fox Research Institute (M.G., Y.W.).
Author Contributions
D.C.W., H.B., M.A.R., and D.S.R. designed, performed, and analyzed all in vitro experiments. D.C.W. and V.S. performed in situ FANCD2 foci formation analysis. F.D. performed bioinformatics analysis. H.C., C.C.C., M.G., and Y.W. performed xenograft experiments and in vivo drug treatment. D.C.W. and D.S.R. wrote the manuscript. D.C.W., H.B., F.D., M.A.R, and D.S.R. edited the manuscript.
Funding
This work was supported by grants from Weill Cornell Medicine, U.S. Department of Defense PC121341, Prostate Cancer Foundation I7-A771, and the Terry Fox Research Institute.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adachi D, Oda T, Yagasaki H, Nakasato K, Taniguchi T, D'Andrea AD, Asano S, Yamashita T. 2002. Heterogeneous activation of the Fanconi anemia pathway by patient-derived FANCA mutants. Hum Mol Genet 11: 3125–3134. [DOI] [PubMed] [Google Scholar]
- Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, Jarosz M, Lipson D, Tagawa ST, Nanus DM, et al. 2013. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol 63: 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, Kossai M, Pauli C, Faltas B, Fontugne J, et al. 2015. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 1: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Bellani M, Liu Y, Seidman MM. 2016. Fanconi anemia: a DNA repair disorder characterized by accelerated decline of the hematopoietic stem cell compartment and other features of aging. Ageing Res Rev 33: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castella M, Jacquemont C, Thompson EL, Yeo JE, Cheung RS, Huang J-W, Sobeck A, Hendrickson EA, Taniguchi T. 2015. FANCI regulates recruitment of the FA core complex at sites of DNA damage independently of FANCD2. PLoS Genet 11: e1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HH, Pritchard CC, Boyd T, Nelson PS, Montgomery B. 2016. Biallelic inactivation of BRCA2 in platinum-sensitive metastatic castration-resistant prostate cancer. Eur Urol 69: 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo SB, Patel AG, Hurley RM, Kaufmann SH. 2013. The elephant and the blind men: making sense of PARP inhibitors in homologous recombination deficient tumor cells. Front Oncol 3: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V. 2015. Update of the human and mouse Fanconi anemia genes. Hum Genomics 9: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Amarachintha S, Wilson AF, Pang Q. 2016. Hyper-active non-homologous end joining selects for synthetic lethality resistant and pathological Fanconi anemia hematopoietic stem and progenitor cells. Sci Rep 6: 22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Rodríguez JA, Spierings EA, de Winter JP, Giaccone G, Kruyt FA. 2005. Identification of multiple nuclear export sequences in Fanconi anemia group A protein that contribute to CRM1-dependent nuclear export. Hum Mol Genet 14: 1271–1281. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, Näf D, Wasik J, D'Andrea AD. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol 19: 4866–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, D'Andrea AD. 2006. Dedicated to the core: understanding the Fanconi anemia complex. DNA Repair 5: 1119–1125. [DOI] [PubMed] [Google Scholar]
- Kachnic LA, Li L, Fournier L, Willers H. 2010. Fanconi anemia pathway heterogeneity revealed by cisplatin and oxaliplatin treatments. Cancer Lett 292: 73–79. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. 2010. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C-C, Li Z, Lopez-Martinez D, Nicholson WV, Vénien-Bryan C, Cohn MA. 2016. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun 7: 12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J, Alon N, Bosnoyan-Collins L, Buchwald M. 1999. Characterization of regions functional in the nuclear localization of the Fanconi anemia group A protein. Hum Mol Genet 8: 1007–1015. [DOI] [PubMed] [Google Scholar]
- Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, et al. 2014. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 74: 1272–1283. [DOI] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, et al. 2015. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka MZ, et al. 2006. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66: 8109–8115. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet 35: 165–170. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G, et al. 2004. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet 36: 1219–1224. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky G, Kotton DN, Fabian AJ, Gray JT, Lee JS, Mulligan RC. 2005. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther 11: 932–940. [DOI] [PubMed] [Google Scholar]
- Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, Vance GH, Alkuraya FS, Hanenberg H, Andreassen PR. 2016. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet 53: 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, et al. 2016. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. New Engl J Med 375: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. 2015. Integrative clinical genomics of advanced prostate cancer. Cell 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528. [DOI] [PubMed] [Google Scholar]
- Schultz N, Lopez E, Saleh-Gohari N, Helleday T. 2003. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res 31: 4959–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swuec P, Renault L, Borg A, Shah F, Murphy VJ, van Twest S, Snijders B, Deans AJ, Costa A. 2016. The FA core complex contains a homo-dimeric catalytic module for the symmetric mono-ubiquitination of FANCI-FANCD2. Cell Rep 18: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100: 2414–2420. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, Mok SC, D'Andrea AD. 2003. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med 9: 568–574. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2015. The molecular taxonomy of primary prostate cancer. Cell 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Twest S, Murphy VJ, Hodson C, Tan W, Swuec P, O'Rourke JJ, Heierhorst J, Crismani W, Deans AJ. 2016. Mechanism of ubiquitination and deubiquitination in the Fanconi Anemia Pathway. Mol Cell 65: 247–259. [DOI] [PubMed] [Google Scholar]
- Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. 2014. Neuroendocrine Prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis—a systematic review and pooled analysis. J Clin Oncol 32: 3383–3390. [DOI] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. 2012. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 44: 910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.