Abstract
The Anseriformes is a well-known and widely distributed bird order, with more than 150 species in the world. This paper aims to revise the classification, determine the phylogenetic relationships and diversification patterns in Anseriformes by exploring the Cyt b, ND2, COI genes and the complete mitochondrial genomes (mito-genomes). Molecular phylogeny and genetic distance analyses suggest that the Dendrocygna species should be considered as an independent family, Dendrocygnidae, rather than a member of Anatidae. Molecular timescale analyses suggests that the ancestral diversification occurred during the Early Eocene Climatic Optimum (58 ~ 50 Ma). Furthermore, diversification analyses showed that, after a long period of constant diversification, the median initial speciation rate was accelerated three times, and finally increased to approximately 0.3 sp/My. In the present study, both molecular phylogeny and diversification analyses results support that Anseriformes birds underwent rapid and recent diversification in their evolutionary history, especially in modern ducks, which show extreme diversification during the Plio-Pleistocene (~ 5.3 Ma). Therefore, our study support that the Plio-Pleistocene climate fluctuations are likely to have played a significant role in promoting the recent diversification for Anseriformes.
Introduction
Adaptive radiation, the evolution of ecological and phenotypic diversity within a rapidly multiplying lineage, has fascinated biologists over the past century [1–3]. Identifying the evolutionary diversification of organisms is a crucial step in understanding their evolutionary history, where the final goal is exploring the factors that might potentially affect the diversification [4]. It was known that molecular phylogenies provide robust framework to study the patterns of speciation and diversification in lineages [5–6]. The branching pattern of a phylogenetic tree can be used to detect changes in speciation through time [7]. This information can also be used to detect speciation bursts and identify historical factors underlying the emergence of ecological and phenotypic divergence within a lineage [8]. Up to now, rapid radiation and burst speciation events have been examined in several taxonomic groups, such as fishes [8, 9] and mammalian [4, 10–12]. Notably, an ancient, rapid radiation event was also observed in the evolutionary history of the Galliformes species [13]. Furthermore, hybridization is a common phenomenon when populations invade new environments and potentially elevates rates of response to selection, which plays an important role in species evolutionary processes, such as adaptive radiations [14]. Particularly, birds show relatively high levels of hybridization [15, 16], such as the high incidence of hybridization in Anseriformes birds (e.g. ducks, geese and swans) [17, 18].
Anseriformes is a large and well-known bird group comprising more than 150 species (approximately 43 genera), including ducks, geese, swans, screamers and the magpie goose [19, 20]. In the past few decades, the Anseriformes species have been among the most studied groups of birds, especially regarding phylogenetic relationships [18, 19, 21–26]. However, the classification of the Anseriformes has been much disputed and revised since the recognition of this group [27–30]. Originally, the order Anseriformes has been considered to be composed of the families Anhimidae (screamers) and Anatidae (ducks, geese, swans and the magpie goose) [21, 31–35]. Currently, the Anseriformes has been divided into three families: Anatidae (ducks, geese and swans), Anhimidae (screamers) and Anseranatidae (the magpie geese) [20, 36–41]. However, some researchers suggest that Dendrocygna species should be considered as an independent family [42, 43]. Besides, the classification of several genera in Anatidae, such as the Amazonetta, Speculanas, Tachyeres and Lophonetta, has been much debated [44–48].
Compared with nuclear DNA, the mitochondrial genome (mito-genome) of animals evolve rapidly [49]. Previous studies proved that the mitochondrial DNA (mtDNA) sequences are a powerful tool for taxonomy, phylogenetic relationships, evolutionary history, etc [10, 28, 50–56]. However, single-gene phylogenies often differ dramatically from studies involving multiple datasets, which suggest that they are often discordance [57]. Therefore, the mito-genome and concatenated genes (mitochondrial gene or nuclear gene) were gradually used to construct reliable phylogeny for determining evolutionary relationships among species or higher taxa [10–13, 57–62]. Currently, the mito-genomes or concatenated mitochondrial genes, such as NADH dehydrogenase subunit 2 (ND2), Cytochrome b (Cyt b), cytochrome oxidase subunit I (COI) and 12S ribosomal RNA (12S rRNA), etc, were widely used for systematics studies in the bird’s evolution [19, 27, 28, 56, 58, 63]. What’s more, with the development of the sequencing, the use of nuclear genes or whole genomics for systematics studies is increasing [11, 25, 26, 64–66].
In the present study, we set out to examine the phylogenetic relationships and revise the classification in the Anseriformes based on mito-genomes (Dataset 1), two concatenated mitochondrial genes (Cyt b and ND2, Dataset 2) and the DNA barcoding gene (COI, Dataset 3). Generally, the diversification pattern of species is not always gradual but can occur in rapid radiations, especially after major environmental changes [67, 68]. In this study, we also want to determine whether Anseriformes birds experienced rapid diversification and explore the potential factors that may have been an influence. Therefore, we performed diversification analyses to further contribute to an understanding of the diversification patterns in Anseriformes.
Materials and methods
Ethical statement
In the present study, all samples were collected from individuals that died naturally and were found in field investigations. Our experimental procedures complied with the current laws on animal welfare and research in China, and were specifically approved by the Animal Research Ethics Committee of Anhui University.
Sample collection and DNA extraction
From 2006 to 2013, muscle samples of nine Anatidae species (Anas acuta, A. poecilorhyncha, A. crecca, A. clypeata, Aythya ferina, Aythya fuligula, Mergus merganser, Tadorna tadorna and Aix galericulata) were collected from the Anqing wetland along the Yangtze River in Anhui Province. Total genomic DNA was extracted using a standard proteinase K / phenol-chloroform protocol, as described by Sambrook & Russell [69]. An EasyPure PCR Purification Kit (TransGene) was used to purify each DNA extraction.
PCR amplification and sequencing
Sixteen pairs of primers (S1 Table) were designed using Primer Premier 5.0 [70] based on the mito-genomes of Anas platyrhynchos (EU009397) [71], A. falcata (NC_023352) [72], Anser fabalis (NC_016922) [27], Dendrocygna javanica (NC_012844) [73] and Anseranas semipalmata (NC_005933) [74]. The product size of the above primer pairs ranged from 1,137 bp to 1,537 bp (S1 Table). Polymerase chain reaction (PCR) was performed in a 50 μL reaction mixture, containing 100 to 200 ng of genomic DNA, 25 μL 2×EasyTaq PCR SuperMix polymerase (TransGen Biotech, containing 1.25U Ex Taq, 0.4mM dNTP, 4mM Mg2+) and 0.4 μM of each primer. Pure molecular biology grade water was added to reach the appropriate volume. Thermal cycling consisted of a denaturation step at 95°C for 5 min, followed by 30 cycles of denaturation (95°C, 30 s), annealing (50–55°C, depending on primer combinations, 40 s) and extension (72°C, 90 s) and a final extension step of 10 min at 72°C. PCR products were purified using an EasyPure PCR Purification Kit (TransGene), and sequenced using previous primers and the BigDye Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems) following the manufacturer’s instructions on an ABI PRISM 3730 automated sequencer. Several different methods (e.g. BLAST search and translation test method) had been adopted to exclude potential nuclear mitochondrial pseudogenes [75].
Sequence analyses
Sequences were assembled by Seqman II (DNAStar, Madison, WI, USA) and examined visually to ensure the accuracy of variable sites identified by the program [76]. Protein-coding genes (PCGs) were identified by comparison with known complete mtDNA sequences of Anseriformes birds using Sequin 11.0. The 22 tRNA genes were identified using the software package tRNA Scan-SE 1.2.1 (http://lowelab.ucsc.edu/tRNAscan-SE/) [77]. The graphical map of the mito-genome was drawn using the online software OrganellarGenomeDRAW (http://ogdraw.mpimp-golm.mpg.de/) [78]. In addition, the DOGMA annotation software was used to check annotated genes [79]. All assembled and annotated mito-genomes have been deposited in GenBank (Accession no. KF312717, KF156760, KF203133, KT345702, KJ710708, KJ722069, KU140667, KU140668 and KJ169568) [80–86]. Other mtDNA sequences used in the analyses were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/pubmed/) (S2, S3 and S4 Tables for a full list accession numbers of sequences). Genetic distances based on the COI gene among species were calculated using the Kimura two parameter (K2P) distance model [87] performed in MEGA5 [88] with 10,000 replications.
Phylogenetic analyses
Phylogenetic trees were reconstructed based on the mito-genomes of 30 Anseriformes species (Dataset 1, S2 Table). In this study, Dataset 1 were estimated under the Maximum Likelihood (ML) and Bayesian Inference (BI) methods, with Gallus gallus (NC_001323) used as an out-group, which were performed with RAxML version 8 [89] and MrBayes 3.2.2 software program [90], respectively.
Before phylogenetic tree constructions, sequence alignment was carried out by using Clustal X 1.8 software [91] and examined visually. To obtain the estimated best fit evolution model for each data set, we performed analyses separately as described above using the MrModeltest 1.0b software program [92] in Paup* 4.0b [93], which was based on the AIC criterion. For ML analysis, node support was calculated with a GTRGAMMA model via rapid bootstrapping with ten runs and 1,000 replications to estimate the best topology. For the BI analysis, two independent runs of four Markov Chains Monte Carlo (MCMC) chains (one cold chain and three hot chains) were simultaneously run under the best fit substitution model (GTR + I + G) for 1,000,000 generations, with sampling conducted every 100 generations until the average standard deviation of split frequencies reached a value less than 0.01. The first 10% of the total trees were discarded as ‘‘burn-in” and the remaining trees were used to calculate 50% majority rule consensus tree and Bayesian posterior probabilities.
Dating analyses
To estimate the precise divergence times within Anseriformes, we dated the divergence time between Anseriformes and Galliformes with multiple out-groups (Columba livia, NC_013978 [94]; Falco peregrinus, NC_000878 [95]; Tinamus guttatus, NC_027260 [96] and Struthio camelus, NC_002785 [96]). All calibration points used in the dating analyses were derived from Jarvis’ study [66] (S5 Table). Then, we used the divergence time between Anseriformes and Galliformes as the calibration point to estimate the divergence within Anseriformes. We applied a Bayesian MCMC method based on the mito-genomes, which employs a relaxed molecular clock approach, and implemented in BEAST 1.7.4 [97, 98]. A relaxed uncorrelated log normal model of lineage variation and a Yule Process prior to the branching rates based on HKY + I + G model were assumed, as recommended by MrModel test 1.0b. Four replicates were run for 1,000,000 generations with tree and parameter sampling that occurred every 100 generations for the first 10% of samples that were discarded as burn-in. All parameters were assessed by visual inspection using Tracer v. 1.6 [99]. To further estimate the divergence times within the Anseriformes, we also used concatenated sequences (ND2 and Cyt b sequences) for 128 species (Dataset 2, S3 Table), and were based on the above calibration points (S5 Table). Compared with Gonzalez’s study [17], seven species (Tachyeres brachypterus, T. leucocephalus, Mergus squamatus, Dendrocygna arcuata, Anhima cornuta, Chauna torquata and Anseranas semipalmata) were added. Relaxed uncorrelated log normal models of lineage variation and the Yule Process were set by basing them on HKY + I + G model as recommended by MrModel test 1.0b.
Tempo and rate shifts of diversification analyses
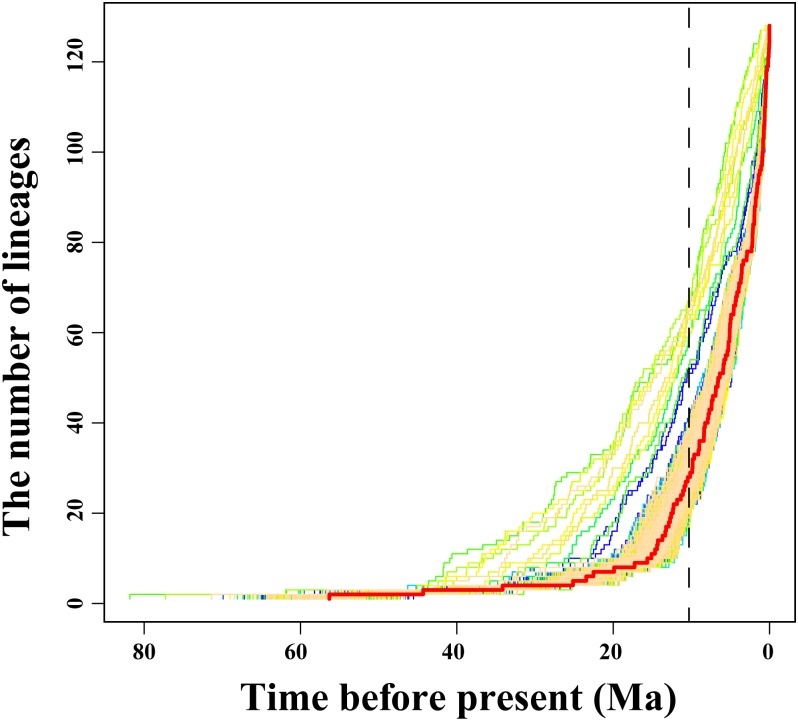
To visualize the temporal accumulation of lineages, a log-transformed lineage-through-time (LTT) plot [100] was performed based on 1,000 trees randomly selected from the BEAST analysis by using APE package (version 4.1) [101] in R language [102]. In this study, the LTT plots were based on the molecular phylogeny of 128 species in Anseriformes (excluding out-groups).
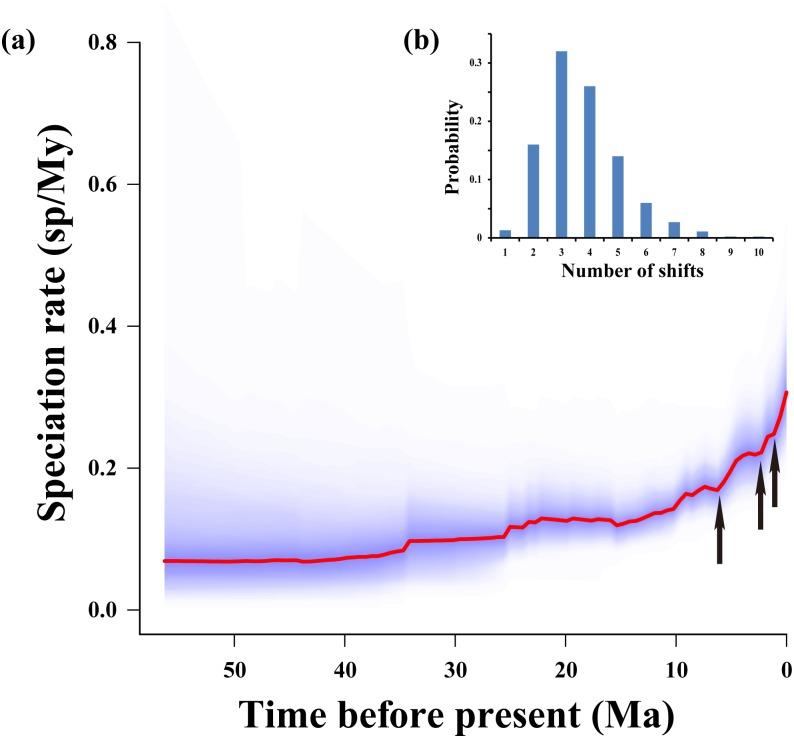
BAMM is a Bayesian approach that uses reversible jump Markov chain Monte Carlo (rjMCMC) sampling to explore shifts in macro-evolutionary regimes assuming they occur across the branches of a phylogeny under a compound Poisson process, and explicitly accommodates diversification rate variation through time and among lineages [103, 104]. Therefore, we used the program BAMM to estimate rates of speciation, conduct rate-through-time analysis of these rates, and identify and visualize shifts in species rates across the molecular phylogeny.
BAMM accounts for non-random and incomplete taxon sampling in the phylogenetic trees by allowing all non-sampled species to be associated with a particular tip or more inclusive clade [103, 104]. Therefore, we assumed that our sampling included 80% of extant Anseriformes species diversity (4 families, 39 genera and 128 species in total; S3 Table). Furthermore, priors for BAMM were generated using setBAMMpriors function, implemented in R package BAMMtools v 2.1.6 [103], by providing the Maximum Clade-Credibility (MCC) tree from BEAST and total species numbers across the order (approximately 160) [41]. Four independent MCMC chains of 20,000,000 generations, sampling event data every 20,000 steps, were run in BAMM and convergence was assessed by computing the effective sample sizes of log likelihoods, as well as the number of shift events present in each sample using the R package coda v. 0.16–1 [105]. After removing 10% of trees as burn-in, we analyzed the BAMM output using BAMMtools and computed the 95% credible rate shift configurations using the summary of posterior distribution of the number of shifts.
Results
Mito-genome organization
In this study, nine mito-genomes were sequenced and annotated, which contain 13 PCGs (including ATP6, ATP8, COI, COII, COIII, ND1, ND2, ND3, ND4, ND4L, ND5, ND6 and Cyt b), two rRNAs (12S rRNA and 16S rRNA), 22 tRNAs and a control region (CR), respectively. The total length of these mito-genomes ranged from 16,599 bp to 16,630 bp. The heavy DNA strand (H-strand) carried most of the genes (12 PCGs, two rRNAs and 14 tRNAs), while ND6 and eight tRNAs were located on the L-strand (S1 Fig). The arrangement of the whole mito-genome of these species were identical to known typical vertebrate patterns [106]. The total length of the 13 PCGs was 11,411 bp, which represented approximately 68.7% of the entire mito-genome in Anatidae. The overall base composition is also similar to other Anseriformes species. For example, A + T content (50.6–51.8%) (S6 Table) is higher than C + G content, which reflects the strong AT bias [107]. What’s more, the relative abundance of nucleotides is C > A > T > G, which showed that Guanine (G) is the rarest nucleotide (S6 Table), similar to other vertebrate animals [48, 49, 107].
Genetic distance
To understand the sequence divergence within the Anseriformes, we calculated genetic distances among different groups based on COI gene for 54 species. In this study, the K2P distances among four families ranged from 0.158 to 0.208, while the average genetic distances were 0.117 ± 0.015 (Table 1, Dataset 3). In addition, the K2P distances among genera within Anatidae ranged from 0.037 to 0.107, while the average genetic distances were 0.103 ± 0.008 (Table 2, Dataset 3). In this study, the genetic distance between Anatidae and Dendrocygnidae is significantly different from genetic distances within the Anatidae.
Table 1. The genetic distances and standard error estimates among Anseriformes species of mtDNA COI based on K2P model.
| 1 | 2 | 3 | 4 | |
| 1. Anatidae | ||||
| 2. Dendrocygnidae | 0.158 ± 0.021 | |||
| 3. Anhimade | 0.208 ± 0.031 | 0.190 ± 0.029 | ||
| 4. Anseranatidae | 0.172 ± 0.026 | 0.161 ± 0.025 | 0.173 ± 0.032 |
1. Anatidae species—Anas acuta, A. bahamensis, A. gibberifrons, A. crecca, A. poecilorhyncha, A. platyrhynchos, A. laysanensis, A. superciliosa, A. falcata, A. penelope, A. strepera, A. discors, A. platalea, A. clypeata, A. querquedula, A. formosa, Amazonetta brasiliensis, Tachyeres pteneres, Lophonetta specularoides, Aythya ferina, A. Americana, A. fuligula, A. marila, A. affinis, Netta rufina, Tadorna tadorna, T. ferruginea, Mergus merganser, M. serrator, M. squamatus, Mergellus albellus, Lophodytes cucullatus, Anser anser, A. brachyrhynchus, A. cygnoides, A. indicus, A. rossii, A. fabalis, A. albifrons, Branta canadensis, B. sandvicensis, B. leucopsis, B. bernicla, Cygnus cygnus, C. columbianus, C. olor, C. atratus;
2. Dendrocygnidae species—Dendrocygna javanica, D. viduata, D. arcuata, D. eytoni, D. bicolor;
3. Anhimade species—Anhima cornuta;
4. Anseranatidae species—Anseranas semipalmata.
Table 2. The genetic distances and standard error estimates among Anatidae species of mtDNA COI based on K2P model.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1. Anas | |||||||
| 2. Aythya | 0.054 ± 0.008 | ||||||
| 3. Tadorna | 0.048 ± 0.009 | 0.054 ± 0.010 | |||||
| 4. Mergus | 0.037 ± 0.006 | 0.055 ± 0.009 | 0.048 ± 0.009 | ||||
| 5. Anser | 0.079 ± 0.011 | 0.107 ± 0.014 | 0.101 ± 0.014 | 0.086 ± 0.012 | |||
| 6. Branta | 0.063 ± 0.009 | 0.093 ± 0.013 | 0.071 ± 0.011 | 0.072 ± 0.010 | 0.043 ± 0.008 | ||
| 7. Cygnus | 0.050 ± 0.008 | 0.074 ± 0.010 | 0.073 ± 0.012 | 0.050 ± 0.009 | 0.061 ± 0.010 | 0.045 ± 0.008 |
1. Anas species–Anas acuta, A. bahamensis, A. gibberifrons, A. crecca, A. poecilorhyncha, A. platyrhynchos, A. laysanensis, A. superciliosa, A. falcata, A. penelope, A. strepera, A. discors, A. platalea, A. clypeata, A. querquedula, A. formosa, Amazonetta brasiliensis, Tachyeres pteneres and Lophonetta specularoides;
2. Aythya species–Aythya ferina, A. Americana, A. fuligula, A. marila and A. affinis;
3. Tadorna species–Tadorna tadorna and T. ferruginea;
4. Mergus species–Mergus merganser, M. serrator, M. squamatus, Mergellus albellus and Lophodytes cucullatus;
5. Anser species–Anser anser, A. brachyrhynchus, A. cygnoides, A. indicus, A. rossii, A. fabalis and A. albifrons;
6. Branta species–Branta canadensis, B. sandvicensis, B. leucopsis and B. bernicla;
7. Cygnus species–Cygnus cygnus, C. columbianus, C. olor and C. atratus.
Phylogenetic reconstructions
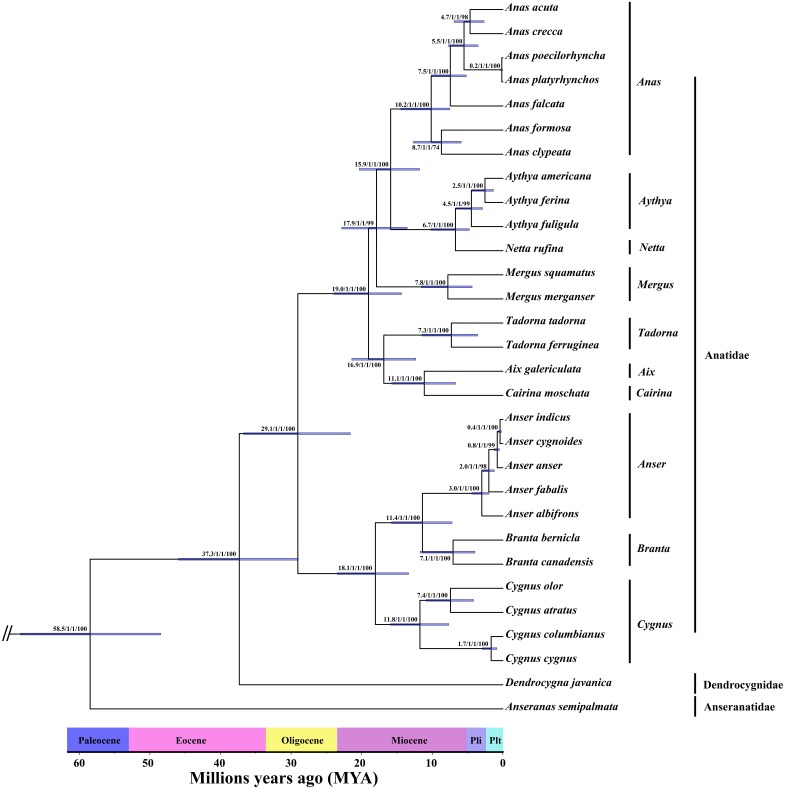
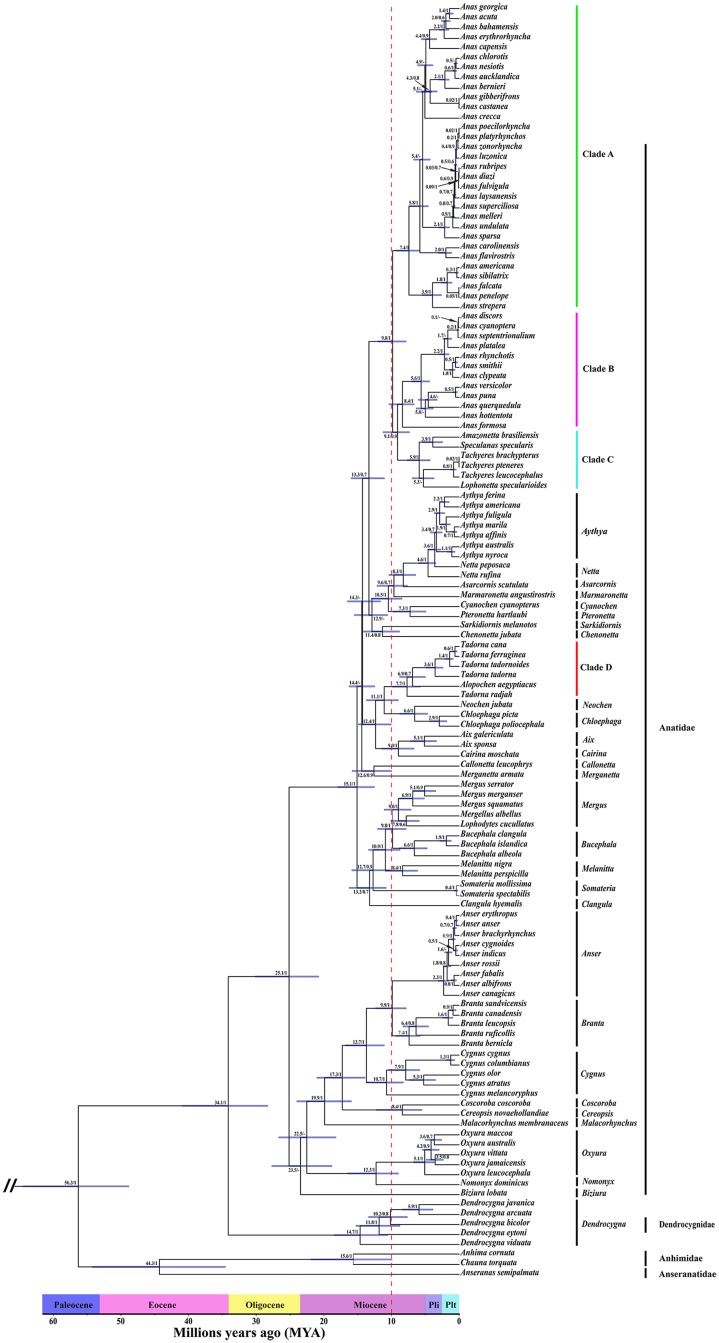
The phylogenetic tree recovered from the ML (S2 Fig) and BI (S3 Fig) analyses of mito-genomes for 30 Anseriformes species resulted in the same topology (Fig 1). In this study, the 30 species clustered into two major lineages, represented by Anatidae–Dendrocygnidae and Anseranatidae, respectively (Fig 1). There are three major lineages in the Anatidae–Dendrocygnidae group (Fig 1). Additionally, topologies recovered from the MCMC method of Cyt b and ND2 for 128 Anseriformes species obtained highly posterior probability values for most nodes (Fig 2), which shared basic topology with the above results and a previous study [18]. Our study revealed that the 128 Anseriformes species also clustered into two major lineages, represented by Anatidae–Dendrocygnidae and Anhimidae–Anseranatidae, respectively (Fig 2). Furthermore, the Anatidae–Dendrocygnidae group comprises three major lineages (Fig 2).
Fig 1. Phylogram showing the phylogenetic relationship in Anseriformes based on the mito-genomes.
The values on nodes include four parts. The first two values indicate the split time and Bayesian posterior probabilities which were calculated by BEAST 1.7.4. The third values were the Bayesian posterior probabilities calculated by MrBayes 3.2.2, and the last values were the Bayesian posterior probabilities calculated by RAxML version 8. Blue bars at nodes show 95% highest posterior density (HPD) of divergence times.
Fig 2. Phylogram showing the phylogenetic relationship in Anseriformes based on two mitochondrial genes.
The values on nodes indicate the split time and Bayesian posterior probabilities which were calculated by BEAST 1.7.4, “-” indicated that the value was less than 70. Blue bars at nodes show 95% HPD of divergence times.
Divergence times
In the present study, we obtained similar divergence time values in the major nodes using a molecular clock based on mito-genomes and two concatenated mitochondrial genes. The divergence time between the Anseranatidae–Anhimidae group and Anatidae–Dendrocygnidae group was at about 58.5 Ma (95% HPD: 48.5–68.4, Dataset 1) (Fig 1 and S4 Fig) or 56.3 Ma (95% HPD: 48.8–64.5, Dataset 2) (Fig 2 and S5 Fig). Moreover, the split time between Anseranatidae and Anhimidae was estimated at about 44.3 Ma (95% HPD: 34.6–54.3, Dataset 2) (Fig 2 and S5 Fig). In addition, the Anatidae has split from Dendrocygnidae at about 37.3 Ma (95% HPD: 29.0–46.0, Dataset 1) (Fig 1 and S4 Fig) or 34.1 Ma (95% HPD: 28.3–40.9, Dataset 2) (Fig 2 and S5 Fig).
Tempo and rate shifts of diversification
The empirical LTT plot displayed that the diversification rate of the Anseriformes had a significant increase during the late Miocene after a long period of constant diversification (Fig 3). In the BAMM analyses, we confirmed convergence of the MCMC chains after discarding the burn-in, according to the result of effective samples size (ESS is 900). In the Anseiformes, the 95% credible set of rate shift configurations with the highest probability included three or more core shifts (Fig 4a). The best configuration detected three shifts within the Anseriformes birds (Fig 4b), suggesting that the mean initial speciation rate was accelerated three times, and finally increased to approximately 0.3 sp/My in their evolutionary history (Fig 4a).
Fig 3. Lineage-through-time (LTT) plots for Anseriformes birds.
The colored lines represent the results of 1000 trees randomly selected from the BEAST analysis. The ref line shows the MCC tree.
Fig 4. Visualizations of diversification rate shifts within Anseriformes.
(a) Speciation-through-time plots utilizing BAMM. Curved red lines represent the median values with the 95% confidence intervals shown in blue. Arrows point to the three significant shifts in rates of speciation; (b) Posterior distribution of the number of rate shifts.
Discussion
Classification implications based on phylogenetic relationships
In the last few decades, the notion of dividing the Anseriformes into three families (Anatidae, Anhimidae and Anseranatidae) was widely prevalent [19, 35–40]. In 1990, Sibley and Ahlquist classified Dendrocygna species into an independent family (Dendrocygnidae) based on DNA-DNA hybridization [42]. However, it was not accepted widely [43]. Based on the tree of two concatenated mitochondrial genes, the Anhimidae clustered together with Anseranatidae, which contains a limited number of species (3 species). Meanwhile, the Dendrocygnidae clustered together with Anatidae, which comprises the majority of species (approximately 125 species). In this study, both trees (mito-genomes, Fig 1 and two concatenated mitochondrial genes, Fig 2) showed that Dendrocygnidae is an independent clade, which diverged in the late Eocene at about 37.3 / 34.1 Ma BP (Figs 1 and 2). In birds, the COI gene is considered as standard for DNA barcoding [108]. For the COI gene, the mean K2P distances within-species, genus and family are 0.43%, 7.93% and 12.71%, respectively [108]. In this study, K2P distances between Dendrocygnidae and the other three families (Anatidae, Anhimade and Anseranatidae) of species ranged from 15.8% to 19.0% (Table 1), which are higher than the average genetic distances within the family. In addition, the mean K2P distance between Anatidae and Dendrocygnidae is 15.8% (Table 1), which is significantly different from the mean K2P distance within the Anatidae (10.3%, Table 2). Therefore, our study provide support for Dendrocygnidae to be an independent family, rather than as a genus within Anatidae.
The crested duck (Lophonetta specularioides) and spectacled duck (Speculanas specularis) were once considered as members of the genus Anas [109–111]. In this study, six species from genera Amazonetta, Speculanas, Tachyeres and Lophonetta clustered in a distinct clade (Clade C, Fig 2), which is the sister group to Clade B (Fig 2). Meanwhile, Clade B is comprised of twelve Anas species (Fig 2). Besides, Clade B and C are sister groups to Clade A, which is comprised of 31 Anas species (Fig 2). Based on phylogenetic relationships, the extant Anas species (Clade A and B) do not form a monophyletic group. Therefore, in systematics, we suggest that placing these six species (Clade C) into the genus Anas would be more appropriate.
The Egyptian goose (Alopochen aegyptiacus) is the only extant species in the genus Alopochen [110, 111]. Based on the phylogenetic analysis, Alopochen aegyptiacus nested tightly together with the Tadorna species in Clade D, rather than being an independent clade (Fig 2). It is obvious that Alopochen aegyptiacus is a member of the genus Tadorna. What’s more, the high incidence of hybridization between Alopochen aegyptiaca and Tadorna tadorna also suggest the close genetic proximity between Alopochen and Tadorna [112]. Therefore, we propose that Alopochen aegyptiacus should be placed in the genus Tadorna.
Ancestral diversification during the Early Eocene Climatic Optimum
Compared to the members of Anhimidae and Anseranatidae, Anatidae and Dendrocygnidae species have well developed feet webs which helped them become successful swimmers [43]. They tend to exploit different ecological niches, which eventually generates ecological adaptation in their evolutionary history. For example, the Anatidae and Dendrocygnidae species exist in various types of lacustrine systems with a worldwide distribution, except for the South Pole, while the Anhimidae (found in South America) and Anseranatidae (Australia, Indonesia and Papua New Guinea) species have a narrower distribution [30, 32, 33, 35]. Our study identified the ancestral diversification between the Anhimidae–Anseranatidae group and Anatidae–Dendrocygnidae group occurred at about 58.5 / 56.3 Ma (Figs 1 and 2) in the late Paleocene. Furthermore, we compared this estimated divergence times with other studies [113–115] using TimeTree (http://timetree.org/) [116], which also proved that these estimates are reliable.
From the late Paleocene (~ 58 Ma) to the early Eocene (~ 50 Ma), the earth’s surface experienced a long-term warming trend that culminated in an extended period of extreme warmth called the Early Eocene Climatic Optimum (EECO) [117–120]. Remarkably, at the boundary between the Paleocene and Eocene epochs (~ 56 Ma), a most intense and abrupt interval of global warming occurred, called the Paleocene–Eocene Thermal Maximum (PETM) [121–124]. During the EECO interval, the mean annual temperature and the mean annual rainfall increased significantly, while these climate changes promoted a major increase in floral and faunal diversity [118, 125, 126]. For example, they have induced diversification in many animal groups, such as insects [127], salamanders [128], bats [10] and the ruminants [12]. In this study, this ancestral diversification in Anseriformes occurred close to the PETM during the EECO. Therefore, we suggest that the warmer climatic conditions during the EECO induced this ancestral diversification in Anseriformes, which reinforced their habit of exploiting different ecological niches.
Rapid and recent diversification during the Plio-Pleistocene
Diversification patterns and other potential driving factors, together with accurate divergence time estimation can be used to predict species change [129]. Therefore, we performed a test to characterize the patterns of diversification of this group by using the molecular phylogeny as a robust framework. Based on the results of divergence time estimation and LTT plot analyses, rapid and recent diversification events can be observed to have occurred in the Anseriformes since 10.0 Ma in the Late Miocene (Figs 2 and 3). Clearly, a higher speciation rate has characterized the rapid and recent diversification in the Anseriformes during the Plio-Pleistocene, while the mean initial speciation rate accelerated three times, and finally increased to approximately 0.3 sp/My (Fig 4). Specifically, the Anatidae comprises the majority of species (approximately 148), which diversified at about 29.1 / 25.1 Ma (Figs 1 and 2). Therefore, we proposed that the Anatidae species might have experienced rapid and recent diversification during the Plio-Pleistocene.
In terrestrial and lacustrine systems, mechanisms of speciation are thought to be important for ecological adaptation [8]. Many speciation events showed that some underlying factors, such as the availability of new resources [2], new habitats [130], or key innovations that promote the exploitation of new resources or habitats, promoted such rapid radiation [131]. In this study, three rapid and recent diversification events have been detected during the Plio-Pleistocene (Fig 4). Among of them, the first acceleration event occurred during the late Pliocene, while the second and third acceleration event occurred during the Pleistocene [132]. During the Plio-Pleistocene intervals, the climate fluctuations induced geographic isolation, habitat fragmentation and changes in resource availability, which acted in concert to produce great species diversity and richness [125, 133–138]. Therefore, Plio-Pleistocene climate fluctuation fostered rapid and recent diversification in many taxonomic groups, such as the invertebrates (e.g. Sternopriscus species, [134]; Kikihia species, [139]; Warramaba species, [140]; Pseudovelia species, [141]); amphibians (e.g. Scinax species, [142]; Hypsiboas species, [143]); reptiles (e.g. Eulamprus species, [144]; Drysdalia species, [145]; Montivipera species, [146]) and mammals (e.g. Myosorex species, [147]; Nannomys, Aethomys, Otomys, Myotomys, Rhabdomys, Mastomys, Saccostomus, Cryptomys and Xerus species, [148]; Mustela species, [149]). Notably, whether the Pleistocene events caused substantial avian diversification has been a matter of long-standing debate [150]. However, many studies proved that the Pleistocene climate cycling played a primary role in driving speciation within many avian species [135, 151–155]. Particularly, many waterfowls (e.g., Mesitornis unicolor, Nipponia Nippon, Pelecanus crispus and Balearica regulorum) have undergone rapid and recent diversification during the Pleistocene [155]. Besides, the Plio-Pleistocene climate fluctuation have also driven the rapid speciation events in Anser species [25]. Therefore, we proposed that the Plio-Pleistocene climate fluctuation have also played a significant role in diversification of Anseriformes birds.
Conclusion
In this study, we sequenced and annotated nine mito-genomes of Anseriformes birds, which ranged from 16,599 bp to 16,630 bp. In addition we have included two mitochondrial genes and reconstructed a strongly supported phylogeny, which covered the majority species (128 species, 39 genera) in Anseriformes. Based on the results of molecular phylogeny and genetic distances, we revised the classification of the Dendrocygna, Amazonetta, Speculanas, Tachyeres, Lophonetta and Alopochen. Furthermore, we strongly suggested that the Dendrocygna species should be considered as an independent family (Dendrocygnidae). During the EECO (from 58 to 50 Ma), the warmer climatic conditions induced the ancestral diversification in Anseriformes, which reinforced their habit of exploiting different ecological niches. At last, our study provided evidence to support that the Plio-Pleistocene climate fluctuation fostered the rapid and recent diversification in Anseriformes, especially in Anatidae.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Genes encoded by the heavy strand were shown outside the circle, and encoded by the light strand were shown inside the circle respectively. The inner ring showed the general GC content of the complete mitochondrial genome sequence.
(TIF)
The nodal numbers are posterior probabilities.
(TIF)
The nodal numbers are posterior probabilities.
(TIF)
(TIF)
(TIF)
Acknowledgments
The authors would like to thank Si’te Luo (Xiamen University, China) for his help in samples collection. We would like to thank Liangheng Yan, Wenliang Zhou, Ling Ding, Bin Wang (Chengdu Institute of Biology, Chinese Academy of Sciences, China), Lan Jiang (Anhui Normal University, China) and Shaoquan Meng (Yulin Normal University, China) for providing helpful advice on the early manuscript. We would like to thank Professor Tariq Ezaz (University of Canberra, Australia) for his linguistic assistance. We are especially thankful to two anonymous reviewers for their professional comments on this manuscript.
Data Availability
All mito-genome sequences files are available from the GenBank (accession numbers: KF312717, KF156760, KF203133, KT345702, KJ710708, KJ722069, KU140667, KU140668 and KJ169568).
Funding Statement
This work was supported by the Anhui Province Higher Education Revitalization Plan, 2014 Colleges and Universities Outstanding Youth Talent Support Program.
References
- 1.Turner GF. The ecology of adaptive radiation. Heredity. 2001;86(6):749–50. [Google Scholar]
- 2.Simpson GG. The major features of evolution. New York: Columbia University Press; 1953. [Google Scholar]
- 3.Osborn HF. The law of adaptive radiation. Am Nat. 1902;36(425):353–63. [Google Scholar]
- 4.Yu W, Wu Y, Yang G. Early diversification trend and Asian origin for extent bat lineages. J Evolution Biol. 2014;27(10):2204–18. [DOI] [PubMed] [Google Scholar]
- 5.Lovette IJ, Bermingham E. Explosive speciation in the New World Dendroica warblers. Proc R Soc Lond B. 1999;266(1429):1629–36. [Google Scholar]
- 6.Johns GC, Avise JC. Tests for ancient species flocks based on molecular phylogenetic appraisals of Sebastes rockfishes and other marine fishes. Evolution. 1998;52(4):1135–46. doi: 10.1111/j.1558-5646.1998.tb01840.x [DOI] [PubMed] [Google Scholar]
- 7.Sanderson M, Donoghue M. Shifts in diversification rate with the origin of angiosperms. Science. 1994;264(5615):1590–3 [DOI] [PubMed] [Google Scholar]
- 8.Rüber L, Tassell JLV, Zardoya R. Rapid speciation and ecological divergence in the American seven-spined gobies (Gobiidae, Gobiosomatini) inferred from a molecular phylogeny. Evolution. 2003;57(7):1584–98. [DOI] [PubMed] [Google Scholar]
- 9.Seehausen O. African cichlid fish: a model system in adaptive radiation research. Proc Biol Sci. 2006;273(1597):1987–98. doi: 10.1098/rspb.2006.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu W, Xu J, Wu Y, Yang G. A comparative study of mammalian diversification pattern. Int J Biol Sci. 2012;8(4):486–97. doi: 10.7150/ijbs.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L, Li YW, Ryder OA, Zhang YP. Analyses of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007;7(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassanin A, Delsuc F, Ropiquet A, Hammer C, van Vuuren BJ, Matthee C, et al. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analyses of mitochondrial genomes. Cr Biol. 2012;335(1):32–50. [DOI] [PubMed] [Google Scholar]
- 13.Shen YY, Liang L, Sun YB, Yue BS, Yang XJ, Murphy RW, et al. A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae. BMC Evol Biol. 2010;10(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19(4):198–207. doi: 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- 15.Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256(5054):193–7. doi: 10.1126/science.256.5054.193 [DOI] [PubMed] [Google Scholar]
- 16.Ottenburghs J, Ydenberg RC, Van Hooft P, Van Wieren SE, Prins HHT. The Avian Hybrids Project: gathering the scientific literature on avian hybridization. Ibis. 2015;157(4):892–4. [Google Scholar]
- 17.Ottenburghs J, van Hooft P, van Wieren SE, Ydenberg RC, Prins HHT. Hybridization in geese: a review. Frontiers in Zoology. 2016b;13(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez J, Duttmann H, Wink M. Phylogenetic relationships based on two mitochondrial genes and hybridization patterns in Anatidae. Journal of Zoology. 2009;279(3):310–8. [Google Scholar]
- 19.Sorenson MD, Cooper A, Paxinos EE, Quinn TW, James HF, Olson SL, et al. Relationships of the extinct moa-nalos, flightless Hawaiian waterfowl, based on ancient DNA. P Roy Soc B-Biol Sci. 1999;266(1434):2187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson SL, Feduccia A. Presbyornis and the origin of the Anseriformes (Aves, Charadrimorphae). Smithsonian Contributions to Zoology. 1980;323:1–24. [Google Scholar]
- 21.Donne-Gouss C, Laudet V, Hänni CA. Molecular phylogeny of Anseriformes based on mitochondrial DNA analyses. Mol Phylogenet Evol. 2002;23(3):339–56. [DOI] [PubMed] [Google Scholar]
- 22.Ruokonen M, Kvist L, Lumme J. Close relatedness between mitochondrial DNA from seven Anser goose species. J Evolution Biol. 2000;13(3):532–40. [Google Scholar]
- 23.Livezey BC. A phylogenetic analyses and classification of recent dabbling ducks (tribe Anatini) based on comparative morphology. Auk. 1991;108(3):471–507. [Google Scholar]
- 24.Livezey BC. Phylogeny and evolutionary ecology of modern seaducks (Anatidae: Mergini). Condor. 1995;97(1):233–55. [Google Scholar]
- 25.Ottenburghs J, Megens HJ, Kraus RH, Madsen O, van Hooft P, van Wieren SE, et al. A tree of geese: A phylogenomic perspective on the evolutionary history of True Geese. Mol Phylogenet Evol. 2016a;101:303–13. [DOI] [PubMed] [Google Scholar]
- 26.Peters JL, McCracken KG, Zhuravlev YN, Lu Y, Wilson RE, Johnson KP, et al. Phylogenetics of wigeons and allies (Anatidae: Anas): the importance of sampling multiple loci and multiple individuals. Mol Phylogenet Evol. 2005;35(1):209–24. doi: 10.1016/j.ympev.2004.12.017 [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Zhou L, Li B, Zhang L. The complete mitochondrial genome of Aix galericulata and Tadorna ferruginea: bearings on their phylogenetic position in the Anseriformes. Plos One. 2013a;9(11):e109701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Zhou L, Zhang L, Luo Z, Xu W. The complete mitochondrial genome of Bean Goose (Anser fabalis) and implications for Anseriformes taxonomy. Plos One. 2013b;8(5):e63334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livezey BC. A phylogenetic classification of waterfowl (Aves: Anseriformes), including selected fossil species. Ann Carnegie Mus. 1997b;66(4):457–96. [Google Scholar]
- 30.Liu G, Zhou LZ, Gu CM. Complete sequence and gene organization of the mitochondrial genome of scaly-sided merganser (Mergus squamatus) and phylogeny of some Anatidae species. Mol Biol Rep. 2012;39(3):2139–45. doi: 10.1007/s11033-011-0961-5 [DOI] [PubMed] [Google Scholar]
- 31.Hoyo JD, Elliott A, Sargatal J. Handbook of the birds of the world, Lynx Edicions, Barcelona, Spain; 1994. [Google Scholar]
- 32.Cracraft J. Toward a phylogenetic classification of the recent birds of the world (Class Aves). Auk. 1981;98(4):681–714. [Google Scholar]
- 33.Howard R, Moore A. A complete checklist of the birds of the world. London: Academic Press Ltd; 1991. [Google Scholar]
- 34.Freethy R. How birds work: a guide to bird biology. Blandford: Blandford Press; 1982. [Google Scholar]
- 35.Delacour J, Mayr E. The family Anatidae. The Wilson Bulletin. 1945;57(1):3–55. [Google Scholar]
- 36.Dickinson EC. The Howard and Moore complete checklist of the birds of the world. Princeton: Princeton University Press; 2003. [Google Scholar]
- 37.Mccracken KG, Afton AD. Data set incongruence and correlated character evolution: an example of functional convergence in the hind-limbs of stifftail diving ducks. Syst Biol. 2000;48(4):683–714. [DOI] [PubMed] [Google Scholar]
- 38.Livezey BC. A phylogenetic analyses of basal Anseriformes, the fossil Presbyornis, and the interordinal relationships of waterfowl. Zool J Linn Soc. 1997a;124(4):397–8. [Google Scholar]
- 39.Livezey BC. A phylogenetic analyses of geese and swans (Anseriformes: Anserinae), including selected fossil species. Syst Biol. 1996;45(4):415–50. [Google Scholar]
- 40.Sraml M, Christidis L, Easteal S, Horn P, Collet C. Molecular relationships within Australasian waterfowl (Anseriformes). Aust J Zool. 1996;44(1):7–58. [Google Scholar]
- 41.Livezey BC. A phylogenetic analyses of recent Anseriform genera using morphological characters. Auk. 1986a;105(4):737–54. [Google Scholar]
- 42.Sibley CG, Ahlquist JE. Phylogeny and classification of birds: a study in molecular evolution. New Haven: Yale University Press; 1990. [Google Scholar]
- 43.MacKinnon JR, Phillipps K, He F. A field guide to the birds of China. Oxford: Oxford University Press; 2000. [Google Scholar]
- 44.Livezey BC. Phylogeny and historical biogeography of steamer ducks (Anatidae: Tachyeres). Syst Zool. 1986b;35(4):458–69. [Google Scholar]
- 45.Corbin KW, Livezey BC, Humphrey PS. Genetic differentiation among steamer-ducks (Anatidae: Tachyeres): an electrophoretic analysis. Condor. 198890: 773–81. [Google Scholar]
- 46.Johnson KP, Sorenson MD. Comparing molecular evolution in two mitochondrial protein-coding genes (cytochrome b and ND2) in the dabbling ducks (tribe Anatini). Mol Phylogenet Evol. 1998;10(1):82–94. doi: 10.1006/mpev.1997.0481 [DOI] [PubMed] [Google Scholar]
- 47.Johnson KP, Sorenson MD. Phylogeny and biogeography of dabbling ducks (genus: Anas): a comparison of molecular and morphological evidence. Auk. 1999;116(3):792–805 [Google Scholar]
- 48.Bulgarella M, Sorenson MD, Peters JL, Wilson RE, McCracken KG. Phylogenetic relationships of Amazonetta, Speculanas, Lophonetta, and Tachyeres: four morphologically divergent duck genera endemic to South America. J Avian Biol. 2010;41(2):186–99. [Google Scholar]
- 49.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141(6):173–216. [DOI] [PubMed] [Google Scholar]
- 50.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27(8):1767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giorgi CD, Saccone C. Mitochondrial genome in animal cells. Cell Biochem Biophys. 1989;14(1):67–78. [DOI] [PubMed] [Google Scholar]
- 52.Galtier N, Nabholz B, Glémin S, Hurst GDD. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 2009;18(22):4541–50. doi: 10.1111/j.1365-294X.2009.04380.x [DOI] [PubMed] [Google Scholar]
- 53.Sorenson MD, Oneal E, Garciamoreno J, Mindell DP. More taxa, more characters: the hoatzin problem is still unresolved. Mol Biol Evol. 2003;20(9):1484–98. doi: 10.1093/molbev/msg157 [DOI] [PubMed] [Google Scholar]
- 54.Van TM, Sibley CG, Hedges SB. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol Biol Evol. 2000;17(3):451–7. [DOI] [PubMed] [Google Scholar]
- 55.Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol S. 2003;34(1):397–423. [Google Scholar]
- 56.Li X, Huang Y, Lei F. Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genomics. 2015;16(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology & Evolution. 2009; 24(6): 332–40. [DOI] [PubMed] [Google Scholar]
- 58.Jiang L, Chen J, Wang P, Ren Q, Yuan J, Qian C, et al. The Mitochondrial Genomes of Aquila fasciata and Buteo lagopus (Aves, Accipitriformes): sequence, structure and phylogenetic analyses. Plos One. 2015;10(8):e0136297 doi: 10.1371/journal.pone.0136297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang N, Kimball RT, Braun EL, Liang B, Zhang Z. Assessing phylogenetic relationships among galliformes: a multigene phylogeny with expanded taxon sampling in Phasianidae. Plos One. 2013;8(5):394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 2007;24(1):269–80. doi: 10.1093/molbev/msl158 [DOI] [PubMed] [Google Scholar]
- 61.Parham JF, Feldman CR, Boore JL. The complete mitochondrial genome of the enigmatic bigheaded turtle (Platysternon): description of unusual genomic features and the reconciliation of phylogenetic hypotheses based on mitochondrial and nuclear DNA. BMC Evol Biol.2006; 6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu AP, Ding W, Zhang ZW, Zhan XJ. Phylogenetic relationships of the avian genus Crossoptilon. Acta Zoologica Sinica. 2005;51(5):898–902. [Google Scholar]
- 63.Alström P, Barnes KN, Olsson U, Barker FK, Bloomer P, Khan AA, et al. Multilocus phylogeny of the avian family Alaudidae (larks) reveals complex morphological evolution, non-monophyletic genera and hidden species diversity. Mol Phylogenet Evol. 2013;69(3):1043–56. doi: 10.1016/j.ympev.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 64.Koju NP, He K., Chalise MK, Ray C, Chen Z, Zhang B, et al. Multilocus approaches reveal underestimated species diversity and inter-specific gene flow in pikas (Ochotona) from southwestern China. Mol Phylogenet Evol. 2016;107:239–45. doi: 10.1016/j.ympev.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 65.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526 (7574):569–73. doi: 10.1038/nature15697 [DOI] [PubMed] [Google Scholar]
- 66.Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–31. doi: 10.1126/science.1253451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venditti C, Meade A, Pagel M. Phylogenies reveal new interpretation of speciation and the Red Queen. Nature. 2010;463(7279):349–52. doi: 10.1038/nature08630 [DOI] [PubMed] [Google Scholar]
- 68.Yoder JB, Clancey E, Des Roches S, Eastman JM, Gentry L, Godsoe W, et al. Ecological opportunity and the origin of adaptive radiations. J Evol Biol. 2010;23(8):1581–96. doi: 10.1111/j.1420-9101.2010.02029.x [DOI] [PubMed] [Google Scholar]
- 69.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Coldspring Harbor Laboratory Press; 2001. [Google Scholar]
- 70.Clarke KR, Gorley RN. Primer v5. Primer-E Ltd. 2001
- 71.Tu JF, Xu JP, Liu HT, Zhao JP, Xing XM, Yang FH. Determination and analyses of complete mitochondrial genome sequence of Peking duck (Anas platyrhychos). Mitochondrial DNA Part A. 2016;27(1):682–3. [DOI] [PubMed] [Google Scholar]
- 72.Pan T, Ren L, Wang H, Che J, Zhang B. Mitochondrial genome of the Anas falcata (Anatidae: Anas). Mitochondrial DNA. 2014;25(2):111–2. doi: 10.3109/19401736.2013.786709 [DOI] [PubMed] [Google Scholar]
- 73.Jiang F, Miao Y, Liang W, Ye H, Liu H, Liu B. The complete mitochondrial genomes of the whistling duck (Dendrocygna javanica) and black swan (Cygnus atratus): dating evolutionary divergence in Galloanserae. Mol Biol Rep. 2010;37(6):3001–15. doi: 10.1007/s11033-009-9868-9 [DOI] [PubMed] [Google Scholar]
- 74.Harrison GA, McLenachan PA, Phillips MJ, Slack KE, Cooper A, Penny D. Four new avian mitochondrial genomes help get to basic evolutionary questions in the late cretaceous. Mol Biol Evol. 2004;21(6):974–83. doi: 10.1093/molbev/msh065 [DOI] [PubMed] [Google Scholar]
- 75.Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J Med Genet. 2008;45(12):769–72. doi: 10.1136/jmg.2008.059782 [DOI] [PubMed] [Google Scholar]
- 76.Burland TG. DNASTAR’s Lasergene sequence analyses software. Bioinformatics methods and protocols. 1999;71–91. [DOI] [PubMed] [Google Scholar]
- 77.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2007;33:686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52(5):267–74. [DOI] [PubMed] [Google Scholar]
- 79.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–5. doi: 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 80.Yan L, Zhang C, Pan T, Zhou W, Hu C, Chang Q, et al. Mitochondrial genome of the Anas acuta (Anatidae: Anas). Mitochondrial DNA. 2015;26(2):297–8. doi: 10.3109/19401736.2013.825783 [DOI] [PubMed] [Google Scholar]
- 81.Zhou W, Zhang C, Pan T, Yan L, Hu C, Xue C, et al. The complete mitochondrial genome of Anas poecilorhyncha (Anatidae: Anas). Mitochondrial DNA. 2015;26(2):265–6. doi: 10.3109/19401736.2013.823191 [DOI] [PubMed] [Google Scholar]
- 82.Hu C, Chang Q, Zhou W, Yan L, Pan T, Xue C, et al. Mitochondrial genome of the Anas crecca (Anatidae: Anas). Mitochondrial DNA. 2015;26(4):625–6. doi: 10.3109/19401736.2013.834434 [DOI] [PubMed] [Google Scholar]
- 83.Sun Z, Wang B, Sun X, Yan L, Pan T, Zhang B. Phylogenetic studies of Anas clypeata (Anatidae: Anas) based on complete mitochondrial DNA sequences. Mitochondrial DNA Part A. 2016;27(6):4320–1. [DOI] [PubMed] [Google Scholar]
- 84.Zhou W, Zhang C, Chang Q, Yan L, Pan T, Zhang B. The complete mitochondrial genome of Aythya ferina (Anatidae: Aythya). Mitochondrial DNA. 2014;27(2):968–9. doi: 10.3109/19401736.2014.926509 [DOI] [PubMed] [Google Scholar]
- 85.Ding L, Zhang C, Chang Q, Yan L, Zhang B. The complete mitochondrial genome of Aythya fuligula (Anatidae: Aythya). Mitochondrial DNA Part A. 2016;27(2):1003–4. [DOI] [PubMed] [Google Scholar]
- 86.Meng S, Wu Z, Pan T, Yan L, Bei Y, Li G, et al. Mitochondrial genome of the Aix galericula (Anatidae: Aix). Mitochondrial DNA Part A. 2016;27(1):318–9. [DOI] [PubMed] [Google Scholar]
- 87.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1981;16(2):111–120. [DOI] [PubMed] [Google Scholar]
- 88.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analyses using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–39. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stamatakis A. RAxML Version 8: A tool for phylogenetic analyses and post-analyses of large phylogenies. Bioinformatics. 2014;30(9):1312–3. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analyses tools. Nucleic Acids Res. 1997;25(25):4876–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nylander JAA. MrModeltest v2: Evolutionary Biology Centre, Uppsala University Sweden. 2004.
- 93.Swofford D. PAUP*. Phylogenetic analysis using parsimony (*and other methods). version 4.0b8. Sinauer Associates, Sunderland, MA. 2001.
- 94.Kan XZ, Li XF, Zhang LQ, Chen L, Qian CJ, Zhang XW, et al. Characterization of the complete mitochondrial genome of the Rock pigeon, Columba livia (Columbiformes: Columbidae). Genet Mol Res. 20109(2):1234–49. doi: 10.4238/vol9-2gmr853 [DOI] [PubMed] [Google Scholar]
- 95.Mindell DP, Sorenson MD, Dimcheff DE. Multiple independent origins of mitochondrial gene order in birds. P Natl Acad Sci USA. 199895(18):10693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.An M, Zhang Z, Li X, Yang S. The complete mitochondrial genome of the White-throated Tinamou, Tinamus guttatus (Tinamiformes, Tinamidae). Mitochondrial DNA Part A. 201627(4):2800–1. [DOI] [PubMed] [Google Scholar]
- 97.Drummond AJ, Suchard MA, Xie D. Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analyses by sampling trees. BMC Evol Biol. 2007;7(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6, http://beast.bio.ed.ac.uk/Tracer. 2014.
- 100.Nee S, May RM, Harvey PH. The reconstructed evolutionary process. Philos T R Soc B. 1994;344(344):305–11. [DOI] [PubMed] [Google Scholar]
- 101.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–90. [DOI] [PubMed] [Google Scholar]
- 102.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2014. [Google Scholar]
- 103.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PloS One. 2014a;9:e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, et al. Bammtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol. 2014b;5(7):701–7. [Google Scholar]
- 105.Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News. 2006;6(1):7–11. [Google Scholar]
- 106.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66(1):409–35. [DOI] [PubMed] [Google Scholar]
- 107.Anderson S, De Bruijn MHL, Coulson AR, Eperon IC, Sanger F, Young IG. Complete sequence of bovine mitochondrial DNA conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982;156(4):683–717. [DOI] [PubMed] [Google Scholar]
- 108.Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. Plos Biol. 2004;2(10):1657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stotz DF, Fitzpatrick FW, Parker TA III, Moskovits DK. Neotropical birds: ecology and conservation. Chicago: University of Chicago Press; 1996. [Google Scholar]
- 110.Sibley CG, Monroe BL. Distribution and taxonomy of birds of the world. New Haven: Yale University Press; 1990. [Google Scholar]
- 111.Sibley CG, Monroe BL. Supplement to distribution and taxonomy of birds of the world. New Haven: Yale University Press; 1993. [Google Scholar]
- 112.De Swardt DH. Egyptian Goose Alopochen aegyptiaca hybridizes with Common Shelduck Tadorna tadorna. Ornithological Observations. 2010;1:63–4. [Google Scholar]
- 113.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32(4):835–45. doi: 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang L, Wang G, Peng R, Peng Q, Zou F. Phylogenetic and molecular dating analysis of Taiwan Blue Pheasant (Lophura swinhoii). Gene. 2014;539(1):21–9. doi: 10.1016/j.gene.2014.01.067 [DOI] [PubMed] [Google Scholar]
- 115.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012; 491(7424):444–8. doi: 10.1038/nature11631 [DOI] [PubMed] [Google Scholar]
- 116.Hedges SB, Dudley J, Kumar S. Timetree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2. doi: 10.1093/bioinformatics/btl505 [DOI] [PubMed] [Google Scholar]
- 117.Zachos JC, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001a;292(5517):686–93. [DOI] [PubMed] [Google Scholar]
- 118.Zachos JC, Dickens GR, Zeebe RE. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature. 2008;451(7176):279–83. doi: 10.1038/nature06588 [DOI] [PubMed] [Google Scholar]
- 119.Bijl PK, Schouten S, Sluijs A, Reichart GJ, Zachos JC, Brinkhuis H. Early Palaeogene temperature evolution of the southwest Pacific Ocean. Nature. 2009;461(7265):776–9. doi: 10.1038/nature08399 [DOI] [PubMed] [Google Scholar]
- 120.Westerhold T, Röhl U. High resolution cyclostratigraphy of the early Eocene–new insights into the origin of the Cenozoic cooling trend. Climate of the Past Discussions. 2009;5(3):309–27. [Google Scholar]
- 121.Gingerich PD. Environment and evolution through the Paleocene–Eocene thermal maximum. Trends Ecol Evol. 2006;21(5):246–53. doi: 10.1016/j.tree.2006.03.006 [DOI] [PubMed] [Google Scholar]
- 122.Röhl U, Westerhold T, Bralower TJ, Zachos JC. On the duration of the Paleocene–Eocene thermal maximum (PETM). Geochem Geophy Geosy. 2007;8(12):229–47. [Google Scholar]
- 123.Zachos JC, Wara MW, Bohaty S, Delaney ML, Petrizzo MR, Brill A, et al. A transient rise in tropical sea surface temperature during the Paleocene-Eocene thermal maximum. Science. 2003;302(5650):1551–4. doi: 10.1126/science.1090110 [DOI] [PubMed] [Google Scholar]
- 124.Tripati A, Elderfield H. Deep-sea temperature and circulation changes at the Paleocene-Eocene Thermal Maximum. Science. 2005;308(5730):1894–8. doi: 10.1126/science.1109202 [DOI] [PubMed] [Google Scholar]
- 125.Woodburnea MO, Gunnell GF, Stucky RK. Climate directly influences Eocene mammal faunal dynamics in North America. P Natl Acad Sci USA. 2009;106(32):13399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Currano ED, Wilf P, Wing SL, Labandeira CC, Lovelock EC, Royer DL. Sharply increased insect herbivory during the Paleocene-Eocene Thermal Maximum. P Natl Acad Sci USA. 2008;105(6):1960–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wilf P, Labandeira CC. Response of plant-insect associations to Paleocene-Eocene warming. Science. 1999;284(5423):2153–6. [DOI] [PubMed] [Google Scholar]
- 128.Vieites DR, Min MS, Wake DB. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. P Natl Acad Sci USA. 2007;104(50):19903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sarker S, Pressey RL, Faith DP, Margules CR, Fuller T, Stoms DM, et al. Biodiversity conservation planning tools: present status and challenges for the future. Annu Rev Env Resour. 2006;31(2):123–59. [Google Scholar]
- 130.Levsen ND, Tiffin P, Olson MS. Pleistocene speciation in the genus populus (Salicaceae). Syst Biol. 2012;61(3):401–12. doi: 10.1093/sysbio/syr120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Farrell BD, Mitter C. Adaptive radiation in insects and plants: time and opportunity. Integr Comp Biol. 1994;34(1):57–69. [Google Scholar]
- 132.Ou X, Zhou S, Lai Z, Zeng L. Discussions on quaternary glaciations and their climatic responding in the Qinghai-Tibetan Plateau. Quaternary Sciences. 2015;35(1):12–28. [Google Scholar]
- 133.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Phil Trans R Soc B. 2004;359:183–95. doi: 10.1098/rstb.2003.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hawlitschek O, Hendrich L, Espeland M, Toussaint EF, Genner MJ, Balke M. Pleistocene climate change promoted rapid diversification of aquatic invertebrates in Southeast Australia. BMC Evol Biol. 2012;12(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huntley JW, Voelker G. A tale of the nearly tail-less: the effects of Plio-Pleistocene climate change on the diversification of the African avian genus Sylvietta. Zool Scr. 2017; doi: 10.1111/zsc.12240 [Google Scholar]
- 136.Favre A, Päckert M, Pauls SU, Jähnig SC, Uhl D, Michalak I. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol Rev. 2015;90(1):236–53. doi: 10.1111/brv.12107 [DOI] [PubMed] [Google Scholar]
- 137.deMenocal PB. Plio-Pleistocene African climate. Science. 1995;270(5233):53–9. [DOI] [PubMed] [Google Scholar]
- 138.deMenocal PB. African climate change and faunal evolution during the Pliocene-Pleistocene. Earth and Planetary Science Letters. 2004;220(2004):3–24 [Google Scholar]
- 139.Marshall DC, Slon K, Cooley JR, Hill KB, Simon C. Steady Plio-Pleistocene diversification and a 2-million-year sympatry threshold in a New Zealand cicada radiation. Mol Phylogenet Evol. 2008;48(3):1054–66. doi: 10.1016/j.ympev.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 140.Kearney M, Blacket MJ. The evolution of sexual and parthenogenetic Warramaba: a window onto Plio–Pleistocene diversification processes in an arid biome. Mol Ecol. 2008;17(24):5257–75. doi: 10.1111/j.1365-294X.2008.03991.x [DOI] [PubMed] [Google Scholar]
- 141.Ye Z, Chen P, Bu W. Terrestrial mountain islands and Pleistocene climate fluctuations as motors for speciation: a case study on the genus Pseudovelia (Hemiptera: Veliidae). Sci Rep-UK. 2016;6:33625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Menezes L, Canedo C, Batalhafilho H, Garda AA, Gehara M, Napoli MF. Multilocus phylogeography of the treefrog Scinax eurydice (Anura, Hylidae) reveals a Plio-Pleistocene diversification in the Atlantic forest. Plos One. 2016;11(6):e0154626 doi: 10.1371/journal.pone.0154626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Koscinski D, Handford P, Tubaro PL, Sharp S, Lougheed SC. Pleistocene climatic cycling and diversification of the Andean treefrog, Hypsiboas andinus. Mol Ecol. 2008;17(8):2012–25. doi: 10.1111/j.1365-294X.2008.03733.x [DOI] [PubMed] [Google Scholar]
- 144.Dubey S, Shine R. Plio-Pleistocene diversification and genetic population structure of an endangered lizard (the Blue Mountains water skink, Eulamprus leuraensis) in south-eastern Australia. J Biogeogr. 2010a;37(5):902–14. [Google Scholar]
- 145.Dubey S, Keogh JS, Shine R. Plio-pleistocene diversification and connectivity between mainland and Tasmanian populations of Australian snakes (Drysdalia, Elapidae, Serpentes). Mol Phylogenet Evol. 2010b;56(3):1119–25. [DOI] [PubMed] [Google Scholar]
- 146.Stümpel N, Rajabizadeh M, Avcı A, Wüster W, Joger U. Phylogeny and diversification of mountain vipers (Montivipera, Nilson et al., 2001) triggered by multiple Plio–Pleistocene refugia and high-mountain topography in the Near and Middle East. Mol Phylogenet Evol. 2016;101:336–51. doi: 10.1016/j.ympev.2016.04.025 [DOI] [PubMed] [Google Scholar]
- 147.Willows-Munro S, Matthee CA. Linking lineage diversification to climate and habitat heterogeneity: phylogeography of the southern African shrew Myosorex varius. J Biogeogr. 2011;38(10):1976–91 [Google Scholar]
- 148.Montgelard C, Matthee CA. Tempo of genetic diversification in southern African rodents: The role of Plio-Pleistocene climatic oscillations as drivers for speciation. Acta oecologica. 2012;42:50–7. [Google Scholar]
- 149.Dawson NG, Hope AG, Talbot SL, Cook JA. A multilocus evaluation of ermine (Mustela erminea) across the holarctic, testing hypotheses of pleistocene diversification in response to climate change. J Biogeogr. 2013;41(3):464–75. [Google Scholar]
- 150.Lovette IJ. Glacial cycles and the tempo of avian speciation. Trends Ecol Evol. 2005;20(2):57–9. doi: 10.1016/j.tree.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 151.Johnson NK, Cicero C. New mitochondrial DNA data affirm the importance of Pleistocene speciation in North American birds. Evolution.2004;58(5):1122–30 [DOI] [PubMed] [Google Scholar]
- 152.Avise JC, Walker D. Pleistocene phylogeographic effects on avian populations and the speciation process. P Roy Soc B-Biol Sci. 1998;265(1395):457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weir JT, Schluter D. Ice sheets promote speciation in boreal birds. Proc Biol Sci. 2004;271(1551):1881–7 doi: 10.1098/rspb.2004.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moyle RG, Filardi CE, Smith CE, Diamond J. Explosive Pleistocene diversification and hemispheric expansion of a “great speciator”. P Natl Acad Sci USA. 2009;106(6):1863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nadachowska-Brzyska K, Li C, Smeds L, Zhang GJ, Ellegren H. Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr Biol. 2015;25(10):1375–80. doi: 10.1016/j.cub.2015.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Genes encoded by the heavy strand were shown outside the circle, and encoded by the light strand were shown inside the circle respectively. The inner ring showed the general GC content of the complete mitochondrial genome sequence.
(TIF)
The nodal numbers are posterior probabilities.
(TIF)
The nodal numbers are posterior probabilities.
(TIF)
(TIF)
(TIF)
Data Availability Statement
All mito-genome sequences files are available from the GenBank (accession numbers: KF312717, KF156760, KF203133, KT345702, KJ710708, KJ722069, KU140667, KU140668 and KJ169568).