Abstract
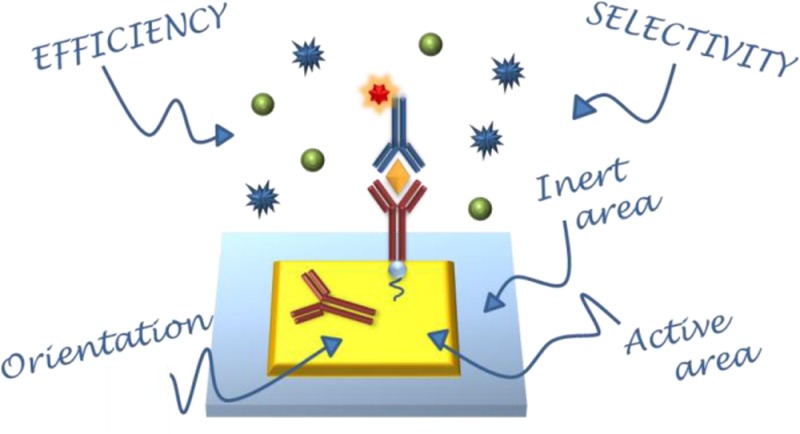
In an ideal plasmonic surface sensor, the bioactive area, where analytes are recognized by specific biomolecules, is surrounded by an area that is generally composed of a different material. The latter, often the surface of the supporting chip, is generally hard to be selectively functionalized, with respect to the active area. As a result, cross talks between the active area and the surrounding one may occur. In designing a plasmonic sensor, various issues must be addressed: the specificity of analyte recognition, the orientation of the immobilized biomolecule that acts as the analyte receptor, and the selectivity of surface coverage. The objective of this tutorial review is to introduce the main rational tools required for a correct and complete approach to chemically functionalize plasmonic surface biosensors. After a short introduction, the review discusses, in detail, the most common strategies for achieving effective surface functionalization. The most important issues, such as the orientation of active molecules and spatial and chemical selectivity, are considered. A list of well-defined protocols is suggested for the most common practical situations. Importantly, for the reported protocols, we also present direct comparisons in term of costs, labor demand, and risk vs benefit balance. In addition, a survey of the most used characterization techniques necessary to validate the chemical protocols is reported.
Keywords: plasmonic sensors, nanostructures, functionalization, biomolecule pattern, characterization
1. Introduction
Current trends that are focused on the miniaturization of devices have led to an ever-growing demand for robust, affordable nanostructure-based technologies that are able to compete with bulkier instruments. Among the different possibilities explored by researchers in the past, the field of plasmonics has been attracting scientific interest for almost three decades, because of the possibilities to squeeze and enhance electromagnetic fields at the subwavelength scale, thus offering applications in a wide range of fields from optoelectronics and quantum information to biomedicine.1 Generally speaking, a biosensor is constituted by a reactive layer able to recognize a specific biomolecule, and a transducer transforming the biochemical input interaction in a physical output signal.1 In this regard, recent advances in nanofabrication have offered possibilities to carefully design the size and shape of nanostructures to finely tune the resonance frequency of surface plasmons and also to confine electromagnetic fields in distinct hot spots with a precision of a few nanometers.2,3 On the other hand, studying the behavior of surface plasmons on roughened surfaces, which can be obtained through scalable, one-step processes, offers the prospect for implementing commercial (low-cost) plasmon-based devices.4 In this view, particular interest has been dedicated to the wide possibilities offered by plasmonics in the field of biosensing. Moreover, the use of nanostructures for sensing confers advantages of a high surface-to-volume ratio and a reactive layer with dimensions similar to biomolecules, thus maximizing the efficiency of the systems.5
Broadly speaking, plasmon-based biosensing approaches can be divided into two categories: the first examines variations in the typical frequency associated with surface plasmon resonance in the presence of an analyte, while the second aims to magnify the intensity of the signals associated with the analyte of interest. A successful example of an approach in the first category is surface plasmon resonance (SPR) sensors, whereas plasmon-enhanced fluorescence (PEF), surface-enhanced Raman scattering (SERS), and surface-enhanced infrared absorption (SEIRA) belong to the second category. For the sake of brevity, we refer the reader to specialized articles and reviews describing these techniques. We note that, although enhanced spectroscopies still have not reached the market, they are extensively and routinely used by researchers. In contrast, SPR and localized surface plasmon resonance (LSPR) systems were commercialized many years ago.6−8 Before starting biosensor design, numerical simulations and theoretical modeling of sensor ideal geometry, composition and sensing technique are usually applied to predict results and to maximize the signals. Modeling strategies are here omitted; for a complete survey on the role of modeling in biosensing design and prediction, the readers are referred to the literature.9,10
One of the most common issues hindering the implementation of plasmonic sensors in biological analyses is the chemical conjugation of the sensor’s active surface in order to increase its selectivity, since the extraction of the analyte of interest from the biological solution by means of consolidated protocols suffers great limitations at low analyte concentrations. An example of the working mechanism of a biosensor device and the pivotal role of the surface functionalization is shown in Figure 1. The mechanisms at the base of plasmonic sensors generally are based on changes of the local refractive index, which can be induced by adsorption of molecules. In the case of analysis of complex biological media, such serum, saliva, or urine, the possibility of nonspecific adsorption and detection of the large variety of molecules constituting the matrix could counterfeit the detection procedure and therefore specific capturing agents are required. Moreover, it is well-known that the plasmonic field decays rapidly as it is drawn away from the surface of the nanostructure. As a consequence, label-free detection of analytes dispersed in solutions at very low concentrations is very challenging, because of the low probability of a molecule attaching or being in close proximity to the metal surface.11 In the majority of cases, binding of molecules to the surface sensor is usually reversible and an equilibrium between molecules attached to the surface and in solution is established. The signal is proportional to the surface coverage, which is dependent on the concentration in the added sample after equilibrium has been established and therefore an uniform coverage is preferred. In such cases, when irreversible binding leads to nonequilibrium conditions, or for very diluted solutions, it should be beneficial to target the functionalization only to the most sensitive parts, namely, the plasmonic hot spot, with the analyte or the active biomolecule necessary to capture the analyte (antibody, aptamers, etc.).12−15 Hence, the functionalization of nanostructures with molecular complexes able to interact with the target analyte concentration drastically improves the sensitivity, defined as the variation of the sensor signal as a function of the variation of the analyte and selectivity, identified as a difference in the response of the device to different analytes. For example, it has been demonstrated that a combination of LSPR and enzyme-linked immunosorbent assay (ELISA) techniques led to an improvement in the sensing efficiency approaching the single molecule limit16,17 Furthermore, radically new methods based on plasmonic metamaterials and hyperbolic metamaterials18−20 are emerging as extremely sensitive biosensors. These new platforms may revolutionize the field of biosensing if effectively combined with the proper surface chemistry able to capture the analytes directly from raw biological fluids.
Figure 1.
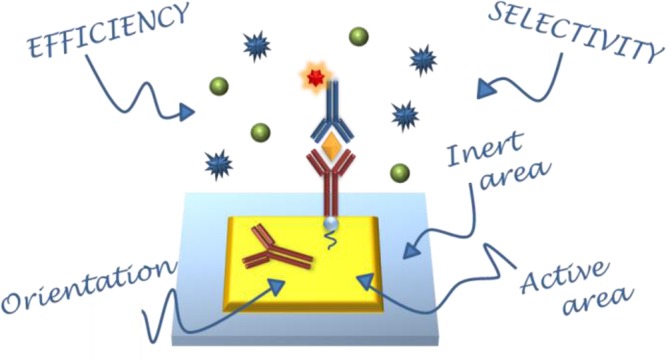
Pictorial description of the most important aspects of sensor functionalization.
This tutorial review aims to present some of the primary tools required for a correct and complete approach to chemically functionalize a plasmonic surface biosensor via top-down technologies. Considerations regarding the use of metal nanoparticles (bottom-up approaches) are avoided in this review, as these materials are usually produced via bottom-up approaches, such as chemical synthesis or laser ablation in liquid, where surface functionalization is essential to prevent aggregation and precipitation effects.21,22 Therefore, the functionalization step is often part of the preparation procedure, and this topic has already been thoroughly investigated elsewhere.23 A hybrid example is represented in the literature by the case of nanoparticles isolated by a ultrathin layer of silica or alumina deposited on a substrate. The presence of the shell prevents their aggregation and separate them from direct contact with the analytes, resulting in significant enhancement of the Raman signal, the so-called SHINERS effect (Shell Isolated Nanoparticle Enhanced Raman Spectroscopy)24−26 and fluorescence, SHINEF (Shell Isolated Nanoparticle Enhanced Fluorescence).27,28
2. THE METAL OF CHOICE
2.1. Motivation
Plasmonic nanoantennas and, more recently, plasmonic metamaterials are among the most promising tools in the field of optical biosensing. Broadly speaking, the techniques used to develop plasmonic nanostructures can be divided into two main categories, namely, top-down and bottom-up approaches. The former mainly rely on lithographic approaches, which enable the fabrication of plasmonic structures and metasurfaces, whereas the latter are based on self-assembly methods for fabricating plasmonic nanoparticles. To effectively work as biosensors, both nanoparticles and plasmonic surfaces must be properly functionalized. However, the functionalization of plasmonic surfaces (typically plasmonic nanoantennas) presents more issues than the functionalization of nanoparticles, which provide high surface-to-volume ratio with excellent biocompatibility using appropriate ligands and can be fully processed in solution. When discussing surface functionalization, the composition of the nanostructures plays a central role. Generally, when working with metal-based substrates, the formation of self-assembled monolayers (SAMs) of molecules with suitable reactive termination is the most common procedure. Considering that biocompatibility is also required when designing a biosensing tool, metals that display plasmonic properties but high reactivity, such as alkali metals, should be avoided.29 In addition, the cost of the raw material should be taken into account.
2.2. Metals, Oxides, and Nitrides
Palladium displayed excellent sensing properties, but it is too expensive for consideration as a possible layer for commercial devices.30 Silver is one of the most valid options for applications in plasmonic biosensing, because it is an inexpensive and efficient substrate that has a high reactivity with thiols for forming SAM.31,32 Nevertheless, its rapid oxidation in air and its slight toxicity to cells are the main drawbacks limiting its application in biomedical fields. Aluminum has also attracted attention recently, because of its favorable optical properties, high natural abundance, low cost, and ease of functionalization.33 The main disadvantage in using aluminum is represented by the difficulties in controlling size and shape during fabrication. Even metal oxides and nitrides have been considered as substrates for plasmonic biosensing, because of their optical properties and biocompatibility; however, they are more difficult to manipulate via nanofabrication techniques.34,35
2.3. Gold
Despite attempts to extend the range of metals suitable for sensing applications, gold still remains a most promising candidate in plasmonic biosensing. Indeed, gold also exhibits good plasmonic properties for wavelengths above the interband transition (λ ≥ 580 nm); being an inert metal, it is not affected by oxidation and it is easy to manipulate using different fabrication techniques. Moreover, gold is compatible with cell culture, and SAMs bound on gold surfaces are stable for long periods after contact with biological media.36 Another advantage of gold is its high reactivity with thiols to form SAMs. This point is of crucial importance for the sensitivity of biological tests: if a good SAM is formed on the metal surface of the reactive layer, the maximum ratio between the number of bound molecules and the superficial area is reached, and the coupled molecules are well-oriented in the biological medium to maximize the binding events with the analyte. The chances of forming a good SAM are dependent both on the surface functionalization technique, which is a topic that will be discussed in detail in the next section of this review, but also on the method of preparation of the substrate, as the preparation method can influence the roughness and crystallinity of the metal surface. Preparing crystal Au(111) textures via physical vapor deposition, followed by thermal annealing, gives rise to the best SAMs,37 while electrodeposition or electroless deposition usually generate rougher Au films, leading to a mixture of structures on the surface, which results in unwanted heterogeneity in molecule orientation. The deposition with a biocompatible primer such as Ti or deposition of Au on mica, instead of glass or silicon as supports, have been demonstrated to be good alternative strategies to obtain almost-single-crystal Au films.38 For the sake of clarity and brevity, the approaches discussed in this review will be limited only to the linking strategies on gold. For more details about linking approaches on other materials, we refer the reader to other review work.36,39,40
3. SURFACE MODIFICATION STRATEGIES
Ideally, a plasmonic sensor should be characterized by an analyte-specific functionalization on its active area and an inert surface elsewhere. In addition, the sensor signal is generally proportional to the efficiency of the binding event, to the surface coverage of the bioactive area (the percentage of the active area that is effectively occupied by the capturing sites), and to the selectivity between this area and the inert area.41 Therefore, in designing a plasmonic sensor, the main issues to be addressed, in terms of surface modification, are the specificity of analyte recognition, the orientation of the immobilized biomolecules acting as the analyte receptor, and the selectivity of surface coverage.
Over the years, many different approaches have been developed for immobilization of biomolecules on the metal surfaces, such as embedding in polymers or membranes,42 sol–gel entrapment,43,44 or layer-by-layer deposition.45−47 The carboxymethylated dextran hydrogel surface has been used for immobilization of aptamers or antibodies.42 Sol–gel process involves the transition of a sol composition from a liquid “sol” to a solid “gel” phase. A sol is first formed by mixing an alkoxide precursor such as tetramethyl orthosilicate (TMOS) or tetraethyl orthosilicate (TEOS) with water, a cosolvent, and an acid or base catalyst. The resulting sol containing a sensing agent (metal particles and bioactive molecules) can be cast as monoliths, thin films on a glass slide, or optical fiber.43 Severe issues exist concerning the encapsulation of biomolecules, because hard reaction conditions are required, which lead to denaturation of biomolecules. New sol–gel processing methods using different precursors, such as organic modified silicate (ORMOSILS) (e.g., methyltriethoxysilane (MTES), propyltrimethoxysilane (PTMS), dimethoxydimethylsilane (DMDMS), etc.) showed promising results in preserving the native activity of biomolecules.44 The layer-by-layer (LBL) techniques are based on the attraction of opposite ionic charges located on metal structures. For the growth of multilayer films, the adsorption occurred as a result of alternating the immersion process of a solid substrate in a cationic and anionic polyelectrolyte solution.45,46 LBL has been used in PEF as a versatile method to tune the distance between fluorophore and metal surface in order to optimize optical response.47 Among all the functionalization methods, self-assembly of molecules is the most diffuse technique in optical biosensors. It was shown that SAM helps in minimizing the nonspecific adsorption problem and also provides stable and oriented immobilization of the analytes.42 In the next section, we summarize solutions based on SAMs that have been proposed in the literature to achieve the requirements of specificity, orientation, and selectivity.
3.1. Specificity
The specificity of the binding event is dependent on the affinity between receptor biomolecules and the specific analyte. Clearly, low affinity reduces the efficiency of the test. The biomolecule/analyte affinity is generally a function of the number of physical and chemical interactions between them. From a physical point of view, it is important that the native conformation of the biomolecule does not change as a consequence of immobilization on the reactive layer.41 From a chemical point of view, the number of weak interactions (van der Waals interaction, dipolar interactions, hydrogen-bond interactions) between the biomolecule and the analyte plays the most important role. For this reason, using multiple recognition approaches by which the analyte is trapped between a primary and a secondary antibody in a sandwich-like configuration is often used to improve molecular recognition.38 Many biological systems have been proposed to recognize specific analytes, and they have been extensively reviewed elsewhere.48−50 The most commonly used biological recognition elements are antibodies or immunoglobulins, which are glycoproteins ∼15 nm × 7 nm × 3.5 nm in size consisting of two polypeptide chains, as depicted in Chart 1a. Each chain is composed by a heavy chain (H), corresponding to the −COOH terminus, and a light chain (L), corresponding to the −NH2 terminus. The two heavy chains are linked together by a disulfide bridge (Fc portion), while the two L chains are structurally standalone (Fab portions of the antibody). The recognition event normally involves one or both of the Fab portions. In the first case, it is possible to chemically reduce the disulfide bond between the two polypeptide chains and split the antibody into two fragments, while still retaining the recognition capacity.38,41,51
Chart 1. General Structures of (a) an Antibody, (b) an Aptamer, and (c) a Sugar.
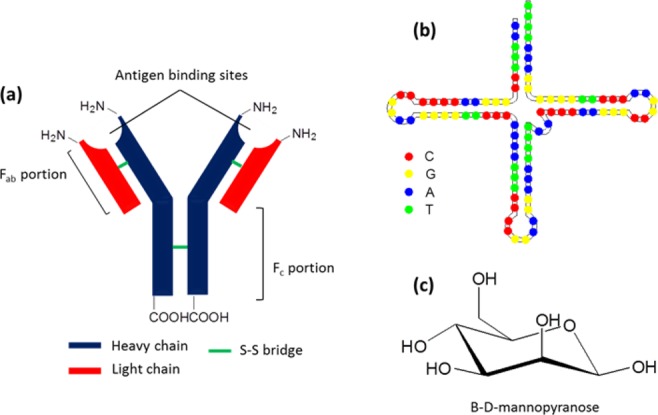
In addition to antibodies, aptamers are also frequently used as a recognition element. Aptamers are single-strained DNA or RNA oligonucleotide sequences capable of specifically binding an analyte with an affinity comparable to antibodies (see Chart 1b). The disadvantage of using aptamers is that their production, generally via synthetic routes, is time and labor consuming, even if they can be more easily and suitably engineered than antibodies.38 Less frequently, but very selectively in some cases, sugars (or post-translationally modified glycoproteins) are used as recognition elements (Chart 1c), especially if the analyte is a protein or enzyme. The disadvantage is that they require a previous modification step to be anchored to the surface.52 Once the recognition element is chosen and independent of its biological nature, the pivotal step in building a new plasmon sensor is the immobilization strategy applied to bring such an element at the right place with the right orientation. Table 1 summarizes the costs vs benefits vs risks of each recognition element.
Table 1. Antibody, Aptamer, and Sugar Costs vs Benefits vs Risks Analysis.
| primary biomolecule | commercial cost ($) | benefits | risks |
|---|---|---|---|
| monoclonal IgG | 100–6000a | • high selectivity | • high costs |
| • high pH and T sensitivity | |||
| DNA aptamer | 1–2 for each single baseb | • good selectivity | • high costs |
| • possibility of engineering | • time and labor consuming | ||
| RNA aptamer | 10–90 for each single baseb | • good selectivity | • high costs |
| • possibility of engineering | • time and labor consuming | ||
| sugar | 1–20c | • low costs | • low selectivity |
| • modification needed |
Commercial costs/mg, from the Thermo Scientific product catalog.
Commercial costs/base, from the IBA product catalog (1 μmol solution).
Commercial costs/mg, from the Sigma–Aldrich product catalog.
3.2. Orientation of Biomolecules: Coupling Methods
Immobilizing the biomolecule on the metal surface with the right orientation is important for at least two reasons. First, many studies have demonstrated that properly oriented biomolecules, i.e., molecules whose binding sites are properly exposed to the sample where the analyte must be detected, have stronger antigen recognition efficiency and avoid specific bindings with other molecules present in the same sample.38,50 Moreover, the orientation influences the biomolecule density on the metal surface.50 Uniformly oriented immobilization ensures high recognition specificity and high sensor sensitivity. Three main coupling strategies can be envisaged to achieve this objective: (i) oriented self-assembly coupling, (ii) covalent coupling via chemical linkers, and (iii) bioaffinity interaction coupling by specific antibody-binding proteins.
3.2.1. Oriented Self-Assembly Coupling
Chemisorption is the oldest and simplest way to immobilize biomolecules on reactive layers. Antibodies adsorbed on Au or Ag surfaces can be realized exploiting the weak interaction between the metal surface and the disulfide bridges that generally exist between cysteine residues on polypeptide chains.41 Similar interactions can also be envisaged with aptamers.53 However, many issues are associated with this approach. First, self-assembly of the biomolecule in its native form does not allow for the orientation to be controlled. Second, a selectivity problem exists between the reactive layer and the inert surrounding area because macromolecules with several different anchor points can create several weak interactions with different materials. As a consequence, biomolecules can be adsorbed in many locations, namely, both on the active area of the sensor and on surrounding areas. Third, practical applications concerning nanostructured devices require several washing steps, in order to remove residues of active biomolecule from the inert area surrounding the active area. This is important, especially for PEF sensors, where the signal quantification is correlated to the difference between dark and brightness areas. Washing can desorb the biomolecules, thus affecting performance and limiting the recyclability or reuse. In the case of antibodies, a simple way to overcome these problems is to split the antibody into two fragments51 by chemically reducing the disulfide bond of the Fc portion (Scheme 1). In this way, the free native thiol groups on the antibody are used to strongly bind the metal surface, thus driving the molecule in a specific orientation. Several studies have evaluated the selection of the best reducing agents to be used for reduction to minimize antibody denaturation and to maximize the immobilization efficiency (see Table 2). In particular, thiols such as 2-mercaptoethylamine (2-MEA), 2-mercaptoethanol (2-ME), and 1,4-dithiotreithol (DTT) have been successful used,54−56 and DTT was the reagent that yielded the best detection sensitivity. The only constraint of such reducing agents is that the agent must be removed from the solution prior to immobilization on the gold surface, because it can compete with the antibody for metal binding. Some antibody fragments can be lost during separation, thus lowering the immobilization yield. Tris(2-carboxyethyl)phosphine (TCEP) was recently proposed as an alternative to thiols as a reducing agent.57 In all reported cases, the ability of the sensors to recognize antigens, the selectivity of recognition and the sensitivity of detection were found to be significantly higher, compared to the same sensors where antibodies were allowed to randomly self-assemble on the metal surface. Nevertheless, although the oriented self-assembly strategy has been extensively proposed for SPR sensors,51,54,55,57 this strategy has not been widely used in nanostructured plasmonic sensor applications. One reason is the fact that chemical modification of the biomolecule is necessary in this strategy. Therefore, if the reaction conditions (pH, solvent, temperature) are not strictly controlled, denaturation can occur. Moreover, the antibody orientation obtained following such a strategy does not maximize the antibody density on the metal surface, which, in nanoscale devices, is more important than in macroscopic sensors. The oriented self-assembly strategy can yield better results with suitable engineered aptamers53 whose structure can be designed a priori in order to optimize the correct orientation during immobilization.
Scheme 1. Oriented Immobilization of Ab Fragments on Gold Surfaces.
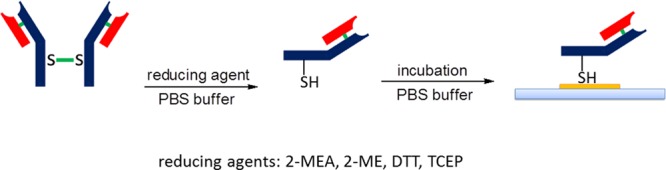
Table 2. Procedures of Typical S–S Reduction for Oriented Immobilization of Ab Fragments on Gold Surfaces.
| reducing agent | commerical cost, Cca ($) | typical procedure | benefits | risks |
|---|---|---|---|---|
| 2-MEA | 90 | 1 mg/mL IgG in PBS and 50 mM 2-MEA are mixed at 37 °C for 1.5 h; after purification, Ab fragments in PBS solution are incubated on an Au chip | • low cost | • competition between 2-MEA and Ab–SH groups |
| • short time | • Ab denaturation | |||
| 2-ME | 3 | 2 mg/mL IgG in PBS and 35 mM 2-ME are mixed at 37 °C for 1 h; after purification, Ab fragments in PBS solution are incubated on an Au chip | • low cost | • competition between 2-MEA and Ab–SH groups |
| • short time | • Ab denaturation | |||
| DTT | 40 | 2 mg/mL IgG in PBS and 40 mM DTT are mixed at 37 °C for 1 h; after purification, Ab fragments in PBS solution are incubated on an Au chip | • low cost | • competition between 2-MEA and Ab–SH groups |
| • short time | • Ab denaturation | |||
| TCEP | 70 | 10 μL of 50 mM of TCEP in PBS and 10 μg/mL IgG in PBS are mixed at room temperature for 1 h; Ab fragments are directly incubated on an Au chip without purification | • low cost | • Ab denaturation |
| • short time | ||||
| • no purification |
Commercial costs per gram or per milliliter, from the Sigma–Aldrich product catalog.
3.2.2. Linker-Mediated Coupling
Covalent linking of biomolecules on a chemically activated metal surface is one of the most widely used methods for immobilizing a recognition element on the sensor reactive layer, improving attachment stability. This route normally requires at least two steps: (a) the creation of a suitable SAM on the metal surface using bifunctional thiols acting as linkers; and (b) the cross-linking between the linker and the complementary native functional groups on the biomolecule.41 From a chemical point of view, there are at least two ways to realize an SAM on a metal surface: “in solution”58−60 and “under vapor”.61,62 For the first method, it has been demonstrated that the success of functionalization, in terms of a well-assembled monolayer on the metal surface, was dependent on many chemical parameters, such as temperature, solvent, pH, and the degree of oxidation of the metal surface. Furthermore, additional physical parameters such as local roughness and crystal orientation may affect the outcome. High temperatures promote the formation of larger ordered domains of molecules on the metal surface.58 The solvent should be chosen in order to minimize its interactions with the thiol acting as the linker. In fact, linkers characterized by long aliphatic and hydrophobic chains create better SAMs if solubilized in nonaliphatic solvents (such as toluene) or in moderately polar solvents (as EtOH).60 Concerning the pH, neutral or gently basic conditions are preferred over acid conditions to favor strong covalent interactions with the metal surface against weak coordination interactions. Moreover, it has been demonstrated that an activation step characterized by gently washing with piranha solution (H2O2:H2SO4 at a 5:2 ratio) or oxygen plasma pretreatment on the gold surface, producing an oxidized gold surface, increases the strength of the S–Au bonds on the surface.46 The reaction time is dependent on the specific thiol used as the linker, and it can vary from hours to days. More recently, dithiocarbamates (N–C–S linkers) have been proposed as an alternative to thiols to form SAMs.63 Dithiocarbamates can be formed in situ via the reaction of carbon disulfide (CS2) and an amine group in solution in the presence of a gold surface.64 Good results in terms of SAM packaging can be achieved by vapor functionalization, which can be performed at ambient pressure61 or under high vacuum conditions.62 This latter technique has been reported as one of the best ways to achieve ordered monolayers; nevertheless, this technique is only rarely used in building sensors, because of the complex experimental setup needed.62
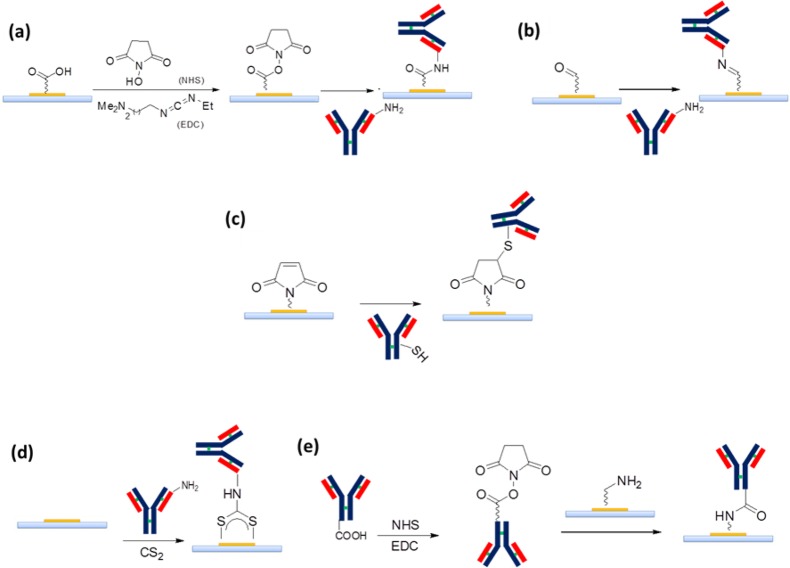
Concerning the strategies used to anchor a biomolecule onto a functionalized support, many interesting methods have been proposed in the last 10 years, although only a few have emerged, because of their flexibility and ease of implementation for nonchemist researchers.63 Normally, such methods allow for native functional groups present on the biomolecule to be exploited for linking the SAM functional groups grafted on the metal surface. Scheme 2 and Table 3 summarize some of the most common chemical cross-linking methods, with particular focus on antibodies immobilization. Instead, suitable modified oligonucleotides can be linked to SAM functional groups with similar classical strategies. Amino and thiol modifications have been routinely used to construct oligonucleotide arrays. Besides, the covalent immobilization of aptamers involves several parameters, each of which must be optimized, such as the chemical stability and complexity of the construct, the simplicity of the derivatization, the loading capacity, and the degree of nonspecific binding of the final product.65 The exposed amine group of lysine residues on antibodies (as well as a suitable 5′NH2- or 3′NH2-modified oligonucleotide65,66) can be used to link metal surfaces functionalized with electrophile groups, such as N-hydroxysuccinimide esters (NHS) to form stable amide bonds (Scheme 2a) or aldehydes to form imines (Scheme 2b). The disadvantage of the NHS strategy is the hydrolytic character of NHS esters, which is responsible for low reaction yields. An additional classical method is to exploit −SH groups on cysteine residues of antibodies (or 5′thiol- or 3′thiol-modified oligonucleotides65,67) by condensing them with a maleimide-functionalized metal surface via Michael reaction to form thioethers (Scheme 2c). Competition with amine groups can be avoided at physiological pH, as amine groups are generally protonated in this condition. Finally, an interesting method to immobilize biomolecules on gold surfaces by exploiting native amine groups is the one-pot reaction with CS2 in water under physiological conditions to form a dithiocarbamate linkage on the gold surface,64,68 where the nitrogen belongs to the biomolecule (Scheme 2d). The carboxylic acid groups of acidic aspartate or glutamate residues can also be used to link gold SAM functionalized with nucleophilic groups (i.e., amine). In this case, the strategy involves transformation of the −COOH group on the antibody into an NHS ester by reacting with 3-(3-dimethylaminopropyl)carbodiimide (EDC) in the presence of NHS (Scheme 2e). In addition to the instability of NHS esters in a water medium and the need to modify the antibody, another risk, in this case, is that the NHS-activated antibody can react with their own peptide residues rather than with the SAM on the metal surface, thus generating undesired antibody clusters in solution.63 An advantage is the high probability that the antibody is attached to the surface with the right orientation, i.e., with the Fc portion immobilized on the support. Indeed, when the other reported methods are used, a heterogeneous population of randomly oriented biomolecules is linked to the metal surface.
Scheme 2. Coupling Strategies to SAM-Modified Au Surfaces Exploiting (a,b) Lysine Residues, (c,d) Cysteine Residues, or (e) Acid Residues on the Biomolecule.
Table 3. Typical Procedures of Coupling Strategies with SAM-Modified Au Surfaces.
| entry | linker | coupling strategy | Cca ($) | typical procedureb | benefits | risks |
|---|---|---|---|---|---|---|
| a | HS–(CH2)n–COOH, i.e., MHDA | EDC/NHS, coupling with lysine residues | 165 | gold slides are incubated overnight 250 μM of MHDA in EtOH and activated by incubation in 2 mM EDC/5 mM NHS solution in MES buffer; after rinsing in PBS, 50 μg/mL of IgG in PBS is added for incubation (30 min, room temperature) | • low cost | • water and air sensitive |
| • multiple steps required | ||||||
| • random orientation | ||||||
| b | HS–(CH2)n–CHO | coupling with lysine residues | 27 | the −NH2 gold chip is immersed in a 5% glutaraldehyde solution in PBS (2 h at room temperature); after PBS rinsing and drying, the chip is incubated with a 100 μg/mL solution of IgG in PBS for 1 h at room temperature | • low cost | • multiple steps required |
| • good stability | • random orientation | |||||
| c | DTSSP | coupling with cysteine or lysine residues | 3000 | gold chip is incubated in a 1.5 mM solution of DTSSP in PBS for 1 h at room temperature; after rinsing, slides are incubated with 40 μg/mL of IgG in PBS solution for 30 min at room temperature | • short time | • water and air sensitive |
| • high cost | ||||||
| • random orientation | ||||||
| d | CS2/NH2–(CH2)n–NH2 | coupling with lysine residues | 0.08 | gold slides are immersed in a mixed solution (1:1, v/v) of 0.1 M CS2 in water and 20 μg/mL of IgG in PBS solution for 2 h at 4 °C | • low cost | • random orientation |
| • short time | ||||||
| • good stability | ||||||
| e | HS–(CH2)n–NH2, i.e., cysteamine | coupling with acid residues activated by EDC/NHS | 40 | 1 mg/mL solution of IgG is activated by incubation in 2 mM EDC/5 mM NHS solution in MES buffer (15 min at r.t.); after quenching using 2-mercaptoethanol (1.2 μL) and purification by desalting column, the solution in PBS is used for incubation (30 min, room temperature) of the functionalized gold chip | • low cost | • water and air sensitive |
| • right orientation | • multiple steps required | |||||
| • Ab clustering |
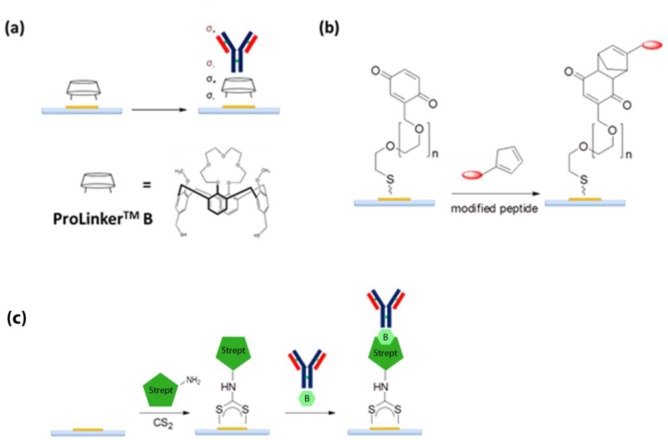
Interesting site-specific chemo-ligation methods for the immobilization of biomolecules on supports have been recently proposed (see Scheme 3 and Table 4). The general concept of such methods is to synthetically introduce specific functional groups to the biomolecules (using the same chemistry of the biomolecule and labeling with fluorophores or tags). For example, cyclopentadienyl groups can be used to activate the molecule toward a Diels–Alder reaction with the metal surfaces functionalized with dienes or quinones.63 On the other hand, research on specific linkers that are able to coordinate the Fc portion of the antibody by host/guest, hydrophobic, or electrostatic interactions has gained interest.75,76 Nevertheless, the large-scale applications of these latter techniques are still minimal, because of the extensive synthetic methods required.
Scheme 3. Site-Specific Coupling of Biomolecules to Au Surfaces: (a) Coupling via Pro-Linker B Linker, (b) Coupling via Diels–Alder Cycloaddition Strategy, and (c) Coupling via Streptavidin–Avidin Strategy Mediated by CS2.
Table 4. Typical Procedures for Site-Specific Coupling of Biomolecules to Au Surfaces.
| entry | linker | coupling strategy | typical procedurea | benefits | risks |
|---|---|---|---|---|---|
| a | Pro-Linker | linker synthesized by multistep synthesis according to ref (73) and commercially available from the authors; coupling employing host–guest, bipolar, and van der Waals interactions with biomolecule functional groups | gold chip was incubated in 3 mM solution ProLinker B solution in CHCl3 for 1 h at room temperature; after rinsing and drying, the chip was incubated in 100 μg/mL PBS IgG solution for 1 h at room temperature | • site-specific coupling | • multistep procedure required |
| • high cost | |||||
| • high time | |||||
| b | Diels–Alder | linker and modified biomolecules synthesized by multistep synthesis according to ref (74) and not commercially available; coupling employs click chemistry between them | gold chip was immersed in the linker solution (1.0 mM) in EtOH at room temperature in a scintillation vial for 12–16 h; the surface was the treated with modified biomolecules (5 mM in water) for 4 h at room temperature | • site-specific coupling | • multistep procedure required |
| • high cost | |||||
| • high time |
3.2.3. Bioaffinity Interaction Coupling
Bioaffinity interaction coupling refers to the oriented immobilization strategy that is based on the creation of several weak interactions between a biomolecule acting as linker and a second biomolecule that is employed for detection of the analyte. The many weak interactions enable a highly specific and strong recognition mechanism. The most common example of bioaffinity interaction coupling is based on the streptavidin–biotin complex in which streptavidin is attached to the support (linker), and biotin is attached to specific antibody sites (Scheme 4).41 In this case, additional steps to immobilize streptavidin and chemical modification of the antibody to introduce the biotin tag are needed (Table 5); nevertheless, the specificity of recognition represents the major strength of this strategy. Streptavidin–biotin coupling is commonly used for oligonucleotides immobilization,67 where streptavidin is covalently linked to a metal surface in order to recognize a biotin-labeled aptamer. Indeed, streptavidin binds biotin with extremely high affinity (dissociation constant Kd ≈ 10–14 mol/L). DNA-direct recognition of an antibody using a single-strained DNA to specifically bind an antibody has also been used as a method to immobilize antibodies for plasmonic sensors.41 The favorite bioaffinity interaction for the immobilization of antibodies in plasmonic sensors is the interaction between them and antibody-binding proteins such as protein G or protein A. Such proteins selectively bind the Fc portion of native antibodies, thus driving them toward the correct orientation on the metal surface and ensuring long-term stability, high purity, and high detection activity against antigens.71 Even if they have overlapping binding sites for the Fc region of the antibodies, there are no structural similarities between protein G and protein A. Both can bind many antibody subtypes from different species, with the only exception of IgM antibodies. A mixture of protein G/protein A is often used to offer the possibility of covering nearly the entire range of antibodies that are linkable to a metal support, thus allowing the design of multiplexed biosensors.77 The only disadvantage is that such proteins must be linked to the metal surface prior to immobilization of the antibodies. Since both proteins G and A have several high affinity binding sites for the Fc portion, the orientation of their linking is less important than in the case of native antibodies, and such setups have been successful realized by classical coupling (see Schemes 4a and 4b) in plasmonic sensors.77,78 Moreover, it is possible to engineer antibody-binding proteins in order to tag them with polycysteine motifs (Scheme 4c) to orient their immobilization.79 The last advantage of using antibody-binding protein is the possibility to purify the antibody needed for the recognition event from a protein mixture directly on the plasmonic surface, without a previous purification step.79 A list of typical procedures is reported in Table 5.
Scheme 4. Bioaffinity Coupling Using (a) Streptavidin–Biotin, (b) Protein A/Protein G Mediated via Classical NHS/EDC Chemistry, (c) CS2 Chemistry, or (d) Polycysteine Tag Engineering.
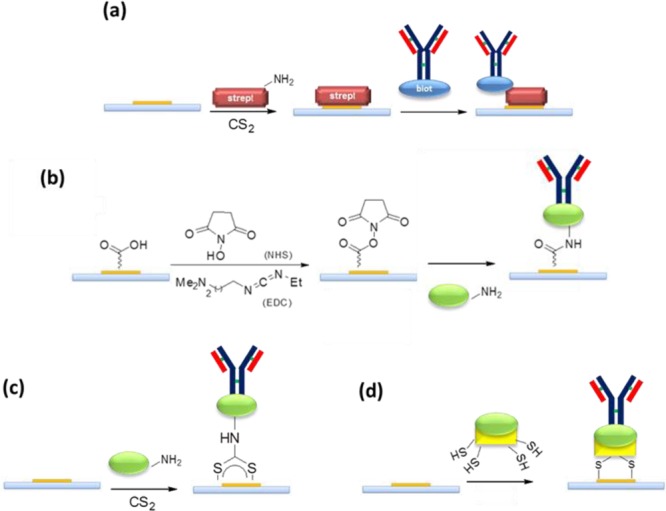
Table 5. Typical Procedures for Bioaffinity Coupling.
| entry | binding protein | Cca ($) | typical procedureb | benefits | risks |
|---|---|---|---|---|---|
| a | streptavidin–biotin | 142 | gold chips are immersed in a mixed solution (1:1, v/v) of 0.1 M CS2 in water and 20 μg/mL of streptavidin in PBS solution for 2 h at 4 °C; after rinsing, the chip was incubated in 100 μg/mL PBS biotinylated-IgG solution for 1 h at room temperature | • very high affinity coupling between linker and biomolecules | • multistep procedure required |
| • high time | |||||
| b | protein A/protein G | 78 | the Au surface is coated with −COOH; after washing, the surface is activated with EDC/NHS mixture in MES buffer and conjugated to 2 mg/mL protein A/G in PBS; the surface is then incubated in 50 μg/mL IgG solution in PBS for 1 h at room temperature | • site-specific orientation | • multiple steps required |
| • low cost | |||||
| c | protein A | 35 | the Au chip is immersed in a mixed solution (1:1, v/v) of 0.1 M CS2 in water and 20 μg/mL protein A in PBS solution for 2 h at 4 °C; after rinsing and drying, slides are incubated with 100 μg/mL IgG in PBS (30 min, room temperature) | • site-specific orientation | |
| • short time | |||||
| • low cost | |||||
| d | engineered protein A (PAG) | c | protein A was engineered by PCR; the Au chip is incubated in 50 μg/mL solution in acetate buffer (4.5 pH) PAG for h at room temperature; after washing, the chip is incubated in 50 μg/mL IgG solution in PBS buffer for 1 h at room temperature | • site-specific orientation | • high cost |
| • high time |
3.3. Selectivity in Biomolecule Patterning
The bioactive area of biosensors should ideally coincide with the sensing area, while all the surroundings regions must be inert to nonspecific adsorption. The latter is of fundamental importance for developing high-sensitivity assays capable of revealing ultralow concentrations of analytes dispersed in highly dense media such as human serum, saliva, urine, or other complex biological fluids. This selectivity requirement means that a well-defined micropattern or nanopattern of active biomolecules must be realized on the sensor surface,38,80 whereas the remaining regions must be passivated (inert to nonspecific binding). Lithographic methods can be used to fabricate surface structures with high spatial resolution, but they are conducted under difficult conditions (solvents and baking) that often can cause biomolecule denaturation. Hence, lithographic patterning is usually performed before biochemical functionalization.79 In the next sections, we describe three additional strategies that have been envisaged in the literature to fabricate such patterns: (a) orthogonal functionalization strategies, (b) tailored wettability to drive molecules toward the region of interest, and (c) inkjet-like methods to create well-defined spots of biomolecules on a continuous or nanostructured metal surface. Of note, microfluidics approaches represent another possible solution that will be intentionally omitted from this review for the sake of brevity. Readers who are interested in this topic can find additional details in other specific reviews.81−84
3.3.1. Orthogonal Functionalization Strategies
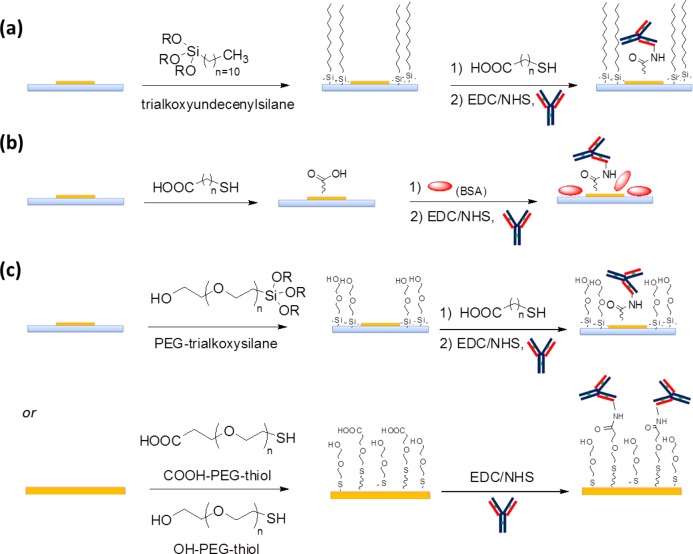
In nanostructured plasmonic sensors, the reactive layer is generally fabricated from a material that differs from the surrounding area. During functionalization, active biomolecules are grafted onto the metal surface and, in principle, the surrounding area must be kept free from any interaction with them. This is achieved by first choosing a linker endowed by a selective functional group for the reactive layer (i.e., sulfur) while covering the inert area by a nonfouling layer. The orthogonality of functionalization is not always obvious, as the chemistry of the two areas are often similar. In addition, many nanostructured substrates may display higher reactivity at the border region between different materials or interdiffusion of different materials, thus complicating the functionalization selectivity. Nevertheless, there are at least three different strategies that can be used to cover the inert area by a nonfouling layer to reach a well-defined pattern of active biomolecules on the reactive area (see Scheme 5).38
Scheme 5. (a and c) Chemical and (b) Bioaffinity Interaction Strategies To Achieve Orthogonal Functionalization in Nanostructured Biosensors.
• Designing the sensor such that the material of the active area is unreactive with the functionalizing agents of the inert area: Au is inert to silanes, while Au can be easily coupled to SiO2. Sensors composed of Au/Si O2 can be easily subjected to orthogonal functionalization by silanization of the SiO2 area with a nonfouling agent,85 followed by thiolization of the reactive layer by a suitable biomolecule linker (Scheme 5a).
• Using antifouling agents: the most diffuse strategy is to use a protein, such as bovine serum albumin (BSA), to form a layer on the inert area by self-assembly (see Scheme 5b). A buffer solution of 0.1%–5% BSA binds both hydrophilic surfaces and hydrophobic surfaces in a highly irreversible manner,86 and this layer prevents the adhesion of antibodies because antibodies do not have high affinity for nonspecific antigens, thus limiting the nonspecific surface bindings.70,75,87,88
• Using polyethylene glycol (PEG) to prevent nonspecific bindings: The ability of PEG to form strong hydrogen bonds with water finally results in the inhibition of the adhesion of other water-soluble molecules, i.e., antibodies,89−91 on the PEG layer. Moreover, it has been calculated that an energy penalty exists for any molecule trying to approach the coated surface of a hydrophilic polymer, because of the low tendency of such polymers for compression, thus preventing the formation of strong van der Waals or electrostatic interactions with biomolecules.92 PEG can be bound to the inert area by covalent coupling, such as with thiol or silane,75,77,93,94 by self-assembly coupling after modification with a protein or a peptide, i.e., poly(l-lysine)-graft-PEG,38,95 or by cross-linked surface chemistry,91 depending on the nature of the inert area (seeScheme 5c).
PEG can also be combined by grafting to methods to other ethylene-glycol-based coatings, i.e., alkanethiol-based oligo(ethylene glycol) (OEG) SAMs.96 In fact, as determined from the work of Prime and Whitesides, OEG-SAMs have a great efficiency in preventing protein absorption,97−99 but they still have some limitations100,101 that could be overcome by a combination of OEG-based SAMs and PEG chains.102,103 As will be widely discussed in section 3.4, the distance between surface and the analyte plays a pivotal role in plasmonic biosensors. Thus, in the sensor design, the antifouling properties, as well as the thickness of the layer, must be considered.
Note that several approaches have been developed as an alternative to the aforementioned methods.104,105 For example, literature reports that hydrophilic neutral polymers different from EG-chains, such as polysaccharides and acrylamides, have been used as antifouling agents.106,107 Masson and co-workers have used peptide-based coatings to minimize the nonspecific absorption of biological media.108,109 Zwitterionic monolayers have also been widely studied as an efficient way to prevent nonspecific adhesion and demonstrated ultralow fouling properties.105,110,111
3.3.2. Biosensors on Superhydrophobic Surfaces
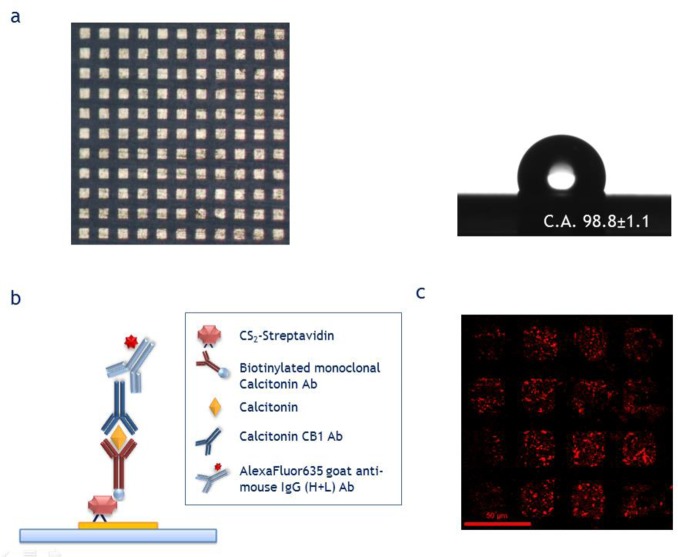
The general concept underlying this approach is to create regions with different wettability, such as hydrophilic domains surrounded by hydrophobic or superhydrophobic domains. When a liquid is in contact with such a surface, molecules will be automatically attracted toward the more hydrophilic domains. An example of a superhydrophobic surface is shown in Figure 2. As shown in Figure 2A, when a drop of water is placed on the surface, it assumes a quasi-spherical shape, rather than spreading indefinitely; in other words, it does not wet the surface. Zarrine-Afsar and co-workers developed a method exploiting increased surface roughness to locally enhance the hydrophilicity in a regular array of wells.112,113 When the substrate is moved against a stationary drop of water from a pipet, the hydrophilic wells capture part of the liquid each time the contact line moves across the edge of the well. The advantage of this method is that it can be used for self-positioning or self-assembly, but it is not suitable for highly diluted solutions, because of the lack of concentration effects. However, this limitation can be overcome by creating surfaces with alternating hydrophobic and hydrophilic microdomains. The concept is shown in Figure 2B: the solution containing molecules of interest is left to evaporate on the surface; intuitively, the more wettable domains will capture the molecules. Indeed, the process is more complicated, and it is essentially due to instabilities created at the boundary between the domains. There are different ways to exploit this effect, and many other methods must still be investigated. For instance, the hydrophilic domains could coincide with the bioactive area of the sensor, and the molecules to be delivered could be those necessary for the functionalization. A practical example is shown in Figures 2C and 2D, in which hydrophilic silver islands are placed on a Teflon substrate (hydrophobic).114 This method was demonstrated to be able to concentrate molecules in regular arrays of wells and analyze them, reaching a limit of detection of 10–100 aM. Furthermore, this method can also be exploited to promote the self-assembly of nanostructures following the layout of the domains. The latter application may be of great interest for investigating or promoting protein crystallization or enhancing spectroscopic signals, such as mass spectrometry (matrix-assisted laser desorption ionization (MALDI)). Notably, the process still works when oleophobic/oleophilic surfaces are considered. We note that even more-extreme devices capable of concentrating the solution at a single point can be conceived. Based on this approach, a few molecules that are dispersed in solution can be delivered to the active area of a single plasmonic nanosensor that is few hundreds of nanometers in size and then revealed via Raman spectroscopy.115
Figure 2.
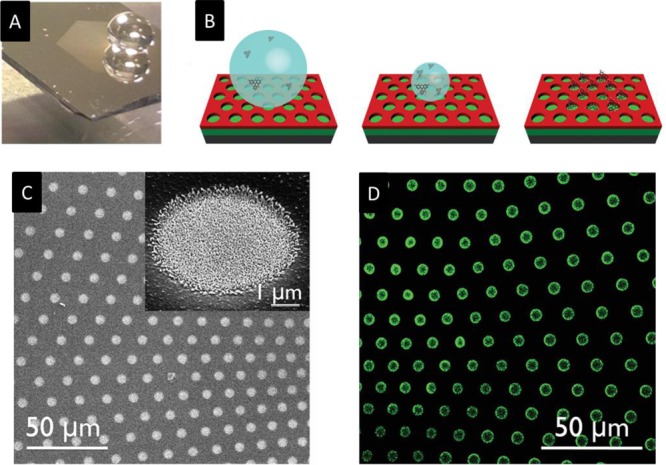
(A) Optical image of a water drop on a superhydrophobic surface. (B) Pictorial description of drop drying on a hydrophobic surface, delivering fluorescein molecules in silver-clustered hydrophilic domains. (C) Scanning electron microscopy (SEM) image showing the geometrical arrangements of Ag small-island films selectively grown inside wells surrounded by superhydrophobic domains. (D) Confocal fluorescence microscopy image in the dye spectral range of the pattern shown in panel (C). (Adapted with permission from ref (114). Copyright 2014, Wiley, Weinheim, Germany.)
Although the investigation of hydrophobic surfaces represents a huge and well-consolidated field of research, the use of this class of devices for biosensing is relatively new. Hence, this approach can be still considered in the embryonal stage. To evaluate the potential of this method, we tested a biosensor designed to measure calcitonin as the antigen, because of its role as a tumor marker for medullary thyroid cancer. The feasibility of the method is described in detail. Polytetrafluoroethylene (PTFE) film was chosen as a basis for the sensor fabrication to guarantee the hydrophobicity of the entire device. The sensing area was defined by gold squares precisely engineered to guarantee the appropriate wettability of the surface. Parameters such as the size of a single square, the distance between adjacent squares, and the sensing area, with respect to the substrate area, were studied. Among the several models tested, a regular pattern of 20-μm-wide squares (40 μm pitch, duty cycle = 0.5) extended on the entire chip area (Figure 3a) provided a good balance in hydrophilic/hydrophobic domains. The substrates were spin-coated with a positive tone resist (Shipley S1813) and thus patterned by optical lithography. After exposure, the samples were developed in MF319 and rinsed with deionized water. Finally, the gold pads where obtained by metal e-beam evaporation and subsequent lift-off in acetone. A thin layer of titanium between the gold layer and the PTFE film worked as an adhesion layer, assuring the stability of the gold pattern and, therefore, preventing degradation of the sensing regions during the functionalization process. To promote the S–Au bonds, the substrates were exposed to an activation step comprising oxygen plasma followed by immersion in soft piranha solution (H2SO4/H2O2). To efficiently immobilize the capturing antibody on the gold surface with a good selectivity, with respect to the surrounding PTFE, a method combining linker-mediated coupling and bioaffinity reaction coupling was exploited. Briefly, the linker composed of CS2 and streptavidin assured a good probability of selecting the gold substrate, because of the spontaneous reaction of carbon disulfide with amine groups to form dithiocarbamates on the gold surface. The substrates were immersed in a mixed solution (1:1, v/v) of 10 μg/mL CS2 in Milli-Q water and 10 μg/mL streptavidin in Milli-Q water for 90 min at room temperature. In the second step, the high affinity and specificity of the recognition between streptavidin and biotin allowed for the efficient and almost irreversible binding of the biotin-conjugated antibody to the linker (Figure 3b); the substrates were incubated in 50 μg/mL biotinylated calcitonin antibody in PBS and 1% BSA solution, used as blocking solution, for 1 h. Note that each step was conducted by immersion of the substrate in the molecule solution and gently mixing for the duration of each step, followed by rinsing in PBS-t (PBS + Tween 0.05%) and drying under mild nitrogen flow. In the third step, the slides were immersed in the analyte solution for 30 min, then rinsed as in the standard procedure and incubated in 50 μg/mL calcitonin monoclonal antibody CB1 for 30 min. Finally, immersion in 500 ng/mL AlexaFluor635 goat antimouse IgG (H+L) antibody solution for 30 min completed the sandwich. Different concentrations of analyte calcitonin, both in PBS and in serum, were measured. The sample analysis was carried out by confocal microscopy (Nikon Model A1+ confocal microscope), this allowed for the overall functionalization outcome to be evaluated while comparing the selectivity between the different domains also for the smallest pattern tested (Figure 3c). The process showed a high selectivity over the different domains under all the mentioned conditions. However, its homogeneity on the pad must still be improved. Via this process, it was possible to detect the presence of calcitonin in serum at a concentration of 1 pg/mL. Thus, the device described here confirms the feasibility of using hydrophobic-engineered substrates as biosensors.
Figure 3.
(a) Gold pattern of the sensor chip on the PTFE film. Best experimental configuration: 40 μm pitch with duty cycle = 0.5, with a static contact angle of ∼100°. (b) Schematic diagram of functionalization of the sensor. (c) Fluorescence confocal microscopy image of the gold-patterned sensor.
3.3.3. Printing Strategies
As an alternative to orthogonal functionalization, several printing methods have been proposed to pattern small-scale spots of biomolecules on the sensor reactive layer. The dimensions, geometric characteristics of the array, and degree of confinement are dependent on the specific technique. Classically, it is possible to distinguish between nonlithographic and lithographic techniques, even if emerging technologies have blurred the boundary of this classification. Classical direct printing consists of the use of automatized inked tips to stain microsized spots on a continuous or pseudocontinuous metal layer.93 All physisorption, chemisorption, or bioaffinity interaction functionalization techniques can occur, depending on which functionalization strategy is preferred. As an example, in the work of Tabakman et al.,93 a microarray printing robot was used to spot a 5 μM antibody solution on a Au/Au island substrate that was previous functionalized with SH-PEG-COOH and activated with EDC/NHS. PEF was used as the sensing mode, and a sandwich with the suitable antigen and secondary antibody was fabricated. The nanoscaled version of direct printing is dip-pen lithography (DPN), a technology that borders between pure lithographic techniques and direct writing, exploiting a suitable “inked” atomic force microscopy (AFM) tip to transport and deposit biomolecules on nanostructured reactive layers.38,80,87 This technique allows the pattern size and geometry to be controlled better than direct printing.80,87 The limit of DPN is the long writing time and the need to precisely control the reaction conditions; for such reasons, even after the development of massive parallelization instruments employing thousands of tips simultaneously, DPN is still not applicable for large patterned surface areas.38,80Nanocontact printing (NCP) is a pure lithographic technique that relies on lithographic fabricated stamp templates to transfer biomolecules onto the reactive layer after their immersion in a molecular “inking” solution. Normally, stamps are made from polymers, such as PDMS.80 Although NCP is a low-cost technique and easily applicable on large superficial areas, it suffers from a lack of control of the pattern, because of diffusion events and imprecision in the realignment step.38 A relatively novel patterning strategy used in nanoplasmonics is nanostencil lithography (NSL). A lithographic fabricated stencil mask characterized by sub-100-nm patterns is used to bring the reactive layer in contact with biomolecules.70 Stencil masks are generally made by a silicon nitride (SiNx) membrane supported by a silicon wafer. Huang el al. fabricated a pyramid-shaped reservoir on a SiNx patterned stencil mask in contact with a PDMS substrate; the biomolecule solution was charged in the reservoir and, after incubation, patterned with different shapes on the PDMS. Fluorescence was used as the sensing mode to reveal patterns.70 Advantages of this technique are its low cost due to mask reusability and the ability to rapidly pattern large superficial areas.70
Molecular imprinting is a nonlithographic technique that exploits polymers to generate imprints of target biological objects on the reactive layer of the sensor. The procedure consists of spotting a prepolymer solution, together with the target molecule and a suitable primer on the metal surface using a robot. The solution is suitably treated to induce polymerization: the functional groups on the prepolymer first react with the functional groups on the target, thus surrounding the target, and then cross-linking between several prepolymer molecules occurs, thus freezing them around the template. After template removal, the remaining cavities have the exact shape of the target biomolecule.72,116 The limit of this technique is that it requires substantial efforts to optimize polymerization reactions to realize only microscaled patterns of cells or big proteins.
The lithographic version of molecular imprinting is photopatterning,117 which is a technique that uses UV light to print the desired patterned image on a surface covered by a photoactivator molecule before binding the biomolecule directly on the illuminated areas.
3.4. How To Choose the Modification Strategy
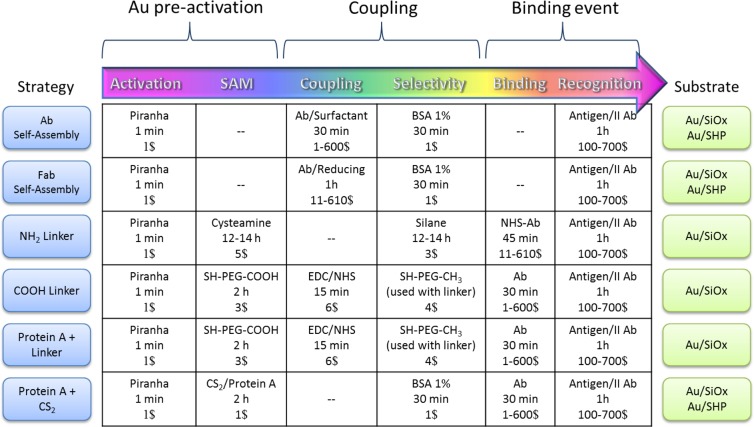
The choice of the modification strategy is dependent on the specific case, and, thus far, no approach has emerged as being superior. However, some guidelines can be found in the literature. As an example, self-assembly immobilization has been extensively used in the oldest SPR sensors, but the low sensitivity associated with this strategy limits its use to nanostructured plasmonic sensors. In contrast, it is possible, in principle, to envisage that oriented self-assembly immobilization would be more suitable for LSPR sensors due to the possibility to selectively orient the biomolecule on small metal areas, with respect to nonmetal inert areas. Oriented self-assembly strategies have found real applications in nanostructured Raman sensors,53 as Raman spectroscopy allows the signal coming from the desired binding event to be selected. Furthermore, the absence of a linker to bring the biomolecule to the reactive surface reduces the overlapping signals that can complicate the spectral interpretation. Most LSPR, PEF, and Raman sensors have been realized using classical covalent linking to the surface, but the sensors require linkers with different lengths. For example, although, for conventional surface plasmon resonance (SPR), the evanescent field can extend to hundreds of nanometers, in the case of nanostructured systems, this value decreases to only a few tens of nanometers.118,119 Therefore, the use of short linkers increases the sensitivity of LSPR sensors, producing a stronger change in reflective mode.52,58,70,75,77,84,94,116 For Raman-based sensors (SERS), the rapid spatial decay of the plasmonic field drastically reduces the detection efficiency for distances of >10 nm120−122 and so it is preferable to avoid the use of linkers or use very short ones. Furthermore, long linkers or linkers with complex functional groups may provide a signal background that hinders the analysis.123 Also, in the case of PEF, field decay effects become consistent for distances of >18–20 nm, but reduced spacing (<4–5 nm) between analyte and the surface can result in quenching of fluorescence;124−126 therefore, linkers with a specific length are required PEF to maximize the enhancement of specific fluorophores.70,88,93 Bioaffinity interaction coupling has been successful employed in LSPR multiplexed plasmonic sensors77 associated with microfluidic devices for patterning and sensing, because of the versatility of antibody-binding protein to interact with a huge class of antibodies and the resistance of the immobilization interaction to continuous flow washing and reaction steps. To the best of our knowledge, protein A and protein G have not been used as linkers for PEF sensors, even though their dimensions are suitable for fluorescence enhancement.71 Regarding patterning strategies, orthogonal functionalization is the easiest strategy for laboratory applications: it has been well-interfaced with all sensing modalities and is associated with all the functionalization strategies previously discussed. The limit of this technique is the lack of robust characterization techniques that can confirm the outcome. Moreover, if orthogonal functionalization is used as a unique selective strategy, no parallel measurements can be performed. In contrast, the integration of orthogonal functionalization protocols in the overall functionalization process where patterning is ensured by printing70,93,116 or microfluidic strategies77,84,94 is currently the most successful way to realize a performing plasmonic sensor. From a practical point of view, different strategies can result in different risks versus benefits, as we described in the step-by-step tables in the previous sections. The average effective time and costs of the most representative functionalization strategies depicted in the previous sections are summarized in Figure 4. We considered a typical functionalization pathway normalized to a 1 cm2 nanostructured device (∼2 mL solutions, 100 μg/mL of Ab) both in the case of typical hydrophilic (Au/SiOx) and superhydrophobic (Au/SHP) substrates.
Figure 4.
Overall time and cost provisions for typical functionalization strategies.
4. CHECKING THE RESULTS: CHARACTERIZATION CONCERNS
The fate of functionalization, both in terms of efficiency and selectivity, strongly influences the sensor sensitivity. Nevertheless, the actual tools available to determine functionalization yields and selectivity on nanostructured biosensors are still inadequate. Indeed, although several techniques have been proposed for biosensor characterization from both chemical and physical aspects, they are only able to provide a qualitative characterization. Step-by-step quantitative characterization and easily achieving nanoparticle functionalization remain major issues in solid nanostructured sensors, with the only exception being the final binding event.
4.1. Qualitative Characterization Techniques
Chemical characterization of functionalized metal layers can be performed using ultrasensing spectroscopic techniques. In particular, high-resolution X-ray photoemission spectroscopy (XPS) and vibrational spectroscopies as SERS and Fourier transform infrared (FTIR) spectroscopy are complementary methods that are largely used to characterize SAM on metal surfaces as well as their coupling with biomolecules. XPS allows analysis of the chemical state of the atomic components of the linkers (i.e., thiols, proteins, DNA) anchored on the reactive layer.127 SAM exposure to X-rays induces the number of emitted photoelectrons from each excited atomic component, depending on its chemical environment. Indeed, the same atom (i.e., C) surrounded by different partners (i.e., H, O, or S) represents a specific XPS peak corresponding to a specific atomic state characterized by a specific shift of the atom valence electron density, toward or away from the bound partner, depending on its electronegativity. The peak intensity is proportional to the presence of a specific atomic state; following this criterion, XPS analysis is also able to determine if an atomic component is covalently bound to the metal surface or only absorbed. As an example, in XPS analysis of a thiol SAM on a metal layer, three different binding energies can be recognized for the S 2p state, depending on whether sulfur is covalently bound to the metal surface as an ordered monolayer, weakly adsorbed on the metal surface with a tilted conformation (namely, a gauche conformation), or only deposited on the surface with a disordered conformation. XPS is a powerful tool for surface investigation and useful for determining the selectivity of the functionalization across different areas of the sample. The limit of the technique is that it becomes less informative as the distance from the surface and the chemical complexity of the functionalization increase. Raman spectroscopy and SERS in particular are very sensitive techniques for characterizing the presence of typical functional groups on the metal surface. The typical spatial resolution is on the order of hundreds of nanometers (in agreement with the Abbe diffraction limit), although combining with scanning probe technologies allows chemical mapping at the nanoscale.128 As the intensity of the SERS signal is dependent on many factors (the absorptivity of the dye molecule, the affinity of that molecule with the metal surface and the goodness of the electronic transfer between them, and the size and shape of the metal structure), the results of SERS analysis are often uncertain.129 Nevertheless, when such parameters are optimized, SERS analysis is useful for characterizing complex molecules as biomolecules or proteins,130 and, therefore, it can be efficiently used to understand the fate of coupling between linkers and primary antibodies on the sensor reactive layer. Also, FTIR spectroscopy can be used to characterize monolayers formed on metallic layer which, acting as mirrors, allow external reflection configuration analysis (FTIR-ERS). The main advantage of FTIR is a marked response to the polarization and incident angle of the exciting radiation and to the orientation of the electric dipoles associated with the films,131,132 thus providing a means for determining the average orientation of molecules on the surface. Moreover, polarization modulation methods can be used to isolate and amplify weak absorption from surface-confined molecules, with respect to ambient phase molecules.133 As an example, polarization-modulated infrared reflection absorption spectroscopy (PM-IRRAS) represents a variation of traditional FTIR with improved sensitivity and orientation specificity, where the incident source is an alternating polarized infrared beam measuring the differential reflectivity of interfacial molecules for the two alternated polarities. Sum-frequency generation (SFG) spectroscopy is a nonlinear optical spectroscopic technique with which one can obtain the vibrational spectrum of interfacial molecules, discriminating them from those in the bulk material. In SFG spectroscopy, two high-intensity laser beams at frequencies ωvis and ωIR overlap at an interface and generate an output beam at frequency ωSFG = ωvis + ωIR in the reflection direction. SFG is forbidden in media with inversion symmetry, such as gases, bulk liquids, amorphous solids, and crystals of achiral molecules, but allowed at interfaces where the inversion symmetry is lost. Therefore, it is surface-specific, with intrinsic selectivity to interfacial contributions.134
Physical characterization tools are able to measure the increase in surface thickness of different areas of the sensor after each functionalization step and can also aid in understanding the fate of functionalization. Ellipsometry and AFM measurements are some of the most diffused techniques that can be used to prove the success of functionalization in nanostructured sensors. Ellipsometry is a technique that can measure layers that are subnanometer to a few micrometers thick, based on the interference generated between the light reflecting from and traveling through the functionalized surface. An advantage of this technique is that it can be used to establish the functionalization success from the first to the last functionalization step, because even very thin surface SAMs can be measured. Moreover, ellipsometric measurements are generally performed on a large surface, and the final thickness is an average of the optical parameters registered from the entire probed area. Consequently, local information on functionalization output is lost.78 In contrast, point-by-point measurements with a spatial resolution on the scale of angstroms are possible by AFM and can be considered complementary to ellipsometry.59,78,87 Therefore, AFM can be a useful tool to qualitatively follow the fate of the different steps of functionalization of the reactive layer, both in terms of efficiency and selectivity, following the height difference between features that have reacted with the functionalizing agent and the features that have not.78,87 A limitation is the long amount of time needed to make measurements. In addition, AFM can be used to determine the force of the interaction of a linker with the surface.135 The common practice is to perform force measurements by using AFM to stretch a linker attached to the surface or to test the mechanical resistance of the SAM.59
4.2. The Issue of Quantification
Quantification of reaction yields and the selectivity of the different functionalization steps of a nanostructured chip are complicated by the need to guarantee sample integrity. Therefore, classical quantification techniques used to establish the total organic carbon present on the surface, such as thermogravimetric analysis (TGA), which are normally applied to nanoparticles, are not applicable in the case of nanostructured surfaces. There are some possible options for quantifying the coupling event of LSPR biosensors. Indeed, the change in intensity of the reflected light in an LSPR experiment, which is directly related to the refractive index (RI) change on the surface, can be plotted against the antibody or antigen concentration used during the coupling or binding event. This gives rise to a calibration curve that can be used to determine the minimum concentration of primary biomolecules needed for a significant response of the sensor or the limit of detection (LOD) of the binding event. Such measurements are still far from precisely quantifying the yield but at least provide information on the linearity of the optical phenomenon, with respect to the concentration of the analyte.136 Concerning the other sensing modalities, quantification concerns exist for the calibration and quantification of the binding event. In PEF, the antigen concentration is correlated with the fluorescence intensity, which is based on the preset microscope conditions (i.e., collection time, gain, position with respect to the nanostructure), subtracting the background signal from the collected fluorescence intensity, and normalizing the signal, with respect to the nanostructure geometry.88 In SERS sensors, preliminary statistical evaluations of the Raman spectra are needed to determine a significant correlation between antigen concentration and the Raman spectra. In summary, analytical methods capable of evaluating and quantifying the fate of each step of functionalization are still critically needed to evaluate the diffusion of such sensors on the market. In conclusion, there remains a lack of quality-control protocols.
5. Conclusions
Developing an efficient plasmonic biosensor requires a multistep methodology that includes design, fabrication, functionalization, and characterization steps. Several different lithographic and nonlithographic techniques are currently available for fabrication. A crucial point in the design is the choice of the materials constituting the reactive and the inert areas of the biosensor, as their diversity will be primary responsible for the selectivity of functionalization. The functionalization strategy can be determined as a function of the composing materials by considering the following points: (a) simplicity of the chemistry used for functionalization; (b) selectivity between the inert and the reactive area and orthogonality between the two functionalization strategies; (c) effective coverage of the metallic surface (plasmonic area) and correct orientation of the active biomolecules; (d) affinity and specificity of the recognition event between the analyte and the biomolecule; and (e) time versus cost balance. Even if several functionalization strategies exist for a metal-to-metal-based biosensor, few studies have been devoted to superhydrophobic surfaces that, according to preliminary results, may improve selectivity and orthogonality in functionalization, thus enabling better sensitivities. Finally, the development of nondestructive characterization tools able to quantify the functionalization yield and selectivity at intermediate levels of sensor assembly is another crucial factor.
Author Contributions
The manuscript was written through contributions made by all authors. All authors have approved the final version of the manuscript.
The research leading to these results received funding from the European Research Council under the European Union’s Seventh Framework Programme (No. FP/2007–2013)/ERC Grant Agreement No. 616213, CoG: Neuro-Plasmonics and under the Horizon 2020 Program, FET-Open: PROSEQO, Grant Agreement No. 687089.
The authors declare no competing financial interest.
References
- Maier S. a.Plasmonics: Fundamentals and Applications, Vol. 677; Springer Science+Business Media LLC: New York, 2004. [Google Scholar]
- Chirumamilla M.; Toma A.; Gopalakrishnan A.; Das G.; Zaccaria R. P.; Krahne R.; Rondanina E.; Leoncini M.; Liberale C.; De Angelis F.; Di Fabrizio E. 3D Nanostar Dimers with a Sub-10-nm Gap for Single-/Few-Molecule Surface-Enhanced Raman Scattering. Adv. Mater. 2014, 26, 2353–2358. 10.1002/adma.201304553. [DOI] [PubMed] [Google Scholar]
- De Angelis F.; Malerba M.; Patrini M.; Miele E.; Das G.; Toma A.; Zaccaria R. P.; Di Fabrizio E. 3D Hollow Nanostructures as Building Blocks for Multifunctional Plasmonics. Nano Lett. 2013, 13, 3553–3558. 10.1021/nl401100x. [DOI] [PubMed] [Google Scholar]
- Raether H.Surface Plasmons on Smooth and Rough Surfaces and on Gratings; Springer Tracts in Modern Physics, Vol. 111; Springer: Berlin, Heidelberg, 1988. [Google Scholar]
- Sannomiya T.; Vörös J. Single Plasmonic Nanoparticles for Biosensing. Trends Biotechnol. 2011, 29, 343–351. 10.1016/j.tibtech.2011.03.003. [DOI] [PubMed] [Google Scholar]
- SensiQ Technologies Home Page; https://www.sensiqtech.com/, accessed Sept. 21, 2016.
- Bio rad Home Page; http://www.bio-rad.com/, accessed Sept. 21, 2016.
- Nicoya Life Sciences Home Page; https://nicoyalife.com/, accessed Sept. 21, 2016.
- Sarid D.; Challener W. A.. Modern Introduction to Surface Plasmons Theory, Mathematica Modeling, and Applications; Cambridge University Press: New York, 2010. [Google Scholar]
- Surface Plasmon Resonance Based Sensors; Homola J., Ed.; Springer Series on Chemical Sensors and Biosensors, Vol. 4; Springer: Berlin, Heidelberg, 2006. [Google Scholar]
- Gentile F.; Coluccio M. L.; Coppede N.; Mecarini F.; Das G.; Liberale C.; Tirinato L.; Leoncini M.; Perozziello G.; Candeloro P.; De Angelis F.; Di Fabrizio E. Superhydrophobic Surfaces as Smart Platforms for the Analysis of Diluted Biological Solutions. ACS Appl. Mater. Interfaces 2012, 4, 3213–3224. 10.1021/am300556w. [DOI] [PubMed] [Google Scholar]
- Feuz L.; Jönsson P.; Jonsson M. P.; Höök F. Improving the Limit of Detection of Nanoscale Sensors by Directed Binding to High-Sensitivity Areas. ACS Nano 2010, 4, 2167–2177. 10.1021/nn901457f. [DOI] [PubMed] [Google Scholar]
- Feuz L.; Jonsson M. P.; Höök F. Material-Selective Surface Chemistry for Nanoplasmonic Sensors: Optimizing Sensitivity and Controlling Binding to Local Hot Spots. Nano Lett. 2012, 12, 873–879. 10.1021/nl203917e. [DOI] [PubMed] [Google Scholar]
- Marie R.; Dahlin A. B.; Tegenfeldt J. O.; Höök F. Generic Surface Modification Strategy for Sensing Applications Based on Au/SiO2 Nanostructures. Biointerphases 2007, 2, 49–55. 10.1116/1.2717926. [DOI] [PubMed] [Google Scholar]
- Dahlin B. A. B.; Jonsson M. P.; Hook F. Specific Self-Assembly of Single Lipid Vesicles in Nanoplasmonic Apertures in Gold. Adv. Mater. 2008, 20, 1436–1442. 10.1002/adma.200701697. [DOI] [Google Scholar]
- Chen S.; Svedendahl M.; Van Duyne R. P.; Käll M. K. Plasmon-Enhanced Colorimetric ELISA with Single Molecule Sensitivity. Nano Lett. 2011, 11, 1826–1830. 10.1021/nl2006092. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Stevens M. M. Plasmonic ELISA for the Ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat. Nanotechnol. 2012, 7, 821–824. 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- Sreekanth K. V.; Alapan Y.; El Kabbash M.; Wen A. M.; Ilker E.; Hinczewski M.; Gurkan U. A.; Steinmetz N. F.; Strangi G. Enhancing the Angular Sensitivity of Plasmonic Sensors Using Hyperbolic Metamaterials. Adv. Opt. Mater. 2016, 4, 1767–1772. 10.1002/adom.201600448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekanth K. V.; Alapan Y.; El Kabbash M.; Ilker E.; Hinczewski M.; Gurkan U. A.; De Luca A.; Strangi G. Extreme Sensitivity Biosensing Platform Based on Hyperbolic Metamaterials. Nat. Mater. 2016, 15, 621–627. 10.1038/nmat4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilio P.; Malerba M.; Toma A.; Zaccaria R. P.; Jacassi A.; De Angelis F. Hybridization in Three Dimensions: A Novel Route toward Plasmonic Metamolecules. Nano Lett. 2015, 15, 5200–5207. 10.1021/acs.nanolett.5b01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio M. L.; Gentile F.; Francardi M.; Perozziello G.; Malara N.; Candeloro P.; Di Fabrizio E. Electroless Deposition and Nanolithography Can Control the Formation of Materials at the Nano-Scale for Plasmonic Applications. Sensors 2014, 14, 6056–6083. 10.3390/s140406056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile F.; Laura Coluccio M.; Candeloro P.; Barberio M.; Perozziello G.; Francardi M.; Di Fabrizio E. Electroless Deposition of Metal Nanoparticle Clusters: Effect of Pattern Distance. J. Vac. Sci. Technol., B: Nanotechnol. Microelectron.: Mater., Process., Meas., Phenom. 2014, 32, 031804. 10.1116/1.4870058. [DOI] [Google Scholar]
- Saha K.; Agasti S. S.; Kim C.; Li X.; Rotello V. M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779. 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. F.; Huang Y. F.; Ding Y.; Yang Z. L.; Li S. B.; Zhou X. S.; Fan F. R.; Zhang W.; Zhou Z. Y.; Wu D. Y.; Ren B.; Wang Z. L.; Tian Z. Q. Shell-Isolated Nanoparticle-Enhanced Raman Spectroscopy. Nature 2010, 464, 392–395. 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- Li J.-F.; Ding S.; Yang Z.; Bai M.; Anema J. R.; Wang X.; Wang A.; Wu D.; Ren B.; Hou S.; Wandlowski T.; Tian Z. Extraordinary Enhancement of Raman Scattering from Pyridine on Single Crystal Au and Pt Electrodes by Shell-Isolated Au Nanoparticles. J. Am. Chem. Soc. 2011, 133, 15922–15925. 10.1021/ja2074533. [DOI] [PubMed] [Google Scholar]
- Chen S.; Meng L.-Y.; Shan H.-Y.; Li J.-F.; Qian L.; Williams C. T.; Yang Z.; Tian Z.-Q. How To Light Special Hot Spots in Multiparticle–Film Configurations. ACS Nano 2016, 10, 581–587. 10.1021/acsnano.5b05605. [DOI] [PubMed] [Google Scholar]
- Aroca R. F.; Teo G. Y.; Mohan H.; Guerrero A. R.; Albella P.; Moreno F. Plasmon-Enhanced Fluorescence and Spectral Modification in SHINEF. J. Phys. Chem. C 2011, 115, 20419–20424. 10.1021/jp205997u. [DOI] [Google Scholar]
- Osorio-Román I. O.; Guerrero A. R.; Albella P.; Aroca R. F. Plasmon Enhanced Fluorescence with Aggregated Shell-Isolated Nanoparticles. Anal. Chem. 2014, 86, 10246–10251. 10.1021/ac502424g. [DOI] [PubMed] [Google Scholar]
- West P. R.; Ishii S.; Naik G. V.; Emani N. K.; Shalaev V. M.; Boltasseva A. Searching for Better Plasmonic Materials. Laser Photonics Rev. 2010, 4, 795–808. 10.1002/lpor.200900055. [DOI] [Google Scholar]
- Sugawa K.; Tahara H.; Yamashita A.; Otsuki J.; Sagara T.; Harumoto T.; Yanagida S. Refractive Index Susceptibility of the Plasmonic Palladium Nanoparticle: Potential as the Third Plasmonic Sensing Material. ACS Nano 2015, 9, 1895–1904. 10.1021/nn506800a. [DOI] [PubMed] [Google Scholar]
- Li W.; Zhang L.; Zhou J.; Wu H. Well-Designed Metal Nanostructured Arrays for Label-Free Plasmonic Biosensing. J. Mater. Chem. C 2015, 3, 6479–6492. 10.1039/C5TC00553A. [DOI] [Google Scholar]
- Dowling D. P.; Donnelly K.; McConnell M. L.; Eloy R.; Arnaud M. N. Deposition of Anti-Bacterial Silver Coatings on Polymeric Substrates. Thin Solid Films 2001, 398-399, 602–606. 10.1016/S0040-6090(01)01326-8. [DOI] [Google Scholar]
- Bisio F.; Proietti Zaccaria R.; Moroni R.; Maidecchi G.; Alabastri A.; Gonella G.; Giglia A.; Andolfi L.; Nannarone S.; Mattera L.; Canepa M. Pushing the High-Energy Limit of Plasmonics. ACS Nano 2014, 8, 9239–9247. 10.1021/nn503035b. [DOI] [PubMed] [Google Scholar]
- Kim J.; Dutta A.; Memarzadeh B.; Kildishev A. V.; Mosallaei H.; Boltasseva A. Zinc Oxide Based Plasmonic Multilayer Resonator: Localized and Gap Surface Plasmon in the Infrared. ACS Photonics 2015, 2, 1224–1230. 10.1021/acsphotonics.5b00318. [DOI] [Google Scholar]
- Naik G. V.; Schroeder J. L.; Ni X.; Kildishev A. V.; Sands T. D.; Boltasseva A. Titanium Nitride as a Plasmonic Material for Visible and near-Infrared Wavelengths. Opt. Mater. Express 2012, 2, 478–489. 10.1364/OME.2.000478. [DOI] [Google Scholar]
- Love J. C.; Estroff L. A.; Kriebel J. K.; Nuzzo R. G.; Whitesides G. M. Self-Assembled Monolayers of Thiolates on Metals as a Form of Nanotechnology. Chem. Rev. 2005, 105, 1103–1169. 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Vaskevich A.; Rubinstein I. Widely-Applicable Gold Substrate for the Study of Ultrathin Overlayers. J. Am. Chem. Soc. 2004, 126, 5569–5576. 10.1021/ja0396448. [DOI] [PubMed] [Google Scholar]
- Reimhult E.; Höök F. Design of Surface Modifications for Nanoscale Sensor Applications. Sensors 2015, 15, 1635–1675. 10.3390/s150101635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalini S.; Bortolotti C. A.; Leonardi F.; Biscarini F. Self-Assembled Monolayers in Organic Electronics. Chem. Soc. Rev. 2017, 46, 40–71. 10.1039/C6CS00509H. [DOI] [PubMed] [Google Scholar]
- Vericat C.; Vela M. E.; Corthey G.; Pensa E.; Cortés E.; Fonticelli M. H.; Ibañez F.; Benitez G. E.; Carro P.; Salvarezza R. C. Self-Assembled Monolayers of Thiolates on Metals: A Review Article on Sulfur-Metal Chemistry and Surface Structures. RSC Adv. 2014, 4, 27730–27754. 10.1039/C4RA04659E. [DOI] [Google Scholar]
- Jung Y.; Jeong J. Y.; Chung B. H. Recent Advances in Immobilization Methods of Antibodies on Solid Supports. Analyst 2008, 133, 697–701. 10.1039/b800014j. [DOI] [PubMed] [Google Scholar]
- Shankaran D. R.; Gobi K. V.; Miura N. Recent Advancements in Surface Plasmon Resonance Immunosensors for Detection of Small Molecules of Biomedical, Food and Environmental Interest. Sens. Actuators, B 2007, 121, 158–177. 10.1016/j.snb.2006.09.014. [DOI] [Google Scholar]
- Wolfbeis O. S. Fiber-Optic Chemical Sensors and Biosensors. Anal. Chem. 2008, 80, 4269–4283. 10.1021/ac800473b. [DOI] [PubMed] [Google Scholar]
- Gupta R.; Chaudhury N. K. Entrapment of Biomolecules in Sol–Gel Matrix for Applications in Biosensors: Problems and Future Prospects. Biosens. Bioelectron. 2007, 22, 2387–2399. 10.1016/j.bios.2006.12.025. [DOI] [PubMed] [Google Scholar]
- Iost R. M.; Crespilho F. N. Layer-by-Layer Self-Assembly and Electrochemistry: Applications in Biosensing and Bioelectronics. Biosens. Bioelectron. 2012, 31, 1–10. 10.1016/j.bios.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Baba A.; Taranekar P.; Ponnapati R. R.; Knoll W.; Advincula R. C. Electrochemical Surface Plasmon Resonance and Waveguide-Enhanced Glucose Biosensing with N-Alkylaminated Polypyrrole/Glucose Oxidase Multilayers. ACS Appl. Mater. Interfaces 2010, 2, 2347–2354. 10.1021/am100373v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K.; Zin M. T.; Ma H.; Sarikaya M.; Huang F.; Jen A. K.-Y. Surface Plasmon Enhanced Fluorescence of Cationic Conjugated Polymer on Periodic Nanoarrays. ACS Appl. Mater. Interfaces 2010, 2, 3153–3159. 10.1021/am100635v. [DOI] [PubMed] [Google Scholar]
- Knopp D.; Tang D.; Niessner R. Review: Bioanalytical Applications of Biomolecule-Functionalized Nanometer-Sized Doped Silica Particles. Anal. Chim. Acta 2009, 647 (1), 14–30. 10.1016/j.aca.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Camarero J. A. Recent Developments in the Site-Specific Immobilization of Proteins onto Solid Supports. Biopolymers 2008, 90, 450–458. 10.1002/bip.20803. [DOI] [PubMed] [Google Scholar]
- Kalia J.; Raines R. T. Advances in Bioconjugation. Curr. Org. Chem. 2010, 14, 138–147. 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyakin A. A.; Presnova G. V.; Rubtsova M. Y.; Egorov A. M. Oriented Immobilization of Antibodies onto the Gold Surfaces via Their Native Thiol Groups. Anal. Chem. 2000, 72, 3805–3811. 10.1021/ac9907890. [DOI] [PubMed] [Google Scholar]
- Yonzon C. R.; Jeoung E.; Zou S.; Schatz G. C.; Mrksich M.; Van Duyne R. P. A Comparative Analysis of Localized and Propagating Surface Plasmon Resonance Sensors: The Binding of Concanavalin A to a Monosaccharide Functionalized Self-Assembled Monolayer. J. Am. Chem. Soc. 2004, 126, 12669–12676. 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- Yang J.; Palla M.; Bosco F. G.; Rindzevicius T.; Alstrøm T. S.; Schmidt M. S.; Boisen A.; Ju J.; Lin Q. Surface-Enhanced Raman Bioassay on Aptamer-Functionalized Mapping. ACS Nano 2013, 7, 5350–5359. 10.1021/nn401199k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniukevic J.; Kirlyte J.; Ramanavicius A.; Ramanaviciene A. Comparison of Oriented and Random Antibody Immobilization Techniques on the Efficiency of Immunosensor. Procedia Eng. 2012, 47, 837–840. 10.1016/j.proeng.2012.09.277. [DOI] [Google Scholar]
- Baniukevic J.; Kirlyte J.; Ramanavicius A.; Ramanaviciene A. Application of Oriented and Random Antibody Immobilization Methods in Immunosensor Design. Sens. Actuators, B 2013, 189, 217–223. 10.1016/j.snb.2013.03.126. [DOI] [Google Scholar]
- Wolf S.Silicon Processing for the VLSI Era; Sierra Nevada Books: Reno, NV, 1990. [Google Scholar]
- Sharma H.; Mutharasan R. Half Antibody Fragments Improve Biosensor Sensitivity without Loss of Selectivity. Anal. Chem. 2013, 85, 2472–2477. 10.1021/ac3035426. [DOI] [PubMed] [Google Scholar]
- Yamada R.; Wano H.; Uosaki K. Effect of Temperature on Structure of the Self-Assembled Monolayer of Decanethiol on Au(111) Surface. Langmuir 2000, 16, 5523–5525. 10.1021/la991394e. [DOI] [Google Scholar]
- Xue Y.; Li X.; Li H.; Zhang W. Quantifying Thiol–Gold Interactions towards the Efficient Strength Control. Nat. Commun. 2014, 5, 4348. 10.1038/ncomms5348. [DOI] [PubMed] [Google Scholar]
- Lee N.-S.; Kang H.-G.; Ito E.; Hara M.; Noh J.-G. Effects of Solvent on the Structure of Octanethiol Self-Assembled Monolayers on Au(111) at a High Solution Temperature. Bull. Korean Chem. Soc. 2010, 31, 2137–2138. 10.5012/bkcs.2010.31.8.2137. [DOI] [Google Scholar]
- Choi Y.; Jeong Y.; Chung H.; Ito E.; Hara M.; Noh J. Formation of Ordered Self-Assembled Monolayers by Adsorption of Octylthiocyanates on Au(111). Langmuir 2008, 24, 91–96. 10.1021/la701302g. [DOI] [PubMed] [Google Scholar]
- Cossaro A.; Dell’Angela M.; Verdini A.; Puppin M.; Kladnik G.; Coreno M.; De Simone M.; Kivimäki A.; Cvetko D.; Canepa M.; Floreano L. Amine Functionalization of Gold Surfaces: Ultra High Vacuum Deposition of Cysteamine on Au(111). J. Phys. Chem. C 2010, 114, 15011–15014. 10.1021/jp104824c. [DOI] [Google Scholar]
- Wong L. S.; Khan F.; Micklefield J. Selective Covalent Protein Immobilization: Strategies and Applications. Chem. Rev. 2009, 109, 4025–4053. 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- Almeida I.; Cascalheira A. C.; Viana A. S. One Step Gold (Bio)functionalisation Based on CS2-Amine Reaction. Electrochim. Acta 2010, 55, 8686–8695. 10.1016/j.electacta.2010.07.084. [DOI] [Google Scholar]
- Devor E. J.; Behlke M. A.. Strategies for Attaching Oligonucleotides to Solid Supports; IDT DNA Report, 2014, pp 1–24.
- Burgener M.; Sänger M.; Candrian U. Synthesis of a Stable and Specific Surface Plasmon Resonance Biosensor Surface Employing Covalently Immobilized Peptide Nucleic Acids. Bioconjugate Chem. 2000, 11, 749–754. 10.1021/bc0000029. [DOI] [PubMed] [Google Scholar]
- Tombelli S.; Minunni M.; Mascini M. Piezoelectric Biosensors: Strategies for Coupling Nucleic Acids to Piezoelectric Devices. Methods 2005, 37, 48–56. 10.1016/j.ymeth.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Leonov A. P.; Wei A. Photolithography of Dithiocarbamate-Anchored Monolayers and Polymers on Gold. J. Mater. Chem. 2011, 21, 4371–4376. 10.1039/c0jm04153j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin A. E.; Coskun A. F.; Galarreta B. C.; Huang M.; Herman D.; Ozcan A.; Altug H. Handheld High-Throughput Plasmonic Biosensor Using Computational On-Chip Imaging. Light: Sci. Appl. 2014, 3, e122. 10.1038/lsa.2014.3. [DOI] [Google Scholar]
- Huang M.; Galarreta B. C.; Artar A.; Adato R.; Aksu S.; Altug H. Reusable Nanostencils for Creating Multiple Biofunctional Molecular Nanopatterns on Polymer Substrate. Nano Lett. 2012, 12, 4817–4822. 10.1021/nl302266u. [DOI] [PubMed] [Google Scholar]
- Bergström G.; Mandenius C. F. Orientation and Capturing of Antibody Affinity Ligands: Applications to Surface Plasmon Resonance Biochips. Sens. Actuators, B 2011, 158, 265–270. 10.1016/j.snb.2011.06.017. [DOI] [Google Scholar]
- Schirhagl R. Bioapplications for Molecularly Imprinted Polymers. Anal. Chem. 2014, 86, 250–261. 10.1021/ac401251j. [DOI] [PubMed] [Google Scholar]
- Lee Y.; Lee E. K.; Cho Y. W.; Matsui T.; Kang I. C.; Kim T. S.; Han M. H. ProteoChip: A Highly Sensitive Protein Microarray Prepared by a Novel Method of Protein Immobilization for Application of Protein-Protein Interaction Studies. Proteomics 2003, 3, 2289–2304. 10.1002/pmic.200300541. [DOI] [PubMed] [Google Scholar]
- Dillmore W. S.; Yousaf M. N.; Mrksich M. A Photochemical Method for Patterning the Immobilization of Ligands and Cells to Self-Assembled Monolayers. Langmuir 2004, 20, 7223–7231. 10.1021/la049826v. [DOI] [PubMed] [Google Scholar]
- Soler M.; Estevez M. C.; Alvarez M.; Otte M. A.; Sepulveda B.; Lechuga L. M. Direct Detection of Protein Biomarkers in Human Fluids Using Site-Specific Antibody Immobilization Strategies. Sensors 2014, 14, 2239–2258. 10.3390/s140202239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y.; Jirasupangkul T.; Yonezawa T. One-Pot Preparation of Cationic Charged Pt Nanoparticles by the Autocatalytic Hydrolysis of Acetylthiocholine. New J. Chem. 2015, 39, 4214–4217. 10.1039/C5NJ00420A. [DOI] [Google Scholar]
- Im H.; Shao H.; Park Y. Il; Peterson V. M.; Castro C. M.; Weissleder R.; Lee H. Label-Free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat. Biotechnol. 2014, 32, 490–495. 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y.; Matos A. I.; Abrantes L. M.; Viana A. S.; Jin G. Antibody Oriented Immobilization on Gold Using the Reaction between Carbon Disulfide and Amine Groups and Its Application in Immunosensing. Langmuir 2012, 28, 17718–17725. 10.1021/la303032f. [DOI] [PubMed] [Google Scholar]
- de Juan-Franco E.; Caruz A.; Pedrajas J. R.; Lechuga L. M. Site-Directed Antibody Immobilization Using a Protein A–Gold Binding Domain Fusion Protein for Enhanced SPR Immunosensing. Analyst 2013, 138, 2023–2031. 10.1039/c3an36498d. [DOI] [PubMed] [Google Scholar]
- Tran H.; Killops K. L.; Campos L. M. Advancements and Challenges of Patterning Biomolecules with Sub-50 nm Features. Soft Matter 2013, 9, 6578. 10.1039/c3sm00149k. [DOI] [Google Scholar]
- He J. L.; Wang D. S.; Fan S. K. Opto-Microfluidic Immunosensors: From Colorimetric to Plasmonic. Micromachines 2016, 7, 29. 10.3390/mi7020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur J. P.; Jonsson A.; Senkbeil S.; Kutter J. P. Recent Advances in Lab-on-a-Chip for Biosensing Applications. Biosens. Bioelectron. 2016, 76, 213–233. 10.1016/j.bios.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Pires N. M. M.; Dong T.; Hanke U.; Hoivik N. Recent Developments in Optical Detection Technologies in Lab-on-a-Chip Devices for Biosensing Applications. Sensors 2014, 14 (8), 15458–15479. 10.3390/s140815458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.; Chung M. T.; McHugh W.; Nidetz R.; Li Y.; Fu J.; Cornell T. T.; Shanley T. P.; Kurabayashi K. Multiplex Serum Cytokine Immunoassay Using Nanoplasmonic Biosensor Microarrays. ACS Nano 2015, 9, 4173–4181. 10.1021/acsnano.5b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand E.; Humblot V.; Landoulsi J.; Petronis S.; Pradier C. M.; Kasemo B.; Svedhem S. Chemical Modifications of Au/SiO2 Template Substrates for Patterned Biofunctional Surfaces. Langmuir 2011, 27, 678–685. 10.1021/la101858y. [DOI] [PubMed] [Google Scholar]
- Jeyachandran Y. L.; Mielczarski E.; Rai B.; Mielczarski J. A. Quantitative and Qualitative Evaluation of Adsorption/desorption of Bovine Serum Albumin on Hydrophilic and Hydrophobic Surfaces. Langmuir 2009, 25, 11614–11620. 10.1021/la901453a. [DOI] [PubMed] [Google Scholar]
- Lee K.-B.; Kim E.-Y.; Mirkin C. a; Wolinsky S. M. The Use of Nanoarrays for Highly Sensitive and Selective Detection of Human Immunodeficiency Virus Type 1 in Plasma. Nano Lett. 2004, 4, 1869–1872. 10.1021/nl049002y. [DOI] [Google Scholar]
- Kandziolka M.; Charlton J. J.; Kravchenko I. I.; Bradshaw J. A.; Merkulov I. A.; Sepaniak M. J.; Lavrik N. V. Silicon Nanopillars as a Platform for Enhanced Fluorescence Analysis. Anal. Chem. 2013, 85, 9031–9038. 10.1021/ac401500y. [DOI] [PubMed] [Google Scholar]
- Trmcic-Cvitas J.; Hasan E.; Ramstedt M.; Li X.; Cooper M. A.; Abell C.; Huck W. T. S.; Gautrot J. E. Biofunctionalized Protein Resistant Oligo(ethylene Glycol)-Derived Polymer Brushes as Selective Immobilization and Sensing Platforms. Biomacromolecules 2009, 10 (10), 2885–2894. 10.1021/bm900706r. [DOI] [PubMed] [Google Scholar]
- Bolduc O. R.; Masson J. Advances in Surface Plasmon Resonance Sensing with Nanoparticles and Thin Films: Nanomaterials, Surface Chemistry, and Hybrid Plasmonic Techniques. Anal. Chem. 2011, 83, 8057–8062. 10.1021/ac2012976. [DOI] [PubMed] [Google Scholar]
- Harbers G. M.; Emoto K.; Greef C.; Metzger S. W.; Woodward H. N.; Mascali J. J.; Grainger D. W.; Lochhead M. J. A Functionalized Poly(ethylene Glycol)-Based Bioassay Surface Chemistry That Facilitates Bio-Immobilization and Inhibits Non- Specific Protein, Bacterial, and Mammalian Cell Adhesion. Chem. Mater. 2007, 19, 4405–4414. 10.1021/cm070509u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger M.; Drobek T.; Spencer N. D. Interaction Forces and Morphology of a Protein-Resistant Poly(ethylene Glycol) Layer. Biophys. J. 2005, 88, 495–504. 10.1529/biophysj.104.045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakman S. M.; Lau L.; Robinson J. T.; Price J.; Sherlock S. P.; Wang H.; Zhang B.; Chen Z.; Tangsombatvisit S.; Jarrell J. a; Utz P. J.; Dai H. Plasmonic Substrates for Multiplexed Protein Microarrays with Femtomolar Sensitivity and Broad Dynamic Range. Nat. Commun. 2011, 2, 466. 10.1038/ncomms1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aćimović S. S.; Ortega M. a; Sanz V.; Berthelot J.; Garcia-cordero J. L.; Renger J.; Maerkl S. J.; Kreuzer M. P.; Quidant R. LSPR Chip for Parallel, Rapid, and Sensitive Detection of Cancer Markers in Serum. Nano Lett. 2014, 14, 2636–2641. 10.1021/nl500574n. [DOI] [PubMed] [Google Scholar]
- Kenausis G. L.; Voros J.; Elbert D. L.; Huang N.; Hofer R.; Ruiz-taylor L.; Textor M.; Hubbell J. A.; Spencer N. D. Poly (l-Lysine)-G-Poly (Ethylene Glycol) Layers on Metal Oxide Surfaces: Attachment Mechanism and Effects of Polymer Architecture on Resistance to Protein Adsorption. J. Phys. Chem. B 2000, 104, 3298–3309. 10.1021/jp993359m. [DOI] [Google Scholar]
- Emilsson G.; Schoch R. L.; Feuz L.; Höök F.; Lim R. Y. H.; Dahlin A. B. Strongly Stretched Protein Resistant Poly(ethylene Glycol) Brushes Prepared by Grafting-To. ACS Appl. Mater. Interfaces 2015, 7, 7505–7515. 10.1021/acsami.5b01590. [DOI] [PubMed] [Google Scholar]
- Prime K.; Whitesides G. M. Self-Assembled Organic Monolayers: Model Systems for Studying Adsorption of Proteins at Surfaces. Science 1991, 252, 1164–1167. 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- Mrksich M.; Whitesides G. M.. Using Self-Assembled Monolayers That Present Oligo(ethylene Glycol) Groups To Control the Interactions of Proteins with Surfaces. In Poly(ethylene glycol): Chemistry and Biological Applications; Harris J. M., Zalipsky S., Eds; ACS Symposium Series, No. 680; American Chemical Society: Washington, DC, 1997; Chapter 23, pp 361–373. [Google Scholar]
- Prime K. L.; Whitesides G. M. Adsorption of Proteins onto Surfaces Containing End-Attached Oligo(Ethylene Oxide): A Model System Using Self-Assembled Monolayers. J. Am. Chem. Soc. 1993, 115, 10714–10721. 10.1021/ja00076a032. [DOI] [Google Scholar]
- Zhao X.-M.; Wilbur J. L.; Whitesides G. M. Using Two-Stage Chemical Amplification To Determine the Density of Defects in Self-Assembled Monolayers of Alkanethiolates on Gold. Langmuir 1996, 12, 3257–3264. 10.1021/la960044e. [DOI] [Google Scholar]
- Schoenfisch M. H.; Pemberton J. E. Air Stability of Alkanethiol Self-Assembled Monolayers on Silver and Gold Surfaces. J. Am. Chem. Soc. 1998, 120, 4502–4513. 10.1021/ja974301t. [DOI] [Google Scholar]
- Lokanathan A. R.; Zhang S.; Regina V. R.; Cole M. A.; Ogaki R.; Dong M.; Besenbacher F.; Meyer R. L.; Kingshott P. Mixed Poly(ethylene Glycol) and Oligo(ethylene Glycol) Layers on Gold as Nonfouling Surfaces Created by Backfilling. Biointerphases 2011, 6 (4), 180–188. 10.1116/1.3647506. [DOI] [PubMed] [Google Scholar]
- Hucknall A.; Rangarajan S.; Chilkoti A. In Pursuit of Zero: Polymer Brushes That Resist the Adsorption of Proteins. Adv. Mater. 2009, 21, 2441–2446. 10.1002/adma.200900383. [DOI] [Google Scholar]
- Banerjee I.; Pangule R. C.; Kane R. S. Antifouling Coatings: Recent Developments in the Design of Surfaces That Prevent Fouling by Proteins, Bacteria, and Marine Organisms. Adv. Mater. 2011, 23, 690–718. 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- Schlenoff J. B. Zwitteration: Coating Surfaces with Zwitterionic Functionality to Reduce Nonspecific Adsorption. Langmuir 2014, 30, 9625–9636. 10.1021/la500057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk Y.; Kato M.; Mrksich M. Self-Assembled Monolayers of Alkanethiolates Presenting Mannitol Groups Are Inert to Protein Adsorption and Cell Attachment. Langmuir 2000, 16, 9604–9608. 10.1021/la0004653. [DOI] [Google Scholar]
- Brink C.; Österberg E.; Holmberg K.; Tiberg F. Using Poly(ethylene Imine) to Graft Poly(ethylene Glycol) or Polysaccharide to Polystyrene. Colloids Surf. 1992, 66, 149–156. 10.1016/0166-6622(92)80131-K. [DOI] [Google Scholar]
- Masson J.; Battaglia T. M.; Cramer J.; Beaudoin S.; Sierks M.; Booksh K. S. Reduction of Nonspecific Protein Binding on Surface Plasmon Resonance Biosensors. Anal. Bioanal. Chem. 2006, 386, 1951–1959. 10.1007/s00216-006-0834-2. [DOI] [PubMed] [Google Scholar]
- Bolduc O. R.; Pelletier J. N.; Masson J. SPR Biosensing in Crude Serum Using Ultralow Fouling Binary Patterned Peptide SAM. Anal. Chem. 2010, 82, 3699–3706. 10.1021/ac100035s. [DOI] [PubMed] [Google Scholar]
- Vaisocherova H.; Yang W.; Zhang Z.; Cao Z.; Cheng G.; Piliarik M.; Homola J.; Jiang S. Ultralow Fouling and Functionalizable Surface Chemistry Based on a Zwitterionic Polymer Enabling Sensitive and Specific Protein Detection in Undiluted Blood Plasma. Anal. Chem. 2008, 80, 7894–7901. 10.1021/ac8015888. [DOI] [PubMed] [Google Scholar]
- Holmlin R. E.; Chen X.; Chapman R. G.; Takayama S.; Whitesides G. M. Zwitterionic SAMs That Resist Nonspecific Adsorption of Protein from Aqueous Buffer. Langmuir 2001, 17, 2841–2850. 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- Zarrine-Afsar A.; Barends T. R. M.; Müller C.; Fuchs M. R.; Lomb L.; Schlichting I.; Miller R. J. D. Crystallography on a Chip. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 321–323. 10.1107/S0907444911055296. [DOI] [PubMed] [Google Scholar]
- Zarrine-Afsar A.; Muller C.; Talbot F. O.; Miller R. J. D. Self-Localizing Stabilized Mega-Pixel Picoliter Arrays with Size-Exclusion Sorting Capabilities. Anal. Chem. 2011, 83, 767–773. 10.1021/ac1021024. [DOI] [PubMed] [Google Scholar]
- Miele E.; Malerba M.; Dipalo M.; Rondanina E.; Toma A.; De Angelis F. Controlling Wetting and Self-Assembly Dynamics by Tailored Hydrophobic and Oleophobic Surfaces. Adv. Mater. 2014, 26, 4179–4183. 10.1002/adma.201400310. [DOI] [PubMed] [Google Scholar]
- De Angelis F.; Gentile F.; Mecarini F.; Das G.; Moretti M.; Candeloro P.; Coluccio M. L.; Cojoc G.; Accardo A.; Liberale C.; Zaccaria R. P.; Perozziello G.; Tirinato L.; Toma A.; Cuda G.; Cingolani R.; Di Fabrizio E. Breaking the Diffusion Limit with Super-Hydrophobic Delivery of Molecules to Plasmonic Nanofocusing SERS Structures. Nat. Photonics 2011, 5 (11), 682–687. 10.1038/nphoton.2011.222. [DOI] [Google Scholar]
- Ertürk G.; Özen H.; Tümer M. A.; Mattiasson B.; Denizli A. Microcontact Imprinting Based Surface Plasmon Resonance (SPR) Biosensor for Real-Time and Ultrasensitive Detection of Prostate Specific Antigen (PSA) from Clinical Samples. Sens. Actuators, B 2016, 224, 823–832. 10.1016/j.snb.2015.10.093. [DOI] [Google Scholar]
- Alvéole Home Page; http://www.alveolelab.com/, accessed Sept. 21, 2016.
- Mazzotta F.; Johnson T. W.; Dahlin A. B.; Shaver J.; Oh S.-H.; Höök F. Influence of the Evanescent Field Decay Length on the Sensitivity of Plasmonic Nanodisks and Nanoholes. ACS Photonics 2015, 2, 256–262. 10.1021/ph500360d. [DOI] [Google Scholar]
- Kedem O.; Tesler A. B.; Vaskevich A.; Rubinstein I. Sensitivity and Optimization of Localized Surface Plasmon Resonance Transducers. ACS Nano 2011, 5, 748–760. 10.1021/nn102617d. [DOI] [PubMed] [Google Scholar]
- Kovacs G. J.; Loutfy R. O.; Vincett P. S.; Jennings C.; Aroca R. Distance Dependence of SERS Enhancement Factor from Langmuir–Blodgett Monolayers on Metal Island Films: Evidence for the Electromagnetic Mechanism. Langmuir 1986, 2, 689–694. 10.1021/la00072a001. [DOI] [Google Scholar]
- Masango S. S.; Hackler R. A.; Large N.; Henry A.-I.; McAnally M. O.; Schatz G. C.; Stair P. C.; Van Duyne R. P. High-Resolution Distance Dependence Study of Surface-Enhanced Raman Scattering Enabled by Atomic Layer Deposition. Nano Lett. 2016, 16, 4251–4259. 10.1021/acs.nanolett.6b01276. [DOI] [PubMed] [Google Scholar]
- Kennedy B. J.; Spaeth S.; Dickey M.; Carron K. T. Determination of the Distance Dependence and Experimental Effects for Modified SERS Substrates Based on Self-Assembled Monolayers Formed Using Alkanethiols. J. Phys. Chem. B 1999, 103, 3640–3646. 10.1021/jp984454i. [DOI] [Google Scholar]
- Li M.; Cushing S. K.; Zhang J.; Suri S.; Evans R.; Petros W. P.; Gibson L. F.; Ma D.; Liu Y.; Wu N. Three-Dimensional Hierarchical Plasmonic Nano-Architecture Enhanced Surface-Enhanced Raman Scattering Immunosensor for Cancer Biomarker Detection in Blood Plasma. ACS Nano 2013, 7 (6), 4967–4976. 10.1021/nn4018284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadeer N. S.; Brennan M. R.; Wilson W. L.; Murphy C. J. Distance and Plasmon Wavelength Dependent Fluorescence of Molecules Bound to Silica-Coated Gold Nanorods. ACS Nano 2014, 8, 8392–8406. 10.1021/nn502887j. [DOI] [PubMed] [Google Scholar]
- Gandra D. N.; Portz C.; Tian L.; Tang D. R.; Xu D. B.; Achilefu S.; Singamaneni S. Probing Distance-Dependent Plasmon-Enhanced Near-Infrared Fluorescence Using Polyelectrolyte Multilayers as Dielectric Spacers. Angew. Chem., Int. Ed. 2014, 53, 866–870. 10.1002/anie.201308516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng A. L.; You M. L.; Tian L.; Singamaneni S.; Liu M.; Duan Z.; Lu T. J.; Xu F.; Lin M. Distance-Dependent Plasmon-Enhanced Fluorescence of Upconversion Nanoparticles Using Polyelectrolyte Multilayers as Tunable Spacers. Sci. Rep. 2015, 5, 7779. 10.1038/srep07779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer K.; Vyalikh D. V.; Gavrila G.; Kade A.; Weigel-Jech M.; Mertig M.; Molodtsov S. L. High-Resolution Photoelectron Spectroscopy of Self-Assembled Mercaptohexanol Monolayers on Gold Surfaces. J. Electron Spectrosc. Relat. Phenom. 2008, 163, 59–64. 10.1016/j.elspec.2008.02.005. [DOI] [Google Scholar]
- De Angelis F.; Das G.; Candeloro P.; Patrini M.; Galli M.; Bek A.; Lazzarino M.; Maksymov I.; Liberale C.; Andreani L. C.; Di Fabrizio E. Nanoscale Chemical Mapping Using Three-Dimensional Adiabatic Compression of Surface Plasmon Polaritons. Nat. Nanotechnol. 2010, 5, 67–72. 10.1038/nnano.2009.348. [DOI] [PubMed] [Google Scholar]
- Merlen A.; Gadenne V.; Romann J.; Chevallier V.; Patrone L.; Valmalette J. C. Surface Enhanced Raman Spectroscopy of Organic Molecules Deposited on Gold Sputtered Substrates. Nanotechnology 2009, 20, 215705. 10.1088/0957-4484/20/21/215705. [DOI] [PubMed] [Google Scholar]
- Das G.; Mecarini F.; Gentile F.; De Angelis F.; Mohan Kumar H. G.; Candeloro P.; Liberale C.; Cuda G.; Di Fabrizio E. Nano-Patterned SERS Substrate: Application for Protein Analysis vs. Temperature. Biosens. Bioelectron. 2009, 24, 1693–1699. 10.1016/j.bios.2008.08.050. [DOI] [PubMed] [Google Scholar]
- Fourier Transform Infrared Spectra; Ferraro J. R., Basile L. J., Eds.; Academic Press: New York, 1978. [Google Scholar]
- Francis S. A.; Ellison A. H. Infrared Spectra of Monolayers on Metal Mirrors. J. Opt. Soc. Am. 1959, 49, 131–138. 10.1364/JOSA.49.000131. [DOI] [Google Scholar]
- Porter M. D. IR External Reflection Spectroscopy: A Probe for Chemically Modified Surfaces. Anal. Chem. 1988, 60, 1143A–1155A. 10.1021/ac00171a720. [DOI] [Google Scholar]
- Volpati D.; Aoki P. H. B.; Alessio P.; Pavinatto F. J.; Miranda P. B.; Constantino C. J. L.; Oliveira O. N. Vibrational Spectroscopy for Probing Molecular-Level Interactions in Organic Films Mimicking Biointerfaces. Adv. Colloid Interface Sci. 2014, 207, 199–215. 10.1016/j.cis.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Yadavalli V. K.; Forbes J. G.; Wang K. Functionalized Self-Assembled Monolayers on Ultraflat Gold as Platforms for Single Molecule Force Spectroscopy and Imaging. Langmuir 2006, 22, 6969–6976. 10.1021/la060320h. [DOI] [PubMed] [Google Scholar]
- Estevez M. C.; Otte M. A.; Sepulveda B.; Lechuga L. M. Trends and Challenges of Refractometric Nanoplasmonic Biosensors: A Review. Anal. Chim. Acta 2014, 806, 55–73. 10.1016/j.aca.2013.10.048. [DOI] [PubMed] [Google Scholar]