Abstract
Aptamers are synthetic, short nucleic acid molecules capable of specific target recognition. Aptamers are selected using a screening method termed Systematic Evolution of ligands by Exponential enrichment (SELEX). We recently have introduced a variant of SELEX called “LIgand-Guided-Selection” (LIGS) that allows the identification of specific aptamers against known cell-surface proteins. Utilizing LIGS, we introduced three specific aptamers against membrane-bound IgM (mIgM), which is the hallmark of B cells. Out of the three aptamers selected against mIgM, an aptamer termed R1, in particular, was found to be interesting due to its ability to recognize mIgM on target cells and then block anti-IgM antibodies binding their antigen. We systematically truncated parent aptamer R1 to design shorter variants with enhanced affinity. Importantly, herein we show that the specificity of the most optimized variant of R1 aptamer is similar to that of anti-IgM antibody, indicating that the specificity of the ligand utilized in selective elution of the aptamer determines the specificity of the LIGS-generated aptamer. Furthermore, we report that truncated variants of R1 able recognize mIgM-positive human B lymphoma BJAB cells at physiological temperature, demonstrating that LIGS-generated aptamers could be re-optimized into higher affinity variants. Collectively, these findings show the significance of LIGS in generating highly specific aptamers with potential applications in biomedicine.
Introduction
Aptamers are synthetic, short nucleic acid molecules capable of specific target recognition.1 Based on their ability to self-assemble via intra- and intermolecular interactions leading to unique three-dimensional conformations, aptamers can specifically bind to a wide range of target molecules. Some of these molecules do not contain endogenous binding sites towards nucleic acid ligands.2 Versatility in synthesis, coupled with facile chemical manipulation, makes aptamers attractive in designing molecular tools for biomedical applications.3,4 Aptamers possess two attributes that contribute to their potential success in designing molecular tools. First, their small, compact structure enables the design of multi-specific molecular modulators without significantly altering pharmacokinetics properties in vivo. Second, their synthetic nature affords compatibility with a variety of functionalities enabling precise manipulation. Aptamers are identified using an in vitro selection method known as Systematic Evolution of Ligands by Exponential enrichment, or SELEX. SELEX isolates and enriches high-affinity binders from a library of nucleic acid molecules against a target.5,6 The process involves three stages: target binding, separation of high- from low-affinity binders, and amplification to multiply copies of binders with the highest affinity.5,6 Finally, a library of nucleic acid molecules is evolved into a pool of high-affinity binders against the target utilized in the selection and finally identified as aptamers.
Recently, much progress has been made to improve the selection of aptamers against complex targets.7,8 For example, cell-SELEX technology was introduced utilizing whole cells, demonstrating the adaptability of SELEX in generating aptamers against cell-surface receptors at their native environment.9–11 In particular, the use of endogenous membrane protein receptors in their native state is preferable to their purified form based on reduced solubility and susceptibility to misfolding.8 Undeniably, such precise targeting is essential in developing therapeutic and diagnostic molecules. To this end, we introduced a variant of SELEX called “LIgand-Guided Selection” (LIGS) that allows the identification of specific aptamers against known (i.e., SELEX) cell-surface proteins.12,13 In particular, LIGS identifies aptamers specific for a predetermined epitope expressed on the cell surface at its native environment. In terms of protocol, LIGS interrupts the selection process of SELEX and introduces a strong, high-affinity bivalent antibody (Ab), which interacts with its cognate epitope to outcompete and replace specific aptamers from an enriched SELEX pool.12,13 Therefore, based on the specificity of a natural pre-existing ligand towards its target, the aptamers identified by LIGS are expected to show higher specificity towards the target ligand than those succeeding as target-specific binders via the typical cell-SELEX route.12,13
Utilizing LIGS, we recently introduced three specific aptamers against membrane-bound IgM (mIgM), which is the hallmark of B cells.12 Out of the three aptamers selected against mIgM, an aptamer termed R1, in particular, was found to be interesting by its ability to recognize mIgM on target cells and then block anti-IgM antibodies binding their antigen. At the same time, however, we found the affinity of R1 is too low to be utilized as a diagnostic tool for cells expressing mIgM. Therefore, we herein report the systematic application of structure-activity relationship (SAR) studies against R1 that, in turn, enabled the design of novel variants of R1 with improved affinity. Moreover, the optimized structure of aptamer R1 variant (R1.2) did not diminish the aptamer’s specificity towards mIgM-expressing panel of B-cell lines, indicating that the functional fold of aptamer R1 was retained, despite the truncations employed. The antibody utilized in selective elution of aptamer R1 binds to both sIgM and mIgM. We found that variant of R1, termed R1.2 also binds to sIgM as well as mIgM demonstrating the specificity of secondary ligands utilized in selective elution of the aptamer governs the aptamer’s epitope specificity. Since the sIgM and mIgM are identical in their amino acid composition, except the constant μ4 (Cμ4) region at the 3’-end of mIgM, demonstration of variant R1.2 binding to both sIgM and mIgM confirms that aptamers can be generated against predetermined epitopes guided by secondary ligands, a hallmark mechanism of LIGS.14 Finally, the most optimized variant of R1 showed binding to mIgM-positive human B lymphoma BJAB cells at physiological temperatures, proving that LIGS-generated aptamers could be re-optimized into higher affinity variants, thus demonstrating the significance of LIGS in generating epitope-specific aptamers with potential applications in biomedicine.
Methods and materials
Cell culture
Cell lines, including Ramos (Burkitt’s lymphoma), BJAB (Burkitt’s lymphoma), CA-46 (Burkitt’s lymphoma), SKLY-16 (B cell lymphoma) and Jurkat.E6 (acute T cell leukemia), were a generous gift from the David Scheinberg Lab and Jason Huse Lab, Memorial Sloan Kettering Cancer Center. Daudi (Burkitt’s lymphoma) and MOLT-3 cell lines (acute lymphoblastic leukemia) were purchased from American Type Culture Collection (ATCC). All cells were cultured using RPMI 1640 medium supplemented with 100units/mL penicillin–streptomycin and 10% fetal bovine serum (heat-inactivated; Invitrogen). All cell lines were routinely assessed for the expression of appropriate CD markers to authenticate the cell line.
DNA synthesis and binding buffers
All DNA reagents needed for DNA synthesis were purchased from Glen Research or ChemGenes. The variants of R1 were chemically synthesized by attaching a fluorophore at the 3’-end using standard solid phase phosphoramidite chemistry on an ABI394 DNA (Biolytics) synthesizer using a 0.2 µmole scale. The completed DNA sequences were de-protected according to the base modification employed and purified using HPLC (Waters) equipped with a C-18 reversed phase column (Phenomenex/Waters/Thermo Fisher). All in vitro experiments were performed using a binding buffer composed of DPBS, 4.5 g/L glucose, 5 mM MgCl2, 100 mg/L, tRNA, and 1 g/L BSA, all from Sigma-Aldrich. The wash buffer was composed of DPBS with 5 mM MgCl2 and 4.5 g/L glucose (Sigma-Aldrich).
Preparation of Solutions and folding conditions
First, 10 µM solutions of R1.1, R1.2 and R1.3 were prepared by dilution of the respective stock solutions with the diluting buffer containing 550 µM KCl. Then, 1µM working solutions of variant R1.1 was prepared by diluting the 10µM solution with binding buffer. The 1µM working solutions of R1.2 and R1.3 were prepared by diluting the 10µM solution using binding buffer containing 0.2M KCl. Random controls were prepared in a manner similar to that of each R1 variant. The folding of random control and aptamer solutions was done by heating at 95°C for 10 minutes and maintaining on ice for 1 hour. The maximum time of one hour for folding was strictly followed because aptamer binding diminishes if the folded aptamer is kept on ice any longer.
Specificity
Specificity assays were conducted with individual aptamers against HPLC-purified random DNA (60mer) control purchased from IDT DNA Technologies. The specificity of aptamer sequences was evaluated by incubating truncated analogues separately with seven different cell lines accompanying the B-cell lines BJAB, Ramos, Daudi, SKLY-16 and CA46, while the negative cell lines were Jurkat.E6 and MOLT-3. These assays were performed by incubating 100 µL of either aptamer (500 nM) or random control with 100,000 cells in 100 µL of cell suspension buffer on ice for 45 minutes, followed by washing twice with 1.5 mL wash buffer each time. Cells were reconstituted in 250 µL wash buffer. Finally, binding was analyzed by flow cytometry by counting 10,000 events for each concentration. Expression of mIgM on all seven cell lines was also analyzed by incubating 100,000 cells in 100 µL volume using a final concentration of 0.5 µg/mL anti-IgM monoclonal antibody (Alexa Fluor 647, mouse anti-Human, Novus Biologicals), followed by flow cytometric analysis. Specificity assays at physiological temperature (37°C) were performed similar to 4°C, except 1.5 × 105 BJAB or Jurkat cells were used, and incubation was performed in a 37°C incubator in a final volume of 200 µL. Cell washing and flow cytometry analysis were performed at room temperature using reagents stored at room temperature.
Determination of binding affinities
At 4°C: Aptamer dilutions and folding were performed as described in the previous section. Binding affinities of R1 variants towards targets cells were determined by using either Ramos (1.0 × 105) or BJAB (0.75 × 105) cells. A range of fluorescently labeled aptamer concentrations was used, and the cells were incubated in either 200 µL or 150 µL of aptamer solutions in binding buffer for 45 minutes on ice. The wash buffer at 4°C (2 ml for Ramos cells and 3ml for BJAB cells) was added for one wash, and the cells were then reconstituted in 250 µL of wash buffer. Aptamer binding towards targets cells was analyzed with flow cytometry for each concentration by recording 5000 events. The calculation of Bmax/2 was done using the same method as described previously.
At 37°C: Binding affinities at physiological temperature (37°C) were determined using conditions similar to those at 4°C, except 1.5 × 105 BJAB cells were used, and incubation was performed in a 37°C incubator in a final volume of 200 µL. Also, cell washing and centrifugation were performed at room temperature using reagents stored at room temperature.
Soluble IgM preparation
Slide-A-Lyzer™ Dialysis Cassettes from Thermo Scientific (cat. no: 66373) were used for buffer exchange of human sIgM. Soluble human IgM was purchased from Sigma (cat. no: 18260) as a 0.8 mg/ml solution in a storage buffer consisting of 0.05 M Tris-HCl, 0.2M NaCl, pH 8.0, and 15mM sodium azide. Human soluble IgM lyophilized from the same storage buffer was purchased from Innovative Research (IR-HUM-GF-LY-20992). One mg of lyophilized sIgM was dissolved in 0.5ml sIgM solution (Sigma, 0.8 mg/ml) to formulate 2.8mg/mL of sIgM. Dialysis was performed at 4 °C overnight in 500 mL of DPBS buffer with constant stirring. The Quick Start Bradford Protein Assay Kit from BioRad (cat. no: 500-0201) was used to determine the concentration of sIgM after buffer exchange, using non-dialyzed soluble sIgM as a standard. We used the standard protocol in a 1 ml cuvette assay in which five concentrations of protein standard (sIgM, Sigma) and the dialyzed sample were incubated for 30 min with 1× dye reagent. Absorbance at 595 nM was recorded using a UV-VIS spectrophotometer (Evolution 300, Thermo Scientific). Standard curve was generated using the 3rd order polynomial trendline (Microsoft Excel), and the concentration of the unknown dialyzed sIgM was calculated using the 3rd order polynomial equation generated from the trendline.
Antigen specificity
Competition against anti-IgM antibody
To determine antigen specificity, a competition experiment between R1.1 and anti-IgM antibody was performed, as described by Zumrut et al. with the following modifications.12,13 A total of 5×105 Ramos cells were incubated in a 400 µL cell suspension buffer with 5µl of Alexa Fluor ® 647 isotype control (Biolegend) or 5ng/µl Alexa Fluor ® 647 anti-IgM antibody (Goat anti human µ-chain, Life Technologies) for 30 min on ice. The cells were then mixed thoroughly, and 50 µL of cells were incubated with 50 µL of 0.5 µM aptamer and random solutions for an additional 30 min on ice to allow competitive binding of the aptamers and antibody against the target. The cells were washed with 2 mL wash buffer and reconstituted in 200 µL of wash buffer for binding analysis using flow cytometry. Three replicates were performed, and percent binding of the aptamer in the presence of anti-IgM antibody was determined by comparing binding without antibody, which was defined as total binding.
Investigation of blocking of anti-IgM antibody binding
For antibody blocking experiments, 4×105 Ramos cells were first incubated with Cy3-labeled R1.2, or random control (1µM), on ice for 45 min. Then, the preincubated cells, with either aptamer or random control, were added into antibody (Alexa Fluor 647-conjugated Affinity Pure F(ab’)2 fragment goat anti-human IgM (Jackson ImmunoResearch)) dilutions ranging from 10 ng/µL to 0.01 ng/µL. After additional incubation for 35 min, the cells were washed once with 2 mL wash buffer and re-suspended in 250 µL wash buffer. The binding of antibody in the presence of R1.2 or random control was analyzed using flow cytometry by counting 5000 events.
Competition with soluble IgM
Antigen specificities of R1 analogues were further analyzed by cell binding assay in the presence of sIgM. BJAB cells (0.5 × 105) were incubated in 50 µL of final volume for 45min in the presence of human sIgM (48.5 µg per tube, at 1µM concentration), or the same amount of BSA as control, against three different FAM-dT-labeled R1.2 concentrations (500 nM, 250 nM and 125 nM). Cells were then washed once using 1 mL wash buffer, reconstituted in 250 µL wash buffer, and analyzed using flow cytometry by counting 5000 events. Also, binding of anti-IgM antibody (Alexa Fluor 647, Goat anti-human µ-chain, Life Technologies), with or without sIgM, was tested by incubating 1.5 × 105 BJAB cells with a final concentration of 2 µ g/ml antibody in the presence of 37 µ g sIgM or BSA as a control.
Determination of Binding Affinity against soluble-IgM
Binding affinity of R1.2 against sIgM was determined by nitrocellulose filter binding assays using 32P labeled R1.2 and random control against serially diluted concentrations of sIgM. One pmole of R1.2 aptamer or random were radiolabeled using T4 PNK (Thermo Scientific, #EK0031) according to manufacturer’s protocol. Labeled aptamer or random DNA was purified using illustra MicroSpin G-25 columns (GE Healthcare, #27-5325-01). The aptamer was then folded in 1xDPBS containing 5mM MgCl2 and 2mM KCl by heating for 10 min at 95 °C and snap-cooled on ice. A final concentration of 0.2 nM labeled R1.2 or random control was used in 50 µL DPBS containing 5mM MgCl2 buffer and incubated with soluble-IgM (1µM to 1nM) for 45 min. Bound R1.2 were separated from unbound aptamer by passing through nitrocellulose and nylon filters under vacuum followed by two subsequent washes with 200 µl DPBS containing 5mM MgCl2. Filters were exposed to phosphor screens overnight which were then imaged using a Storm Molecular Imager Phosphorimager (GE Healthcare, Piscataway, NJ). The image was analyzed by ImageQuant software, and non-specific interaction of aptamer with the filter membrane (no sIgM) was subtracted from aptamer signal at each sIgM concentration. The data was plotted using GraphPad Prism software to obtain the dissociation constant.
Results
Truncation and analysis of affinity
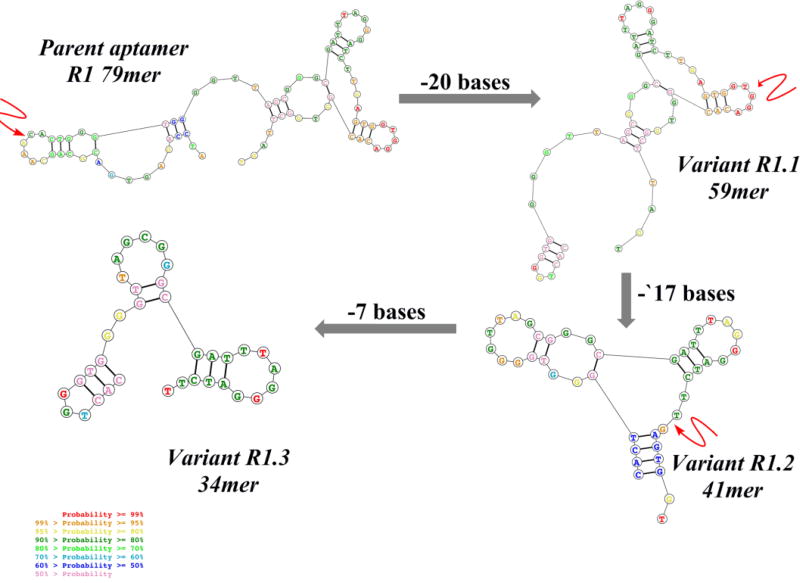
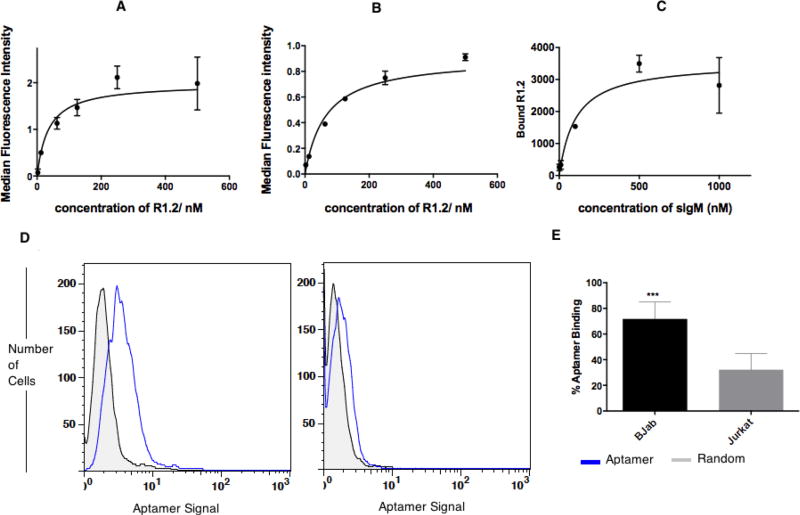
The reported original parent aptamer R1 is 79 bases in length with an affinity of 315 nM. By removing the nucleic acid segment corresponding to the forward primer of R1, termed R1.1 (59mer), a truncated variant was also introduced.12 Here, variant R1.1 was further evaluated for affinity (73.2± 13.7 nM) against BJAB, a cell line derived from Burkitt’s lymphoma, which is also known to express high levels of mIgM (affinity curve, Supplementary (S) Fig. S1a). Based on the observed improvement in affinity upon truncation of full-length R1 aptamer, the binding of R1.1 with BJAB cells was evaluated at physiological temperature of 37°C. Interestingly, an affinity of 186 ± 31.6 nM was observed for R1.1 (Fig.S1b) against mIgM-positive BJAB cells at 37°C, suggesting that the functional secondary structure of R1.1 had been retained and was stable at both 4°C and 37°C, despite the removal of 20 bases from the 5’-end. It is well known that aptamers tend to show highest affinity at the temperature utilized during the selection step and that change in temperature could potentially lower affinity towards its target. Therefore, the affinity of R1.1 at physiological conditions proved acceptable. However, we reasoned that affinity could be further improved by designing multivalent analogues. Therefore, we further truncated a second variant based on R1.1 by removing an additional 17 bases from the 3’-end to generate R1.2 (Scheme 1). Variant R1.2 showed a slightly improved affinity of 35.5±8.94 nM at 4°C compared to R1.1 (Fig.1). Also, at physiological temperature, variant R1.2 showed a trend similar to that of R1.1, with a slightly increased affinity of 65.6±5.88 nM, suggesting that R1.2 might be the most optimized version of parent R1 aptamer (Fig.1A–B). Binding analysis of R1.2 against control Jurkat cells showed slightly higher nonspecific binding or uptake, however, specific binding is still significantly higher suggesting that R1.2 had also retained its specificity at 37°C (Fig. 1D and E). We next evaluated the possibility of removing bases from the 3’-end without disrupting the region of the aptamer molecule which involves functional fold (Scheme 1), and, thus, a third variant R1.3 was developed by truncating 7 bases from the 3’-end of R1.2. However, aptamer variant R1.3 only showed an affinity of 134±23.8 nM (Fig. S1c) at 4°C, which was approximately four-fold less than that of R1.2. The reduced affinity of R1.3, suggested that additional truncation of R1.2 at the 3’-end resulted in destabilizing the functional fold of the aptamer, thereby lowering its affinity.
Scheme 1.
Truncation of parent aptamer R1 to generate variants with shorter lengths. Twenty bases were removed from parent aptamer R1 to design R1.1, followed by 17 bases from the 3’- end of R1.1 to design R1.2. Finally, 7 more bases were removed from the 3’-end of R1.2 to generate R1.3. Arrow indicates truncated position in each sequence. A complete list of sequences can be found in Scheme S1. The 2-dimensional structures were obtained from http://rna.urmc.rochester.edu/RNAstructureWeb/index.html.
Figure 1.
Analysis of R1.2 against mIgM-positive BJAB cells. Binding affinities of R1.2 were determined by using BJAB (0.75 × 105 for 4°C and 1.5 × 105 for 37°C) cells incubated with 2.5nM to 500nM of R1.2, or random control, solutions for 45 minutes on ice (A) or 37°C (B) incubator. After washing with 3ml wash buffer, cells were reconstituted in 250 µL of wash buffer, and aptamer binding was analyzed with flow cytometry for each concentration by recording 5000 events. (C) Binding affinity of R1.2 against sIgM using nitrocellulose filter binding assay-utilizing 32P labeled R1.2 against 1µM–1nM sIgM. Serially diluted concentrations of sIgM were incubated with 32P-labelled R1.2 or random DNA for 45-minutes on ice. The bound versus unbound molecules were separated on nitrocellulose filter device by washing twice by 200 µL of wash buffer. Bound R1.2 was quantified utilizing a phosphorimager. The calculation of Bmax/2 was done as described elsewhere.12,13 (D) R1.2 shows specific binding to BJAB cells at 37°C. (E) Bar diagrams represents overall conclusion from six independent binding assays. (*** : P ≤ 0.001, obtained using student’s T-test.)
Next, we analyzed whether variant R1.2 could recognize soluble IgM (sIgM). Two IgM forms are present in humans.15 Membrane form (mIgM), which is expressed on the cell membrane, and the soluble form, or sIgM, is produced by B-cells.15–17 Interestingly, both mIgM and sIgM have nearly identical amino acid composition, the only difference being that mIgM contains an additional 42 amino acids.14 This additional 42 amino acid sequence has been shown to span from the juxtaposition to the membrane at the C-terminal of the heavy µ chain to the transmembrane region.14 The anti-IgM antibody utilized to selectively elute aptamer R1 binds to both versions of mIgM.18 Therefore, the affinity of the most optimized variant R1.2 towards sIgM was evaluated. In doing so, variant R1.2 or the corresponding random DNA was labeled with 32P, and purified labeled molecules were incubated with range of concentrations of sIgM on ice. Unbound sequences subsequently separated using a nitrocellulose filter, and binding of R1.2 to sIgM was analyzed using a phosphorimager. Analysis of binding of labeled variant R1.2 showed an affinity of 102±42.4 nM towards sIgM (Fig. 1C, and control random Fig. S1E), suggesting that R1.2 does indeed recognize sIgM but with a slightly lower affinity than that towards mIgM.
Analysis of specificity against IgM-positive and IgM-negative cell lines
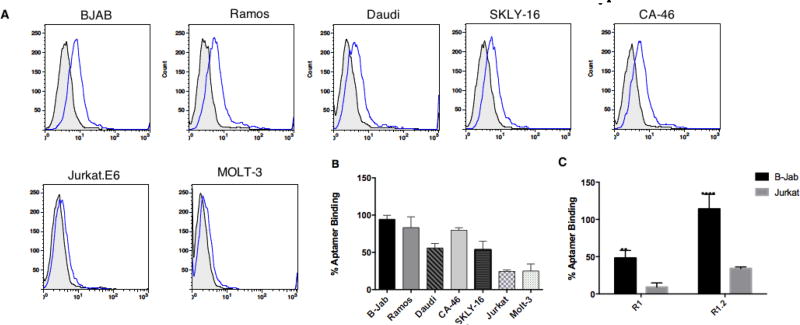
We evaluated the specificity of the truncated variants R1.1, R1.2 and R1.3 utilizing B-cell lines known to express mIgM. Two cell lines lacking mIgM were used as controls. Using 3’-FAM-dT-labeled aptamers and a corresponding random DNA sequence to address background sticking, the binding of each variant with cell lines was quantified using flow cytometry. We also performed a positive control using fluorescently labeled anti-IgM antibody against the cells to ensure that the cell lines used in this assay were indeed positive or negative for mIgM (Fig. S2a). Evaluation of whether the truncation of the full-length aptamer led to an increase in background binding revealed no significant increase in non-specific binding of variant R1.2 (Fig. 2C). While variant R1.2 show ~25% increment of background compared to full-length aptamer, the specific binding towards BJAB cells is also increased by ~66% suggesting that specificity of the aptamer is retained despite the truncation. In addition, all cell lines that tested positively bound to anti-IgM antibody were also positive for all variants of parent aptamer R1 (Fig. 2 for specificity of R1.2; Fig. S2b for specificity R1.1; and Fig. S2c for specificity of R1.3).
Figure 2. Binding analysis of Aptamer R1.2 against mIgM-positive B-cell lines and mIgM-negative T-cell lines.
(A) Histograms of R1.2 binding against B- and T- cell lines. The assays were performed by incubating either aptamer, or random control, on ice for 45 minutes, followed by washing twice with 1.5 mL wash buffer each time. Binding was analyzed by flow cytometry by counting 10,000 events for each concentration. (B) Overall conclusion from three independent R1.2 binding assays for each cell line. The % of aptamer binding was calculated as described elsewhere.13,19 (C) Comparison of R1 and R1.2 binding to targeting BJAB cells and non-targeting Jurkat cells.
Competition of variant R1.2 with soluble IgM
Soluble IgM is present in high concentrations in human plasma, and concentrations of sIgM can be altered by conditions such as bacterial or viral infection. That is, such infections can lead to immune response by B-cells, triggering secretion of sIgM. Since both mIgM and sIgM are essentially identical, anti-IgM antibody is also known to interact with both forms of IgMs, a characteristic we analyzed using flow cytometry. To evaluate the binding of anti-IgM to both sIgM and mIgM, we added an excess sIgM to the binding buffer in place of BSA, and BJAB cells were suspended in this modified buffer prior to incubating with anti-IgM antibody. Subsequently, the binding, or lack thereof, of fluorescently labeled anti-IgM antibody was investigated against BJAB cells (Fig.S3). As expected, the anti-IgM antibody showed diminished binding towards BJAB cells when sIgM was present in the binding buffer compared to the control, suggesting that anti-IgM binds to both the soluble and membrane IgM (Fig.S3).
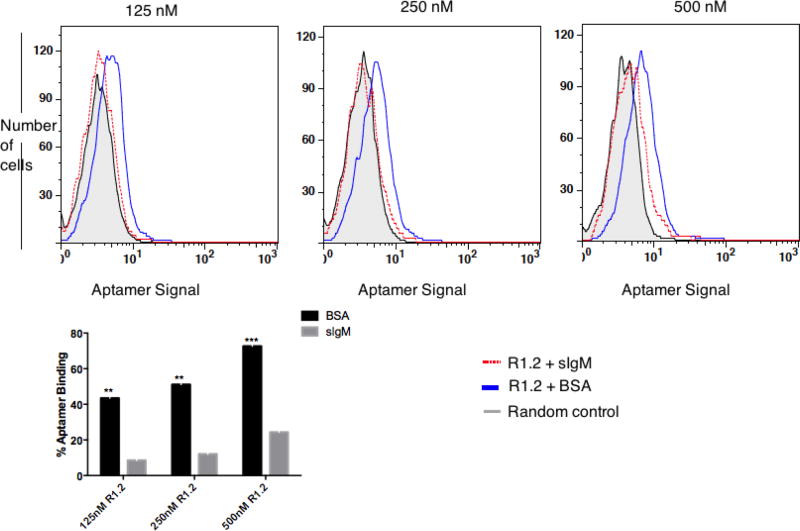
Using a similar approach, we next evaluated whether aptamer R1.2 could sustain its specific recognition of mIgM in the presence excess sIgM. To perform this assay, the binding of high-affinity variant R1.2 in three different concentrations was tested against mIgM-positive BJAB cells in the presence excess sIgM (1 µM) or BSA (Fig. 3). If aptamer R1.2 were found to bind both soluble and membrane IgM, we would then observe diminished fluorescence intensity on BJAB cells compared to cells without sIgM. We observed a ~80 % decrease in the binding to BJAB cells at 125nM R1.2, ~76% decrease in the binding of R1.2 at 250nM and, finally, a 66% decrease in the binding of R1.2 at 500nM when sIgM was present in the binding buffer, suggesting that variant R1.2 does indeed bind to both IgM forms and the difference in affinity towards mIgM and sIgM seems to play a minimal role when an excess sIgM is present, challenging potential applications of R1.2 as a therapeutic delivery agent in vivo (Fig. 3). Furthermore, the diminished fluorescence intensity (compare blue and red histograms in Fig. 3) indicates that aptamer R1.2 is distributed between both mIgM and sIgM, leading to lower fluorescence signal on the cells, suggesting that the epitope of R1.2 is exclusive to both sIgM and mIgM and no co-receptor molecules on the cell membrane stabilize the binding of R1.2 leading to higher affinity.
Figure 3. Binding analysis of R1.2 with BJAB cells in the presence of 1µM sIgM.
Antigen specificity of R1.2 was further analyzed by cell binding assay in the presence of sIgM. BJAB cells (0.5 × 105) were incubated for 45min in the presence of human soluble IgM (48.5 µg per tube), or the same amount of BSA as a control, against three different FAM-dT-labeled R1.2 concentrations (500nM, 250nM and 125 nM). Cells were then washed once using 1 ml of wash buffer, followed by analysis of R1.2 binding by flow cytometry. Bar diagram is the overall conclusion from two independent experiments. Asterisks represent adjusted p values from Sidak’s multiple comparisons test based on comparing BSA vs. sIgM data for each concentration. (**: P ≤ 0.01; *** : P ≤ 0.001)
Competition of variant R1.1 with anti-IgM antibody
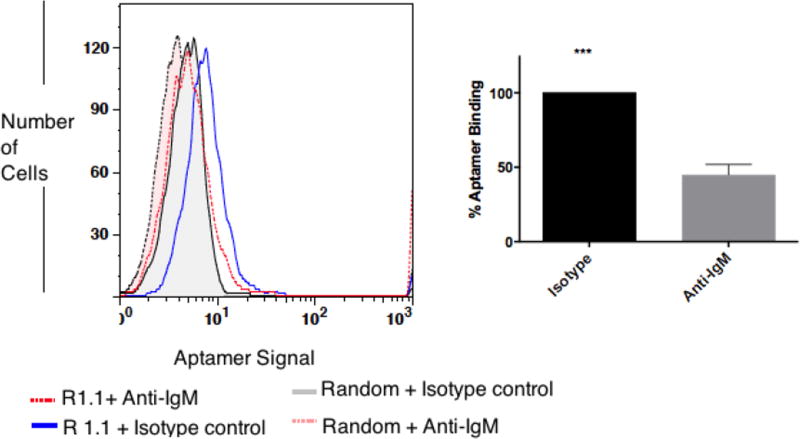
We previously showed that parent aptamer R1 competes with anti-IgM antibody.12 It was hypothesized that the affinity of R1 would be substantially lower than that of anti-IgM antibody. Therefore, in the case of an aptamer (R1) competing with the same antigen or antigen near an antibody binding site, the addition of a high concentration of anti-IgM antibody should decrease the binding of aptamer R1. We investigated whether the reasonably improved variant R1.1 would also compete with the anti-IgM antibody. To make this determination, we performed an experiment similar to that of previously reported by utilizing an isotope control antibody. Results showed about 55±7.1 % decrease in aptamer binding in the presence of high concentration of anti-IgM antibody (5ng/µl) (Fig. 4). Interestingly, we did not observe such competitive displacement of variants R1.2 and R1.3, which might be explained by the shorter aptamer sequence having a more favorable fit with the binding epitope.
Figure 4. Anti-IgM antibody outcompetes R1.1.
In the presence of isotype control, R1.1 binds to BJAB cells, but not when anti-IgM is present. Bar diagram on the right is overall conclusion from three independent competition experiments. % of binding values in the presence of anti-IgM antibody compared to isotype control was calculated as ), where specific binding = Aptamer Mean Fluorescence Intensity (MFI) − Random MFI. (***: P ≤ 0.001, obtained by two-tailed t-test)
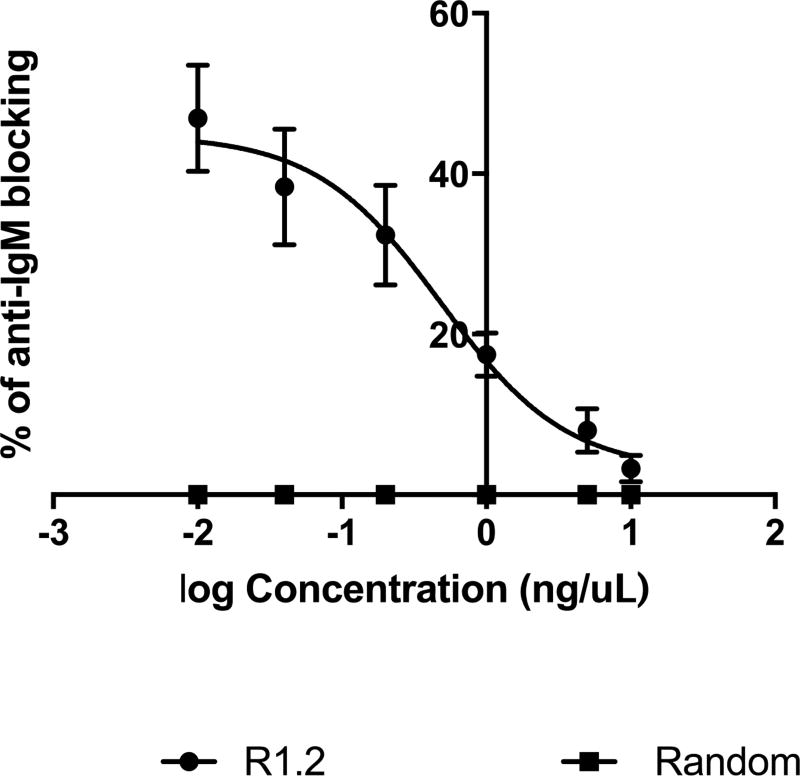
We reported that R1.1 blocks anti-IgM binding, but only when lower concentrations of anti-IgM were used. Therefore, a reverse competition experiment (blocking) utilizing low concentration of anti-IgM was conducted to evaluate whether variant R1.2 could also block anti-IgM antibody binding to BJAB cells. Utilizing six different concentrations of F(ab’)2 fragment goat anti-human IgM, ranging from 10 ng /µL to 0.01 ng/µL, the binding of anti-IgM was evaluated against BJAB cells, which were pre-incubated with 1µM R1.2, or random control. Results showed that aptamer R1.2 did block the binding of the antibody (Fig. 5). At low concentrations of F(ab’)2 (<1ng/µL of anti-IgM), variant R1.2 showed its highest blocking of anti-IgM of binding by about 46.94 ± 6.62%. We observed no blocking (0% of blocking) by randomized control. The percentage of blocking decreased as a function of increasing antibody concentration; suggesting that when antibody concentration is below its Kd towards mIgM, binding of aptamer R1.2 is prominent and can block anti-IgM binding to BJAB cells, whereas at high anti-IgM concentrations, antibody binding predominates (Fig. 5).
Figure 5. Binding of antibody with BJAB cells preincubated with R1.2 or random control.
In the presence of random control, antibody bound to the cells by 100%; hence, anti-IgM blocking is zero. In contrast, the presence of aptamer inhibited anti-IgM binding to BJAB cells when lower concentration of anti-IgM was used. For each antibody concentration, % blocking of antibody binding by the aptamer compared to random DNA was calculated as . Results from three independent experiments were plotted against log antibody concentration. (MFI: Median Fluorescence Intensity)
Discussion
Nucleic acid aptamers are versatile synthetic ligands with high potential for the development of molecular tools for a variety of biomedical applications.20–23 Aptamers have been selected by traditional SELEX since 1990 by coupling combinatorial library screening with in vitro evolution.5,6 Among many variants of SELEX, our group recently introduced a derivative of “complex target SELEX” which we termed Ligand-guided Selection, or LIGS.12,13 The core principle of LIGS is rooted in introducing higher affinity secondary competing ligands to either induce a conformational switch of the receptor to destabilize the aptamer-receptor complex or exploit the fundamental differences in concentrations of individual molecules in a combinatorial library as an avenue to selectively elute highly specific aptamer molecules from a partially evolved SELEX library. Using this method, we have selected highly specific aptamers against mIgM utilizing anti-IgM antibody interacting with mIgM as the secondary ligand. Membrane IgM is the major subunit of the BCR complex uniquely expressed on B-cells and B-cell NHL.18 However, since these selected aptamers generally showed lower affinities, this study aimed to optimize the structure of one of the LIGS-generated aptamers to determine if such optimization would lead to aptamers against mIgM with higher affinity, but without compromising specificity. To do this, we first truncated aptamer R1 from both 3’ and 5’ ends, removing primer regions, which were shown to play a limited role in stabilizing the structure. After systematic truncation of aptamer R1, the affinity and specificity of its variant and those that followed were evaluated against cells expressing target mIgM. Typically, DNA and RNA aptamer lengths are in the range of 75–90 bases, with fixed primer regions contributing to 50% of the length. The randomized region of an aptamer is shown to be contributing to the functional fold of the aptamers. Therefore, subsequent systematic structure-activity-relationship studies are essential to optimize the length of the aptamer. The first truncation of parent aptamer R1 was done by removing 20 bases contributed from the forward primer yielding variant R1.1. The subsequent affinity analysis suggested that the forward primer did not contribute to stability of the functional fold. We then truncated 17 bases from the reverse primer region, and subsequent affinity analysis suggested that removed bases did not affect the binding motif of the aptamer. Finally, further reduction of 7 bases from 3’ end, led decrease in affinity suggesting the double-stranded segment in variant R1.2 (scheme 1) may be stabilizing the binding motif of the aptamer. Truncation of aptamers to enhance their affinity has been widely applied.24,25 For example, an aptamer discovered against PTK7 utilizing live cell-SELEX was truncated and then modified to yield high affinity second-generation aptamers.26 Also, an aptamer against transferrin receptor was subjected to minimization to yield better variants. The “minimized” variant resulted in an aptamer with higher affinity, indicating that the untruncated version might have contained a low concentration of functional fold, leading to its lower overall affinity.27 In the present paper, the binding of variants R1.1 and R1.2 to mIgM-positive BJAB cells at physiological temperatures indicates that the functional fold of the aptamer had been retained, despite the truncations employed or the change in temperature. The two-fold decrease in affinity compared to 4°C could have resulted from the change of the aptamer or the change of lateral angle of mIgM as a function of increasing temperature.18 Nevertheless, the affinity of R1.2 could be further improved by designing bivalent or multivalent analogues as shown before.18
All variants were analyzed against five B-cell lymphoma cell lines and two non-B-cell lines. The B-cell lines appeared to show different expression levels of mIgM based on anti-IgM antibody staining, and all variants showed a similar trend, indicating that both aptamers and antibody had identical binding patterns. With specificity and affinity having been confirmed, epitope specificity analysis revealed that truncated versions of R1 also retained their epitope specificity. Binding analysis of R1.1 in the presence anti-IgM antibody showed that aptamer binding was diminished by 50%, suggesting that 1) R1.1 was displaced by antibody, 2) antibody destabilized R1.1, or 3) aptamer variant R1.1 could not bind its target as a result of steric constraints of the antibody. At the same time, however, we did not observe competitive displacement of variant R1.2 or R1.3, indicating that the compact nature of the shorter variants can recognize their epitope and that anti-IgM cannot displace these shorter variants. Interestingly, when variant R1.2 was used at higher concentrations (1µM) and pre-incubated with BJAB cells prior to adding anti-IgM, R1.2 blocked antibody binding. This blocking effect is prominent at lower antibody concentrations, suggesting that anti-IgM can no longer recognize its epitope when R1.2 is bound to the cells. However, we did not observe this effect with higher concentrations of anti-IgM antibody. We selected the parent aptamer from a partially evolved pool using the same antibody. Therefore, the difference in concentration might have played a role in the binding kinetics. The bivalent nature of an antibody favors high affinity as a consequence of lower entropic penalty caused by the binding than that of the monovalent version. However, when the aptamer is at high concentration and the antibody is at concentrations lower than its affinity constant, aptamer binding appears to predominate over that of the antibody.
The binding evaluation of variant R1.2 against sIgM revealed that the affinity of R1.2 towards sIgM is approximately 3-fold lower than the affinity towards mIgM. It is well known that aptamers are highly specific towards the fold of the protein that was used in the SELEX method. While, this aptamer could bind to an epitope common to both soluble and membrane IgM, there might be slight structural variants between mIgM and sIgM leading slightly different affinities towards sIgM and mIgM. Of the two forms of IgM, sIgM is secreted by B-cells during differentiation. It has been shown that both sIgM and mIgM contain the same mRNA coding up to the fourth-constant region Cμ4.14 Since the anti-IgM antibody specific for both mIgM and sIgM was used in selective elution of the aptamer during selection, it is possible that the aptamer binds to a region of mIgM distal to its Cμ4 region. Membrane IgM is a key molecule of the BCR complex uniquely expressed in B-cells.16 Because of this unique expression, anti-IgM antibody can be used to detect B-cells. Soluble IgM is present in normal human serum with an approximate range of concentration of 45–150 mg/dL, while mIgM is exclusively expressed in B-cells and B-cell lymphoma and leukemia.28,29 During the early stage of infection, the concentration of sIgM is elevated owing to its main role in primary immune response.30 Also, immune deficiency disorders, such as meningitis, pneumonia and gram-negative sepsis, can lead to the suppression of sIgM antibody production.15 Therefore, the use of molecular probes based on variants of R1.2 could be attractive for the detection of both sIgM and mIgM ex vivo.
In conclusion, we have systematically truncated parent aptamer R1, as identified through the novel Ligand-Guided-Selection, to enhance affinity. Specificity analysis using B-cell lines demonstrated that the specificity of the truncated version was not compromised. Furthermore, binding analysis using sIgM showed that aptamer variant R1.2 bound to both soluble and membrane IgM, indicating that the specificity of the ligand utilized in selective elution of the aptamer determines the specificity of the aptamer. To enhance affinity in future studies, dimeric aptamers will be designed, and ligand-induced receptor internalization will be evaluated. Also, aptamer variant R1.2 will be further evaluated for its utility as a diagnostic agent to measure infectious levels of virus/bacteria or measure levels of sIgM and as a potential diagnostic tool for B-cell lymphoma.
Supplementary Material
Acknowledgments
Authors gratefully acknowledge funding from NIGMS grant 5SC3GM105578. Authors are indebted for Dr. Matthew Levy and the members of his laboratory, at the Department of Biochemistry, Albert Einstein College of Medicine, 1301 Morris Park Ave, Bronx, New York, 10461 for their assistance with radioactive labeling and nitrocellulose binding assays.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Current opinion in chemical biology. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Eaton BE, Gold L, Zichi DA. Let's get specific: the relationship between specificity and affinity. Chemistry & biology. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 3.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinical chemistry. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 4.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nature reviews. Drug discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Accounts of chemical research. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 8.Mallikaratchy P. Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens. Molecules. 2017;22 doi: 10.3390/molecules22020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clinical chemistry. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 11.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zumrut HE, Ara MN, Fraile M, Maio G, Mallikaratchy P. Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins. Nucleic acid therapeutics. 2016 doi: 10.1089/nat.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zumrut HE, Ara MN, Maio GE, Van NA, Batool S, Mallikaratchy PR. Ligand-guided selection of aptamers against T-cell Receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells. Analytical biochemistry. 2016;512:1–7. doi: 10.1016/j.ab.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980;20:303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- 15.Ehrenstein MR, Notley CA. The importance of natural IgM: scavenger, protector and regulator. Nature reviews. Immunology. 2010;10:778–786. doi: 10.1038/nri2849. [DOI] [PubMed] [Google Scholar]
- 16.Fuentes-Panana EM, Bannish G, Monroe JG. Basal B-cell receptor signaling in B lymphocytes: mechanisms of regulation and role in positive selection, differentiation, and peripheral survival. Immunological reviews. 2004;197:26–40. doi: 10.1111/j.0105-2896.2004.0105.x. [DOI] [PubMed] [Google Scholar]
- 17.Geisberger R, Lamers M, Achatz G. The riddle of the dual expression of IgM and IgD. Immunology. 2006;118:429–437. doi: 10.1111/j.1365-2567.2006.02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic acids research. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumrut HE, Ara MN, Fraile M, Maio G, Mallikaratchy P. Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins. Nucleic acid therapeutics. 2016;26:190–198. doi: 10.1089/nat.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. Journal of the American Chemical Society. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 21.Sullenger B, Woodruff R, Monroe DM. Potent Anticoagulant Aptamer Directed against Factor IXa Blocks Macromolecular Substrate Interaction. J Biol Chem. 2012;287:12779–12786. doi: 10.1074/jbc.M111.300772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khedri M, Rafatpanah H, Abnous K, Ramezani P, Ramezani M. Cancer immunotherapy via nucleic acid aptamers. International immunopharmacology. 2015;29:926–936. doi: 10.1016/j.intimp.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Famulok M, Mayer G. Aptamer modules as sensors and detectors. Accounts of chemical research. 2011;44:1349–1358. doi: 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- 24.Eaton BE, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi DA. Post-SELEX combinatorial optimization of aptamers. Bioorganic & medicinal chemistry. 1997;5:1087–1096. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 25.Maio GE, Enweronye O, Zumrut HE, Batool S, Van NA, Mallikaratchy PR. Systematic Optimization and Modification of a DNA Aptamer with 2’-O-Methyl RNA Analogues. Chemistry Select. 2017;2:2335–2340. doi: 10.1002/slct.201700359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem : a European journal of chemical biology. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 27.Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furst DE. Serum immunoglobulins and risk of infection: how low can you go? Semin Arthritis Rheum. 2009;39:18–29. doi: 10.1016/j.semarthrit.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Molecular immunology. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Manson JJ, Mauri C, Ehrenstein MR. Natural serum IgM maintains immunological homeostasis and prevents autoimmunity. Springer seminars in immunopathology. 2005;26:425–432. doi: 10.1007/s00281-004-0187-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.