Abstract
We recently introduced a screening technology termed ligand-guided selection, (LIGS), to selectively identify target-specific aptamers from an evolved cell-SELEX library. Cell-SELEX utilizes a large combinatorial single-stranded oligonucleotide library and progressively selects DNA ligands against whole cells with variable DNA-binding affinities and specificities by repeated rounds of partition and amplification. LIGS exploits the partition step and introduces a secondary, pre-existing high-affinity monoclonal antibody (mAb) ligand to outcompete and elute specific aptamers towards the binding target of the antibody, not the cell. Here, using anti-CD3ε mAb against the cluster of differentiation 3 (CD3ε), as the guiding ligand against one of the domains of the T-Cell Receptor (TCR) complex expressed on Jurkat.E6 cells, we discovered three specific aptamers against TCR complex expressed on an immortalized line of human T lymphocyte cells. In sum, we demonstrate that specific aptamers can be identified utilizing an antibody against a single domain of a multidomain protein complex in their endogenous state with neither post- nor pre-SELEX protein manipulation.
Keywords: Aptamer, evolution, monoclonal antibody, Ligand, CD3ε, T-cell receptor
Introduction
DNA aptamers are small synthetic nucleic acid strands that specifically bind to a target molecule with high affinity1,2. The method of aptamer selection known as SELEX (Systematic Evolution of Ligands by Exponential enrichment) was originally introduced by two independent groups1,2. SELEX screens short single-stranded oligonucleotides against a variety of target ligands via an iterative and evolutionary process of continuous enrichment to identify target-specific binders. A typical SELEX library is vastly heterogeneous with a large number of distinct nucleic acid molecules (~approximately 1013 molecules). Each molecule folds into a unique secondary structure, which leads to a distinct geometrical shape. Depending on shape complementarity and noncovalent electrostatic or hydrophobic interactions, a few nucleic acid sequences can specifically bind to the desired target. Subsequently, bound sequences are separated and amplified using Polymerase Chain Reaction (PCR) to generate an evolved library. The process is repeated until high-affinity binders are enriched, resulting in a homogeneous library with high-affinity nucleic acid aptamers against the target of interest.
SELEX has resulted in generating a significant number of aptamers against targets ranging from small molecules to whole cells; however, translational applications of aptamers have been limited3. Therefore, steps to improve SELEX have been introduced, for example, cell-SELEX, which was introduced as a method to select aptamers against membrane receptors in their endogenous state4. In addition, a bead-based selection method has been introduced to increase selection diversity aimed at generating therapeutic aptamers5. To expand target specificity, “internalizing cell-SELEX technology” and hybrid-SELEX have also been introduced. Hybrid-SELEX incorporates the enrichment of the SELEX library against the purified protein target first, followed by cell-SELEX, utilizing cells that express the same protein, while cell-internalization-SELEX is designed to select aptamers towards RNA molecules capable of internalizing into cells6,7. To increase the clinical practicality of aptamer selection, development of methods to identify aptamers able to specifically recognize predetermined epitopes in their endogenous state with no prior- or post SELEX sample manipulations on receptor proteins would be most desirable.
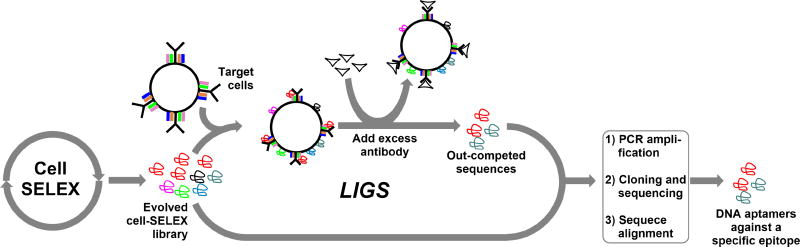
To address this, we recently developed a method called Ligand-guided Selection, (LIGS), which selects aptamers that specifically bind a predetermined epitope expressed on the target cell surface8. LIGS takes advantage of the partition step in cell-based SELEX and introduces a secondary, pre-existing high-affinity ligand, in effect a monoclonal antibody (mAb), to outcompete and elute specific aptamers binding to the receptor target of the antibody, not the cell. Conventional SELEX is designed to winnow out low-affinity binders through a competitive process whereby high-affinity binders move on by repeated rounds of partition and amplification through the selection process. We hypothesized that the addition of a secondary stronger specific ligand in excess against a specific epitope of interest will selectively outcompete specific aptamers competing to bind to the same epitope or a related epitope from an evolved pool pre-incubated with the whole cell.. Therefore, at a partial enrichment stage in the cell-SELEX iterative process, LIGS interrupts the process and exploits this competitive selection by introducing a stronger, known high-affinity ligand against a specific protein receptor (epitope) target of interest in order to 1) directly outcompete and replace aptamers specific towards the target of interest and, more importantly in terms of LIGS, 2) elute aptamers resulting from conformational changes induced through the interaction of the secondary ligand with its target epitope at its endogenous state. Therefore, based on the specificity of a natural pre-existing ligand towards its target and the conformational changes induced through antibody-protein receptor binding, the aptamers identified by LIGS are expected to show higher specificity towards the target protein or a protein co-expressed with the target protein compared to those aptamers evolved as binders through cell-SELEX (Scheme 1).
Scheme 1. LIGS.
Conventional cell-SELEX is first employed against target cells until a partial enrichment of DNA aptamer library is achieved. Next, the partially enriched library is divided into fractions. The first fraction is PCR-amplified, cloned and sequenced. These sequences are enriched towards target cells. An excess of mAb is then introduced on the second fraction, which is preincubated with target cells to selectively outcompete and elute potential aptamers that would tend to bind to the cognate epitope less strongly compared to mAb. The sequences outcompeted by antibody are next PCR-amplified, cloned, and sequenced. By virtue of antibody-cognate epitope binding, these LIGS-generated sequences are specific towards the target surface protein of the antibody. Finally, sequences obtained from DNA sequencing of both fractions are aligned using the ClustalX.2 program, and based on set criteria, specific aptamer candidates against respective epitopes on the target cells are identified.
Here, we utilized LIGS to identify aptamers against CD3ε expressed on Jurkat.E6 cells from a partially enriched SELEX library. Using high-affinity anti-CD3 antibody against a specific epitope on the CD3ε chain as the secondary ligand, we successfully identified three specific aptamers against CD3ε, one of the domains of the T-cell Receptor (TCR) complex expressed on T lymphocytes. CD3ε is one of the ectodomains of the TCR complex expressed on T-cells. The TCR complex is a multidomain, transmembrane protein consisting of a αβ heterodimer and both CD3εγ and CD3εδ ectodomains. The main αβ heterodimer consists of a variable and constant domain, while the CD3ε domain is conserved and non-glycosylated, making CD3ε an attractive target for aptamer development9.
Methods and Materials
Cell lines, Jurkat.E6 (T lymphocyte) and Ramos (Burkitt’s lymphoma), were generously provided by David Scheinberg and Morgan Huse, Memorial Sloan Kettering Cancer Center. All cells were cultured in RPMI 1640 medium supplemented with 100 units/mL penicillin–streptomycin and 10% fetal bovine serum (heat-inactivated; Invitrogen). Cell lines were validated by flow cytometric assays utilizing antibodies against surface markers uniquely expressed on each cell line.
Buffer compositions
Washing buffer was composed of 1×DPBS containing 4.5g Glucose/1 L and 5 ml of 1M MgCl2/1L. DNA Binding Buffer (DB) was composed of 1×DPBS containing 4.5g Glucose/1 L, 5 ml of 1M MgCl2/1L, and 100mg/1L tRNA. Cell Suspension Buffer (CSB) was composed of 1×DPBS containing 4.5g Glucose/1 L, 5 ml of 1M MgCl2/1L, 100mg/1L tRNA, and 2g/1L BSA.
Phosphoramidites
All of the DNA reagents needed for DNA synthesis were purchased from Glen Research or ChemGenes. The DNA oligo sequences were chemically synthesized with a FAM-dT at the 3’-end using standard solid phase phosphoramidite chemistry on an ABI394 DNA (Biolytics) synthesizer using a 0.2 µmole scale. Aptamer candidates were synthesized in house using a solid phase DNA synthesizer according to the manufacturer’s protocol (Applied Biosystems, Inc. Model 394). The completed DNA sequences were deprotected using conditions required for modifications and purified using HPLC (Waters) equipped with a C-18 reversed phase column (Phenomenex). DNA concentration was determined by a UV-VIS spectrophotometer (Thermo Scientific; Evolution 300) and stored in DNA Binding Buffer (DB) at −20°C.
Cell-SELEX procedure
We routinely conducted PI staining of the cells and flow cytometric analysis of CD3ε expression utilizing PE-labeled anti-CD3ε antibody (BD Pharmingen mouse anti-human) along with an isotype control (mouse IgG1 BioLegend) to ensure high-quality cells expressing CD3ε prior to performing each round of SELEX.
The ss-SELEX DNA library in DB buffer was heated at 95°C for 5 minutes and “snapcooled” in ice for 30min prior to selection. Cells were washed three times with the wash buffer to remove cell debris and apoptotic cells and subsequently re-suspended in 100 µL of a cell suspension buffer prior to incubation with 100 µL of an ss-DNA library for 40 minutes on ice. The first round of selection was done with 10 × 106 cells and 100 nmol of ss-DNA SELEX library.
Cells that bound to the library were washed with wash buffer (12 mL) to remove weak or nonspecifically bound DNA strands in the first round. The bound DNA library was eluted by heating at 95 °C for 10 minutes in 200 µL DNAse-/RNAse-free water. A two-step polymerase chain reaction (PCR) was employed to expand the evolved library as reported elsewhere.10 A single-stranded DNA was made using avidin agarose beads (Pierce) and desalted using NAP-10 columns (GE) as described by Sefah et al.11 For subsequent SELEX, we followed Zumrut et al8.
Ligand-guided-Selection Protocol
Ligand Competition: As introduced in Zumrut et al., LIGS was used to selectively elute aptamers against TCR complex8. Briefly, the enriched 16th library of ss-DNA cell-SELEX was folded by heating and subsequent cooling for 20 minutes. Then, 10 × 105 cells were incubated with 250 nM 16th cell-SELEX-round ss-DNA of 25 µL for 40 minutes in ice and washed twice with1 mL and 0.5 mL wash buffer. Pretreated Jurkat.E6 cells with the 16th SELEX-pool were suspended in 50 µL of binding buffer and then incubated with (2.5 µL) of APC mouse anti-human CD3 antibody (BD Pharmingen; cat. no. 555342) 40 min on ice to compete and elute the potential aptamer candidates. Following incubation, the eluted 16th fraction obtained through competition and found in the supernatant was collected and amplified by PCR. A two-step PCR was performed as described in Zumrut et al8. First, the whole fraction resulting from LIGS was amplified using 10-PCR cycles. Then, a second PCR was employed, and the number of cycles was optimized to obtain adequate yields necessary for the cloning step. To ensure the presence of CD3ε expressed on Jurkat.E6 cells, 10 ×105 cells were incubated in parallel with an APC mouse antihuman CD3 antibody (BD Pharmingen; cat. no. 555342) or isotype control (APC mouse IgG1-k, BioLegend; cat. no. 400121). 1 µl antibody/ isotype was added per 1×105 cells and incubated at 4 °C for 30 min in cell suspension buffer. After incubation, all samples were washed and then analyzed by FACSCalibur™ (Cytek) by counting 10000 events. Two different SELEX libraries were generated: 1) the DNA pool from the SELEX-16th round specifically enriched against Jurkat.E6 cells and 2) the competitively eluted fraction of the SELEX-16th round using ligand competition. These were both cloned into a bacterial cloning system using a TOPO TA cloning kit (Invitrogen), and positive colonies were subsequently sequenced by the DNA sequencing core facility at Albert Einstein College of Medicine.
Specificity assays
Assays conducted with individual aptamers were analyzed against a HPLC-purified FAM-dT-labeled random DNA (60mer) control with the sequence nnnnnnnnnnnnnnnnnn nnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnt purchased from IDT DNA technologies.
The bindings of the aptamer sequences were evaluated by incubating Jurkat.E6 cells or Ramos cells (75 × 103) with a series of concentrations of FAM-dT-labeled aptamer in 100 µL of binding buffer on ice for 45 minutes. The cells were then washed with 1.5 mL of wash buffer at 4°C and reconstituted in 300 µL of wash buffer. The binding of the constructs was analyzed using flow cytometry by counting 5000 events for each concentration.
Determination of the apparent dissociation constant of aptamers
Using binding buffer, six different working concentrations of the aptamer and control library were prepared: 1) 1000 nM, 2) 500 nM, 3) 250nM, 4) 125 nM, 5) 20.8 nM, and 6) 3.46 nM. Cells were prepared for flow cytometry analysis by washing three times with wash buffer. 75 × 103 cells were incubated with each aptamer concentration and random library for 40 min on ice. After washing cells with 1.5 ml of wash buffer, the cells were analyzed with the FACSCalibur™ flow cytometer by counting 5000 events. FlowJo software was used to determine median fluorescence intensity for each concentration of aptamer sample and random control. Median fluorescence intensity of random control was subtracted from corresponding median fluorescence intensity of each aptamer concentration. This assay was done in triplicate. Bmax/2 was calculated using the method described in Sefah et al.11
Competition assay with individual aptamer molecules
Fluorescently labeled 1 µM aptamer (50 µL) was incubated on ice with 75 × 103 Jurkat.E6 cells for 45 minutes. Then anti-CD3ε HIT3a clone or anti-TCRαβ was added and incubated for an additional 45 minutes. At the end of incubation, cells were washed with 1.5 mL of wash buffer and reconstituted in 300 µL of wash buffer. Binding of aptamer and antibody was analyzed by flow cytometry.
Results and Discussion
Cell-SELEX against Jurkat.E6 cells followed by LIGS
To begin, the expression of CD3ε on Jurkat.E6 cells was confirmed by flow cytometry using a fluorescently labeled anti-CD3ε antibody (Supporting Information; Figure S1). The selection library consisted of 45 randomized nucleotides flanked by two primers, as reported in Tang et al.,10 but PCR conditions were further optimized to ensure high PCR efficiency of the library. The first round of cell-SELEX employed approximately 7 million Jurkat.E6 cells to ensure that all potential binders were retained. A total of 5 million cells were then used during the second round of selection, but the number of cells used in cell-SELEX was decreased to 2.5 million in subsequent rounds to increase the stringency of the selection. Again, we followed the cell-SELEX protocol described in Tang et al.,10 who showed that aptamers could be enriched towards a single cell type without incorporating negative selection. Since LIGS is designed to selectively elute specific aptamers using a secondary pre-existing ligand, anti-CD3ε mAb, in this example, we hypothesized that off-target sequences would not hinder the selective elution of specific aptamers. We monitored the enrichment of the library first at the 5th round of cell-SELEX, continuing with five more rounds to enrich potential aptamer sequences. After round 10 of cell-SELEX, we monitored the progress of the selection every 2 rounds (Supporting Information: Figure S2). We observed a significant enrichment against Jurkat.E6 cells at round 16 compared to earlier rounds based on FACS analysis (Figure 1a). At this point, cell-SELEX was stopped, and LIGS was introduced to a fraction of round 16 from cell-SELEX. We interrupted cell-SELEX at the very early stage of enrichment, as indicated by low fluorescence shift for pool 16 (Figure 1a). In such partially enriched library, it was hypothesized that concentrations of individual sequences would be very low, below their Kd, and thus well suited for competitive elution.
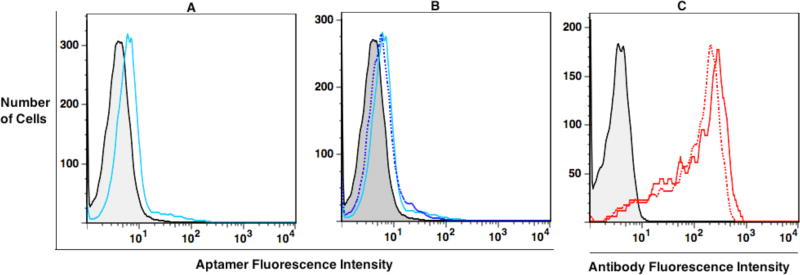
Figure 1.
Flow cytometry of evolved library and LIGS. Fluorescence intensity on X-axis indicates the binding of fluorescently labeled evolved round 16 pool, or anti-CD3ε HIT3a clone. (A) Analysis of binding of cell-SELEX round 16 (blue line) against Jurkat.E6 cells. The background binding was analyzed utilizing a random DNA pool from the 0th round (gray). (B) Cell-SELEX round 16 incubated with Jurkat.E6 cells and addition of anti-CD3ε HIT3a clone. Supernatant was collected after introduction of LIGS, and cells were analyzed for binding for round 16 of cell-SELEX after adding antibody. Fluorescence intensity corresponding to round 16-bound Jurkat.E6 cells was decreased (dashed blue line) in comparison to round 16 without antibody, suggesting that some sequences had, indeed, been outcompeted by antibody-receptor binding (C) Analysis of anti-CD3ε HIT3a clone binding to Jurkat.E6 cells (solid red line absence, and the dashed red line presence of cell-SELEX round 16.)
Accordingly, for the first step of LIGS, a total of 1× 105 Jurkat.E6 cells were prewashed with wash buffer and incubated 40 minutes with 6.25 pmols of round 16 of cell-SELEX. After incubation, cells were washed twice, first with 1 mL wash buffer and then 0.5 mL wash buffer, to remove unbound DNA molecules. Next, cells were reconstituted in 50 µL cell binding buffer, and 2.5 µL of anti-CD3ε HIT3 clone were added. Competitive elution of CD3ε–specific aptamers by the antibody was allowed for 40 minutes on ice. Following incubation, cells were spun down, and supernatant containing competitively eluted aptamers was collected. Cells were analyzed after LIGS to confirm the interaction of anti-CD3ε HIT3a clone with CD3ε on Jurkat.E6 cells (Figure 1c). We also, observed slightly decreased fluorescence intensity on the round 16 X-axis of cell-SELEX binding to Jurkat.E6 after LIGS (Figure 1b; compare solid blue versus dashed blue lines), suggesting that some of the potential specific DNA aptamer sequences may have been replaced by the addition of the antibody.
Supernatant containing competitively eluted potential DNA molecules from LIGS were then PCR-amplified. To ensure that all copies of competitively eluted potential DNA aptamers were adequately amplified, a two-step PCR process was conducted.
Finally, two libraries were cloned into bacterial vector using TOPO TA cloning and subjected to DNA sequencing: round 16 of cell-SELEX, consisting of sequences enriched towards Jurkat.E6 cells and competitively eluted pool, consisting of sequences specific for CD3ε.
Analysis of sequences generated from LIGS
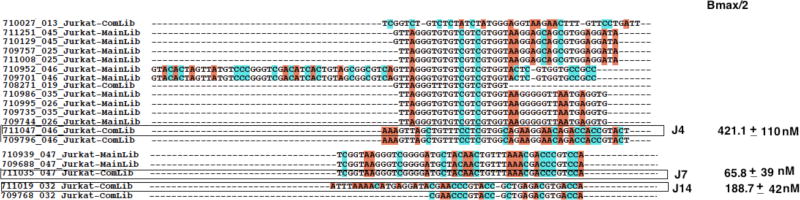
Sequences were analyzed using ClustalX.212, and results revealed an enrichment pattern similar to that of an evolved cell-SELEX library (Supporting Information; Sequence Alignment, Table 1). Specifically, analysis revealed multiple copies of the same sequences, or repeated motifs shared in common, but interrupted by segments of DNA bases unique to each sequence. Next, alignment was performed on the competitively eluted sequences from LIGS and sequences from round 16 of cell-SELEX containing all sequences evolved towards Jurkat.E6 cells. Three homologous patterns were observed between the two libraries: 1) repetition of the same sequences within the pool unique to the respective libraries; 2) repetition of the same sequences in both competitively eluted library and round 16 of cell-SELEX library; and 3) repetition of sequences with common motifs in both libraries (Supporting Information; Table 1). In the case of 3), even though sequences were derived from two different pools, they shared a common motif, differing only by a few bases. (Figure 2, aptamer J7). We hypothesized that specifically enriched sequences towards the Jurkat.E6 cell line would dominate the library and that after subsequent cloning and DNA sequencing steps, these sequences would still predominate such that round 16 of cell-SELEX library would contain all sequences enriched towards Jurkat.E6 cells. Very importantly, however, the sequences obtained from LIGS would favor sequences selectively eluted by anti-CD3 antibody binding to CD3ε epitope, or one of the components of TCR. Therefore, we focused on the sequences that repeatedly appeared within a family with common motifs from the two different pools. Since the objective of this study is aimed at selecting the aptamers with most binding specificity based on LIGS, only the sequences competitively eluted by anti- CD3ε antibody sharing common motifs within the library, or with round 16 of cell- SELEX library, were selected for synthesis.
Figure 2.
Sequence alignment of three hits: J14, J7 and J4. The sequences from Ligand-guided Selection and sequences from Cell-SELEX round 16 were aligned using ClustalX2. MainLib = sequences obtained from Cell-SELEX-round 16; ComLib = sequences obtained from competitively eluted library (LIGS). The three specific aptamer candidates, including J4, J7 and J14, were evaluated based on set criteria for selection and calculated Bmax/2.
Analysis of Specificity
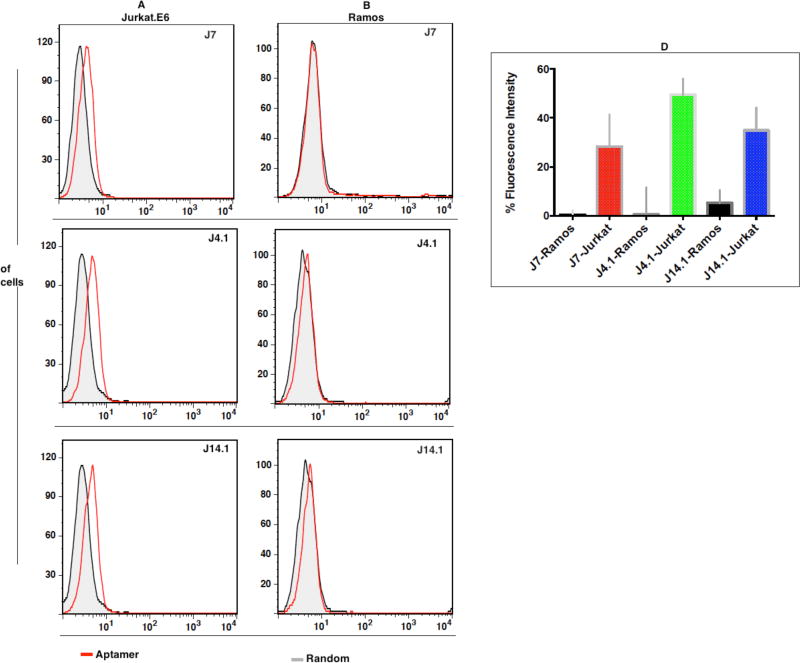
A total of 27 individual sequences were synthesized with FAM-dT at the 3’-end using standard phosphoramidite solid-state synthesis, followed by reversed phase HPLC purification. We first investigated aptamer specificity by analyzing the binding of individual fluorescently labeled aptamers with Jurkat.E6, using Burkitt’s lymphoma cell line Ramos as the negative control cell line. Burkitt’s lymphoma, which is from the B-cell lineage, does not express TCR-CD3 complex; therefore, sequences that do not bind to Ramos cells would be specific for TCR complex13. Interestingly, out of 27 tested sequences (Supporting Information; Figure S3), three sequences, J4, J7 and J14, showed specificity against Jurkat.E6 cells, but not control Ramos cells.
Twenty-four tested sequences from the competitively eluted library either bound to both Jurkat.E6 cells and Ramos cells or did not bind to either cell line. Sequences not binding to either cell line could be nonspecific background sequences from the partially evolved cell-SELEX pool or they might be sequences with high off-rates contaminating the LIGS pool. The sequences binding to both cell lines might be targeting receptors common to both cell lines. Following this validation step, we focused on the three positive hits for further analysis of affinity and antigen specificity.
Analysis of affinity of J7, J4 and J14.1
During post-SELEX structure-activity relationship studies, it has been shown that truncation of full-length aptamer is essential to optimize fold and increase affinity14. Therefore, in order to maximize the most favorable fold of our LIGS aptamers, we systematically truncated from the 3’ and 5’ ends of J4 and J14 (Supporting Information; truncated J4.1 and J14.1 in Table 2) for use in later studies. All three sequences were analyzed in triplicate for their binding constant against Jurkat.E6 cells. We observed considerably high Bmax/2 for J14.1, suggesting that J14.1 approached high specificity against Jurkat.E6 cells, but not affinity, while J4 and J7 showed comparable binding affinities to aptamers generated from cell-SELEX in other reports,10 suggesting that aptamers J4 and J7 approached affinity and specificity towards CD3-positive Jurkat.E6 cells (Supporting Information; Figure S3). This example and that of Zumrut et al. show much less aptamer affinity than aptamers selected from cell-SELEX. This could be explained by the interruption of cell-SELEX at a partially enriched stage, in accordance with the standard LIGS procedure, thus blocking the complete evolution of aptamers. Therefore, we are currently investigating degree of enrichment as a function of affinity of sequences eluted using LIGS.
Analysis of binding specificity of J4.1, J14.1 and J7 towards CD3ε
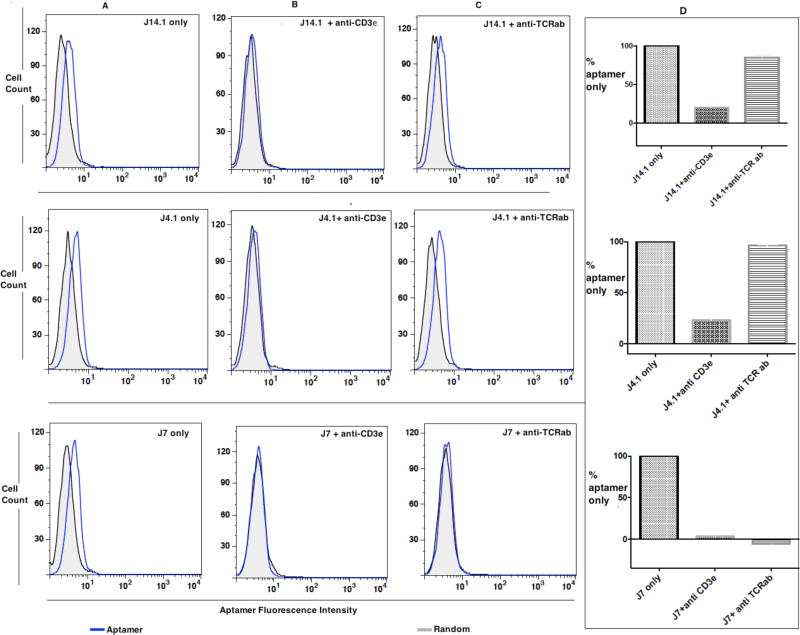
Next, we investigated the specificity towards the epitope on CD3ε by utilizing competitive binding experiments against anti-CD3ε antibody, which was used in LIGS. Anti-TCRαβ antibody was employed as a control to investigate if aptamer binding would be affected by adding the epitope-specific secondary anti-CD3ε HIT3a clone. Antibody displacement assays have previously been used as a valid tool to confirm epitope-specific aptamers selected against whole cells.15–17 Here, 50 pmols of aptamer and equal number of pmols of random sequences were incubated with 75,000 cells for 40 min at 0 °C. Then, either anti-CD3ε HIT3a clone or anti-TCRαβ antibody in excess was added to allow competitive binding for an additional 40 minutes. Cells were subsequently washed and analyzed for aptamer binding using flow cytometric assay (Fig. 4).
Figure 4.
Epitope identity. Flow cytometric competitive binding analysis of J4, J7 and J14.1 without (A) or with anti-CD3ε (B) or with anti-TCRαβ antibody (C) and overall conclusion from data presented in A, B and C from two independent experiments normalized to the aptamer binding to Jurkat.E6 cells with no added antibody (D). Each FITC-labeled random control or J4.1, J7 or J14.1 (1 µM) was incubated for 40 minutes on ice with 75 × 103 Jurkat.E6 cells. Then, binding buffer or anti-CD3ε HIT3a clone (panel B) or anti-TCRαβ antibody (panel C) was added and incubated for an additional 40 minutes. The cells were washed with 1.5 mL of wash buffer and the binding of respective aptamer analyzed by flow cytometry. Aptamer fluorescence intensity on X-axis indicates the binding of each aptamer. Thus, increment of fluorescence intensity can be directly compared to baseline random control as an indicator of aptamer binding. Aptamer fluorescence intensity on the X-axis shifted to background when anti-CD3ε HIT3a was added to all three aptamers pre-incubated with Jurkat.E6 cells, and binding of J7 and J14.1 was affected when anti-TCRαβ antibody was added. On the other hand, no difference in fluorescence intensity was observed for random control (gray-black); therefore, competitive binding experiments against aptamer J7, J4.1 and J14.1 using anti- CD3ε antibody showed that the stronger binder to CD3ε, effectively displaces all three aptamers. Binding of corresponding antibody is shown in Figure S5 in Supporting Information.
The displacement assay, as illustrated in Figure 4, suggested the variability and loss of binding of LIGS aptamer candidates when either anti-CD3ε or anti-TCRαβ antibody, as secondary ligands, was added to aptamer-bound Jurkat.E6 cells. In the presence of both anti-CD3ε antibody and anti-TCRαβ antibody, aptamer J7 completely lost its binding affinity, as indicated by the decrease in fluorescence shift for aptamer binding on the X-axis. To account for this, it is possible that conformational changes induced by anti- TCRαβ or anti-CD3ε upon binding the TCR-CD3 complex weakened the J7-TCR complex, resulting in displacement of the aptamer. In contrast, when anti-CD3ε, but not anti-TCRαβ, antibody was added, binding of aptamer J4.1 was lost, suggesting that aptamer J4.1 bound to an epitope unique to CD3ε in a manner independent of anti-TCRαβ. Finally, aptamer J14.1 lost binding affinity when anti-CD3ε was added and only slightly when anti-TCRαβ antibody was added. Fluorescence shift on the X-axis is very high by the addition of antibodies, most likely because one antibody contains multiple fluorophores, leading to higher signal (Supporting Information; Figure S5). Taken together, all three aptamers, including J7, J14.1, and J4.1, showed binding affinity to Jurkat E6 cells.
Importantly, however, when anti-CD3ε was added, all three aptamers lost their binding to Jurkat.E6 cells as the aptamer fluorescence intensity on the X-axis shifted to background suggesting the specificity of aptamers generated by LIGS towards CD3ε. While, pre-incubated aptamer J7 was displaced by both anti-CD3ε and anti-TCRαβ. Therefore, these findings prove that LIGS can be utilized to identify aptamers specific to a predetermined epitope, or closely related epitope, from a multiple-domain complex, in this case TCR complex, on the target cell, Jurkat.E6 cells.
Aptamers are synthetic molecules; therefore, their shelf-life is longer, and they are stable against heat and compatible with a variety of solvents19. Initially, aptamers were selected using SELEX against purified proteins in solution. However, these aptamers have shown limited applicability by their failure to identify endogenous protein targets in vitro and in vivo, as noted above20. The breakthrough cell-SELEX11,21 allows the selection of aptamers towards membrane receptor targets in their native state at their endogenous levels with no prior requirement for the overexpression of a protein. Nevertheless, proteomic identification of the receptor protein ligand of aptamers generated from cell-SELEX is a challenge. With such limitation, therapeutic and diagnostic applications of aptamers remain challenging. Therefore, to address this challenge, we have introduced LIGS, a simple technique to selectively separate aptamers binding to a specific epitope using a secondary ligand specific to the same epitope. From a fundamental point-of view, LIGS technology pushes separation efficiency to a remarkably high level. That is, the competition strategy allows us to separate out a few aptamer molecules that bind to a specific site of a specific receptor molecule in its endogenous state from a complex library evolved against a whole cell. Since the aptamers selected using LIGS are selectively eluted based on the interaction of the secondary ligand with its target at its endogenous state, LIGS-generated aptamers will have higher potential in identifying the same receptor in a clinical setting. Moreover, apart from selecting aptamers against epitopes in a multidomain protein complex, LIGS can be applied to a number of platforms, including peptide libraries. LIGS-generated aptamers can also be selected toward a small-molecule ligand-binding site, utilizing small-molecule ligand-receptor interaction as a guide.
In conclusion, by using an antibody against one of the domains of multi-domain complex, we have shown that specific aptamers could be selectively eluted, demonstrating the significance of LIGS in generating highly specific nucleic acid ligands toward a broader range of receptor molecules already characterized as surface markers. LIGS can be differentiated from other SELEX strategies because it selectively outcompetes a set of already partially enriched cell-SELEX aptamers against a predetermined epitope at their endogenous native state by ligand-receptor, i.e., antibody-protein receptor, interactions. This approach can be extended to a number of combinatorial screening platforms, including phage display libraries and small-molecule libraries.
Supplementary Material
Figure 3.
Binding of three specific aptamers identified in the validation assay. Aptamer binding to Jurkat.E6 cells (3A) and Ramos (3B). Sequence analysis was done by using a 1µM solution of respective aptamers against 75–100 × 104 cells which were incubated for 1 hour at 4 °C and subsequently washed twice with wash buffer prior to flow cytometry for binding analysis. Overall conclusion from the data presented in 3A and 3B from three different independent experiments (3C) with Y axis = [(MFI aptamer-MFI random/MFI aptamer)*100]
Acknowledgments
Authors are grateful for NIH-NIGMS SCORE SC3 GM105578 grant and Lehman College startup funds.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Jiang F, Liu B, Lu J, Li F, Li D, Liang C, Dang L, Liu J, He B, Badshah SA, Lu C, He X, Guo B, Zhang XB, Tan W, Lu A, Zhang G. Progress and Challenges in Developing Aptamer-Functionalized Targeted Drug Delivery Systems. International journal of molecular sciences. 2015;16:23784–23822. doi: 10.3390/ijms161023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He W, Elizondo-Riojas MA, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, Volk DE, Durland RH, Englehardt J, Cavasotto CN, Gorenstein DG. X-aptamers: a bead-based selection method for random incorporation of drug like moieties onto next-generation aptamers for enhanced binding. Biochemistry. 2012;51:8321–8323. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiel WH, Thiel KW, Flenker KS, Bair T, Dupuy AJ, McNamara JO, 2nd, Miller FJ, Giangrande PH. Cell-internalization SELEX: method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol Biol. 2015;1218:187–199. doi: 10.1007/978-1-4939-1538-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumrut HE, Ara MN, Fraile M, Maio G, Mallikaratchy P. Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins. Nucleic acid therapeutics. 2016 doi: 10.1089/nat.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinherz EL. alphabeta TCR-mediated recognition: relevance to tumor-antigen discovery and cancer immunotherapy. Cancer Immunol Res. 2015;3:305–312. doi: 10.1158/2326-6066.CIR-15-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Analytical chemistry. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 11.Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nature protocols. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 12.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 13.Bojarczuk K, Bobrowicz M, Dwojak M, Miazek N, Zapala P, Bunes A, Siernicka M, Rozanska M, Winiarska M. B-cell receptor signaling in the pathogenesis of lymphoid malignancies. Blood Cells Mol Dis. 2015;55:255–265. doi: 10.1016/j.bcmd.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic acids research. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Molecular & cellular proteomics : MCP. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Jiang G, Li W, Qiu K, Zhang M, Carter CM, Al-Quran SZ, Li Y. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. Journal of hematology & oncology. 2014;7:5. doi: 10.1186/1756-8722-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boltz A, Piater B, Toleikis L, Guenther R, Kolmar H, Hock B. Bi-Q specific aptamers mediating tumor cell lysis. J Biol Chem. 2011;286:21896–21905. doi: 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 20.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv. 2015;33:1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clinical chemistry. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.