Abstract
The long noncoding RNA (lncRNA) H19 is a maternally expressed imprinted gene that plays important roles in tumorigenesis, progression, and metastasis. However, the association between polymorphisms on H19 and breast cancer (BC) susceptibility has remained obscure. In this case–control study, we assessed the interaction between two lncRNA H19 single-nucleotide polymorphisms (SNPs) (rs217727 C>T, rs2839698 C>T) and the risk of BC in a Chinese Han population. In total, 1,005 BC cases and 1,020 healthy controls were enrolled in this study. Correlations between genotypes and BC risk were evaluated by multivariate logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs). False-positive report probability calculation was also utilized to identify false-positive associations. We observed that the rs217727 T variant was consistently significantly associated with an increased risk of BC in both codominant and dominant models (CT vs CC, OR 1.25, 95% CI 1.03–1.51; TT vs CC, OR 1.56, 95% CI 1.15–2.09; CT + TT vs CC, OR 1.31, 95% CI 1.09–1.57), and all associations remained significant after Bonferroni correction (P<0.025). Subsequent stratified analyses also revealed that associations between BC risk and rs217727 genotypes were more profound in patients with estrogen receptor-positive, human epidermal growth factor receptor 2 (HER2)-negative, and hormone receptor-positive–HER2-negative molecular subtypes (all passed the threshold for Bonferroni correction, P<0.005). These findings extend available data on the association of H19 polymorphisms and BC susceptibility. Based on these results, we encourage further large-scale studies and functional research to confirm our findings and better elucidate the underlying biological mechanisms.
Keywords: breast cancer, H19, lncRNA, polymorphisms, genetic susceptibility
Introduction
Breast cancer (BC) is one of the most frequently diagnosed malignancies and the leading cause of death from cancer in women worldwide.1 For the year 2016, it was estimated that in the US approximately 246,660 female patients would be diagnosed with BC and 40,450 would die from it.2 In China, the incidence of BC has increased rapidly in recent years and become the most common cancer for women in major cities.3,4 The development of BC is a complex multistep process involving both environmental factors and genetic variations. It is well established that age, obesity, previous benign breast disease, positive family history of BC, and female menstrual and reproductive status are associated with the development of BC.5–7 For genetic factors, numerous single-nucleotide polymorphisms (SNPs) in low-penetrance susceptibility genes8–17 have been identified to be associated with an elevated risk of BC, suggesting a significant contribution of inherited factors in BC susceptibility. Therefore, the identification of additional potential SNPs in low-penetrance genes could have a great impact on risk estimation for BC and provide earlier application of proper therapeutic strategies to decrease its mortality rate.
In recent years, long noncoding RNAs (lncRNAs), a novel kind of RNA, have attracted extensive attention for their wide range and complex regulatory functions in human diseases. lncRNAs are defined as transcribed RNA molecules that are longer than 200 nucleotides and not translated into proteins.18 Although their functions were not originally clear, lncRNAs are now known to play critical roles in carcinogenesis, including transcriptional, posttranscriptional, and epigenetic regulation of cancer-related genes, thereby resulting in the cell-cycle progression, apoptosis, invasion, and migration.19,20 The H19 lncRNA is located on human chromosome 11p15.5, encoding a 2.3 kb long, spliced, and polyadenylated ncRNA that plays important roles in embryonic development and growth control.21,22 It acts as an imprinted gene expressed from the maternal chromosome.23 Moreover, differentially methylated regions (DMRs), which lie upstream of H19, were found to be critical in the regulation of H19 gene expression.23,24 DMRs are commonly considered CpG-rich and frequently meet the criteria for CpG islands. Therefore, it is likely that some DMRs are related to genetic or epigenetic modifications of tissue-specific imprinted genes.25,26
Barrow et al27 revealed that the aberrant events and increased variation in imprinted gene methylation were more frequent in invasive BC and more associated with negative estrogen receptor (ER) and progesterone receptor (PR) status. Accumulating evidence has demonstrated that H19 lncRNA is abnormally expressed and promotes cancer-cell proliferation in many tumors, such as BC and hepatocellular, esophageal, and bladder cancers,28–31 suggesting an oncogenic function. SNPs locating on lncRNA H19 have also been identified to regulate its expression and function. For example, the CT + TT genotype of rs217727 and rs2839698 is significantly associated with an increased risk of gastric cancer.32 An elevated risk of BC and bladder cancer has also been discovered in the TT carriers for H19 rs217727,33,34 while for the rs2839698 CT genotype, this carrier has been reported to be associated with a decreased risk for non-muscle-invasive bladder cancer.35 Therefore, we conducted this case–control study of 1,005 BC patients and 1,020 healthy controls to evaluate the interactions between two H19 lncRNA tag SNPs (rs217727 and rs2839698) and the risk of BC in a southeast China Han population. Moreover, we also assessed relationships between tag SNPs and traditional risk factors, as well as specific molecular subtypes of BC defined by ER, PR, and human epidermal growth factor receptor 2 (HER2) status.
Materials and methods
Ethics statement
This study and consent procedure was approved by the ethics committee of the Affiliated Union Hospital of Fujian Medical University. All participants provided written informed consent to be included in the study.
Study participants and specimen collection
This study was a hospital-based case–control study of 1,005 BC patients and 1,020 healthy controls. All subjects were genetically unrelated Chinese residents of Fujian Province. BC cases (aged 21–79 years) were consecutively recruited from the Affiliated Union Hospital of Fujian Medical University from May 2010 to April 2016. Eligible patients were histopathologically confirmed with primary BC without restriction of histological type or age. Cancer-free controls were frequency-matched to cases on age (±3 years) and randomly selected from unrelated community residents attending routine health checkups in the same hospital. Each subject was interviewed face to face by a trained interviewer to obtain information on demographic factors, menstrual and reproductive history, breastfeeding, and previous benign breast-disease history, as well as family history of BC. Approximately 3 mL venous blood was collected from each participant into a test tube containing EDTA. Detailed information on clinicopathological characteristics of BC patients was collected from medical records. Specific data for ER, PR, and HER2 status were obtained by immunohistochemistry from each patient’s pathology report.
DNA extraction and genotyping
Genomic DNA was extracted from peripheral-blood samples of patients and controls using a commercially available kit (whole-blood DNA extraction kit; BioTeke, Beijing, China) according to the manufacturer’s instructions. Genotype analyses for the two H19-lncRNA tag SNPs were performed by a 2×48-Plex SNP scan kit (G0104K; Genesky Biotechnologies, Shanghai, China). Primers and probes were all designed and synthesized by Thermo Fisher Scientific (Waltham, MA, USA). DNA samples were ligated and amplified by polymerase chain reaction (PCR) following the standardization protocol recommended by the manufacturer. Ligation products were obtained with an ABI3730XL sequencer and raw data analyzed by GeneMapper 4.1 software (Thermo Fisher Scientific).
To ensure the accuracy of genotyping, all analyses were performed without knowledge of case or control status. In addition, about 10% of samples were randomly selected from both cases and controls. Direct sequencing (BGI Sequencing, Beijing, China) was utilized for genotype confirmation, and the result was 100% concordant.
Quantitative real-time reverse-transcription (RT)-PCR analysis of H19 mRNA expression levels
Expression levels of H19 mRNA were examined by quantitative RT-PCR in 256 paired tissue samples of BC patients and corresponding normal tissue. Total RNA was extracted from frozen tumors and corresponding normal tissue using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocols. RNA was reverse-transcribed into cDNA with a PrimeScript RT reagent kit (Takara, Kusatsu, Japan). All assays were performed with an SYBR Ex Taq kit (Takara), and relative H19-expression levels were calculated with β-actin by the 2−ΔCt method.
Statistical analysis
Differences between case and control groups in demographic characteristics and traditional risk factors were evaluated with Student’s t-test (for continuous variables) and χ2 test (for categorical variables). Hardy–Weinberg equilibria for genotype distribution were assessed by a goodness-of-fit χ2 test to compare observed genotype frequencies with expected ones among control subjects. Associations among genotypes, BC risks, and risk factors were evaluated using computing ORs and its 95% CIs from multivariate logistic regression analysis, adjusted for age, body mass index (BMI), age at menarche and menopause, menopausal status, number of pregnancies, and family history of BC. Power analysis for this study was performed using Quanto version 1.2.4, and the disease risk in the Chinese population was 268.6 per 100,000. In addition, false-positive report probability (FPRP) was calculated to identify FP associations between H19 polymorphisms and BC for all significant genetic effects observed in this study. FPRP was set with prior probabilities of 0.001, 0.01, 0.1, and 0.25, with OR 1.5 in the dominant model, and a probability <0.4 was considered noteworthy. All statistical analyses were two-sided, and P<0.05 was considered significant. All P-values were corrected for multiple comparisons according to the Bonferroni method. All statistical analyses were performed with SPSS version 20.0 for Windows (IBM, Armonk, NY, USA).
Results
Characteristic of study subjects
Distributions of selected characteristics between BC cases and control subjects are shown in Table 1. There were 1,005 cases and 1,020 controls involved in this study. No significant differences were observed between cases and controls in age, age at menopause, menopausal status, or previous benign disease (P>0.05). However, compared with healthy controls, BC patients were more likely to have higher mean BMI, later age at menarche and first live birth, fewer pregnancies, and greater frequency of BC family history (P<0.05). Among the 1,005 BC cases, 678 (67.5%) were ER-positive, 618 (61.5%) PR-positive, and 275 (27.4%) HER2-positive.
Table 1.
Distributions of selected characteristics in breast cancer cases and cancer-free controls
| Characteristics | Cases (n=1,005), n (%) |
Controls (n=1,020), n (%) |
P-value |
|---|---|---|---|
| Age at diagnosis, years (mean ± SD) | 46.5±10.2 | 46.7±11.1 | 0.646 |
| Body mass index, kg/m2 (mean ± SD) | 23±3.2 | 22.5±2.6 | <0.001 |
| Age at menarche, years (mean ± SD) | 15.5±1.7 | 15.2±1.6 | 0.001 |
| Age at menopause, years (mean ± SD) | 49.8±3.6 | 49.9±3.5 | 0.598 |
| Age at first live birth, years (mean ± SD) | 25±3.5 | 24.2±3.4 | <0.001 |
| Menopausal status | 0.059 | ||
| Premenopausal | 656 (65.3) | 668 (65.5) | |
| Postmenopausal | 342 (34) | 333 (32.6) | |
| Unnatural menopausea | 7 (0.7) | 19 (1.9) | |
| Pregnancies, n | <0.001 | ||
| ≤2 | 570 (56.7) | 404 (39.6) | |
| >2 | 435 (43.3) | 616 (60.4) | |
| Hormone therapy | 0.013 | ||
| Yes | 45 (4.5) | 25 (2.5) | |
| No | 960 (95.5) | 995 (97.5) | |
| Previous benign breast disease | 0.054 | ||
| Yes | 41 (4.1) | 26 (2.5) | |
| No | 964 (95.9) | 994 (97.5) | |
| Family history of breast cancer | <0.001 | ||
| Yes | 75 (7.5) | 12 (1.2) | |
| No | 930 (92.5) | 1,008 (98.8) | |
| ER status | |||
| Positive | 678 (67.5) | ||
| Negative | 327 (32.5) | ||
| PR status | |||
| Positive | 618 (61.5) | ||
| Negative | 387 (38.5) | ||
| HER2 status | |||
| Positive | 275 (27.4) | ||
| Negative | 730 (72.6) |
Note:
Included hysterectomy operation and other status.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Effects of tag SNPs in H19 and BC risk
Relative locations of the two selected tag SNPs are summarized in Figure 1. Observed genotype frequencies in the two SNPs were both in agreement with those expected from the Hardy–Weinberg equilibrium (P=0.968 for rs217727 and P=0.871 for rs2839698, respectively, shown in Table 2). Genotype distributions of rs217727 and rs2839698 are displayed in Table 3. We used computing OR and 95% CI to evaluate associations of H19 SNPs with BC risk in codominant and dominant models. Results of multivariate logistic regression analyses revealed that for rs217727, the carriers of the CT or TT genotype had distinctly an increased risk of BC compared with CC carriers (adjusted OR 1.25, 95% CI 1.03–1.51 and adjusted OR 1.56, 95% CI 1.15–2.09, respectively). When we combined CT and TT genotypes to construct a dominant model, significantly increased risk was also discovered in CT + TT genotypes, with an adjusted OR of 1.31 (95% CI 1.09–1.57). Moreover, P-values for CT and TT genotypes of rs217727 in the codominant model and CT + TT genotype of rs217727 in the dominant model all remained significant, even after Bonferroni correction (P<0.025). Associations for variant genotypes in rs217727 were also dose independent, with a P-trend of 0.006. In the power analysis, we had power of 0.835 to detect an OR of 1.31 (1.09–1.57) with a frequency of 59.9% for rs217727. However, no significant differences in genotype distribution were found in codominant (CT versus TT, CC versus TT) or dominant (CT + CC versus TT) models for rs2839698.
Figure 1.
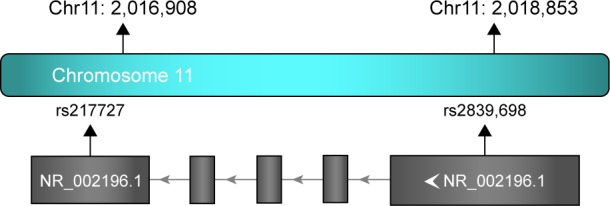
Relative position of rs217727 and rs2839698 in H19.
Table 2.
Hardy–Weinberg equilibrium (HWE) tests for H19 long noncoding RNA polymorphisms
| SNP | Reference allele/risk allele | HWE | P-valuea |
|---|---|---|---|
| rs217727 | C/T | 0.063 | 0.968 |
| rs2839698 | C/T | 0.275 | 0.871 |
Note:
For HWE test.
Table 3.
Genotype frequencies of H19 long noncoding RNA polymorphisms in breast cancer cases and controls
| SNP | Genotype | Cases (n=1,005), n (%) | Controls (n=1,020), n (%) | P-valuea | Adjusted OR (95% CI)b | P-value (trend)c |
|---|---|---|---|---|---|---|
| rs217727 | ||||||
| C/T | CC | 403 (40.1) | 465 (45.6) | 1 | ||
| CT | 471 (46.9) | 450 (44.1) | 0.023* | 1.25 (1.03–1.51) | ||
| TT | 131 (13) | 105 (10.3) | 0.004* | 1.56 (1.15–2.09) | 0.006 | |
| CT + TT | 602 (59.9) | 555 (54.4) | 0.004d,* | 1.31 (1.09–1.57) | ||
| rs2839698 | ||||||
| C/T | CC | 452 (45) | 484 (47.5) | 1 | ||
| CT | 440 (43.8) | 432 (42.3) | 0.554 | 1.06 (0.88–1.28) | ||
| TT | 113 (11.2) | 104 (10.2) | 0.5 | 1.11 (0.82–1.51) | 0.234 | |
| CT + TT | 553 (55.0) | 536 (52.5) | 0.466d | 1.07 (0.89–1.28) |
Notes:
P<0.025,
significant after Bonferroni correction;
adjusted by age, body mass index, age at menarche and menopause, menopausal status, number of pregnancies, and family history of breast cancer where appropriate;
for genotypes between cases and cancer-free controls;
two-sided χ2 test for differences in frequency distribution of combined genotypes (dominant model) between cases and controls.
Abbreviation: SNP, single-nucleotide polymorphism.
Stratified analysis of H19 polymorphisms and BC risk
We further analyzed the effects of the rs217727 and rs2839698 genotypes on the risk of BC in the dominant model among different subgroups of demographic characteristics and reproductive factors. As indicated in Table 4, the corrected P-value cutoff after Bonferroni correction was P<0.0035. For the T carriers of rs217727 (CT + TT genotypes), elevated risks of BC were more likely to be evident in subgroups of younger patients (age <40 years, OR 1.57, 95% CI 1.11–2.22), higher BMI individuals (BMI ≥23 kg/m2, OR 1.31, 95% CI 1.2–1.87), premenopausal women (OR 1.32, 95% CI 1.05–1.65), and subjects with later menarche (OR 1.34, 95% CI 1.02–1.75), later menopause (OR 1.71, 95% CI 1.05–2.8), earlier age at first live birth (OR 1.39, 95% CI 1.06–1.82), and fewer pregnancies (two or fewer, OR 1.31, 95% CI 1–1.7). However, none of these subgroups passed the threshold for Bonferroni correction (P<0.0035). No significant heterogeneity was detected within any of the subgroups either. As to the C carriers of rs2839698 (CT + CC genotypes), no significant positive associations or heterogeneity were observed.
Table 4.
Stratified analyses of H19 long noncoding RNA polymorphisms and breast cancer susceptibility
| rs217727
|
rs2839698
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (CC/CT + TT) |
Controls (CC/CT + TT) |
P-valuea | OR (95% CI)b | P-valuec | Cases (CC/CT + TT) |
Controls (CC/CT + TT) |
P-valuea | OR (95% CI)b | P-valuec | |
| Age, years | ||||||||||
| <40 | 108/176 | 140/148 | 0.011 | 1.57 (1.11–2.22) | 0.251 | 130/154 | 137/151 | 0.972 | 0.99 (0.71–1.4) | 0.606 |
| ≥40 | 295/426 | 325/407 | 0.08 | 1.21 (0.98–1.51) | 322/399 | 347/385 | 0.394 | 1.1 (0.89–1.36) | ||
| Body mass index, kg/m2 | ||||||||||
| <23 | 264/286 | 201/269 | 0.059 | 1.31 (0.99–1.74) | 0.987 | 263/287 | 262/329 | 0.912 | 1.02 (0.77–1.34) | 0.586 |
| ≥23 | 244/347 | 159/255 | 0.03 | 1.31 (1.2–1.87) | 221/249 | 190/224 | 0.313 | 1.13 (0.89–1.44) | ||
| Age at menarche, years | ||||||||||
| <16 | 217/297 | 247/284 | 0.115 | 1.23 (0.95–1.58) | 0.655 | 236/278 | 238/293 | 0.604 | 0.94 (0.73–1.2) | 0.138 |
| ≥16 | 186/305 | 218/271 | 0.036 | 1.34 (1.02–1.75) | 216/275 | 246/243 | 0.903 | 1.25 (0.96–1.63) | ||
| Menopausal status | ||||||||||
| Premenopausal | 267/389 | 133/209 | 0.016 | 1.32 (1.05–1.65) | 0.97 | 292/364 | 311/357 | 0.601 | 1.06 (0.85–1.32) | 0.963 |
| Postmenopausal | 309/359 | 148/185 | 0.108 | 1.31 (0.94–1.81) | 159/183 | 164/169 | 0.699 | 1.07 (0.77–1.47) | ||
| Age at menopause, years | ||||||||||
| ≤50 | 94/137 | 71/95 | 0.684 | 1.09 (0.71–1.69) | 0.226 | 115/116 | 85/81 | 0.671 | 1.1 (0.71–1.69) | 0.642 |
| >50 | 54/100 | 76/91 | 0.03 | 1.71 (1.05–2.8) | 70/84 | 77/90 | 0.813 | 0.94 (0.59–1.52) | ||
| Age at first live birth, years | ||||||||||
| <25 | 164/268 | 248/306 | 0.016 | 1.39 (1.06–1.82) | 0.533 | 201/231 | 269/285 | 0.498 | 1.1 (0.84–1.43) | 0.963 |
| ≥25 | 216/315 | 192/227 | 0.13 | 1.23 (0.94–1.6) | 235/296 | 197/222 | 0.437 | 1.11 (0.85–1.44) | ||
| Pregnancies, n | ||||||||||
| ≤2 | 240/330 | 199/205 | 0.047 | 1.31 (1–1.7) | 0.968 | 245/325 | 185/219 | 0.482 | 1.1 (0.85–1.43) | 0.765 |
| >2 | 163/272 | 266/350 | 0.045 | 1.3 (1–1.68) | 207/228 | 299/317 | 0.768 | 1.04 (0.81–1.34) | ||
Notes:
P<0.0035;
adjusted by age, body mass index, age at menarche and menopause, menopausal status, number of pregnancies, and family history of breast cancer where appropriate;
heterogeneity test.
Subsequently, in order to determine whether the associations between rs217727 and rs2839698 genotypes and BC risk were modified by specific molecular subtypes, we conducted case-only stratified analysis according to ER, PR, and HER2 status (Table 5). Compared with the CC genotype of rs217727, CT + TT genotypes were found to be associated with patients who were ER-positive (OR 1.34, 95% CI 1.09–1.64) and HER2-negative (OR 1.4, 95% CI 1.14–1.71). Differences were more profound in hormone receptor (HR)-positive–HER2-negative patients (OR 1.45, 95% CI 1.16–1.81), but not in triple-negative BC individuals (OR 1.25, 95% CI 0.89–1.75). However, for rs2839698 variants, no significant association was discovered according to ER, PR, or HER2 status. All positive associations remained significant after Bonferroni correction (P<0.005).
Table 5.
Associations of H19 long noncoding RNA polymorphisms and specific molecular subtypes for breast cancer patients
| rs217727
|
rs2839698
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases (CC/CT + TT) |
P-valuea | OR (95% CI)b | P-valuec | Cases (CC/CT + TT) |
P-valuea | OR (95% CI)b | P-valuec | |
| ER status | ||||||||
| Positive | 268/410 | 0.004* | 1.34 (1.09–1.64) | 0.577 | 291/387 | 0.134 | 1.17 (0.95–1.43) | 0.111 |
| Negative | 135/192 | 0.135 | 1.22 (0.94–1.58) | 161/166 | 0.418 | 0.9 (0.7–1.16) | ||
| PR status | ||||||||
| Positive | 247/371 | 0.006 | 1.34 (1.08–1.66) | 0.675 | 259/359 | 0.075 | 1.21 (0.98–1.49) | 0.088 |
| Negative | 156/231 | 0.072 | 1.25 (0.98–1.59) | 193/194 | 0.313 | 0.88 (0.7–1.12) | ||
| HER2 status | ||||||||
| Positive | 118/157 | 0.651 | 1.07 (0.81–1.41) | 0.71 | 138/137 | 0.364 | 0.88 (0.67–1.16) | 0.126 |
| Negative | 285/445 | 0.001* | 1.40 (1.14–1.71) | 313/416 | 0.186 | 1.14 (0.94–1.39) | ||
| HR+ HER2− | 208/336 | 0.001* | 1.45 (1.16–1.81) | 231/313 | 0.169 | 1.16 (0.94–1.45) | ||
| TNBC | 68/100 | 0.198 | 1.25 (0.89–1.75) | 77/91 | 0.759 | 1.05 (0.75–1.47) | ||
Notes:
P<0.005,
significant after Bonferroni correction;
adjusted by age, body-mass index, age at menarche and menopause, menopausal status, number of pregnancies, and family history of breast cancer;
for heterogeneity test.
Abbreviations: CI, confidence interval; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; OR, odds ratio; PR, progesterone receptor; TNBC, triple-negative breast cancer.
False-positive report probability validation
FPRP analysis was further utilized to determine whether significant findings were really noteworthy or because of chance (Table 6). When the assumption of prior probability was set at 0.01, the association between rs217727 CT + TT genotypes and BC risk remained noteworthy for all subjects (FPRP 0.197). For stratified analysis, significantly elevated BC risks were also detected in cases who were ER-positive, HER2-negative and HR-positive–HER2-negative (FPRP 0.341, 0.114, and 0.141, respectively).
Table 6.
False-positive report probability values for associations between breast cancer risk and genotypes in stratified factors
| Genotype | Stratified factors | Positive OR (95% CI)a | P-valueb | Prior probability
|
|||
|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | ||||
| rs217727 | |||||||
| CT/CC | All subjects | 1.25 (1.03–1.51) | 0.023 | 0.06 | 0.161 | 0.678 | 0.955 |
| TT/CC | All subjects | 1.56 (1.15–2.09) | 0.004 | 0.021 | 0.061 | 0.419 | 0.879 |
| CT + TT/CC | All subjects | 1.31 (1.09–1.57) | 0.004 | 0.007 | 0.022 | 0.197 | 0.712 |
| ER+ | 1.34 (1.09–1.64) | 0.004 | 0.015 | 0.045 | 0.341 | 0.84 | |
| HER2− | 1.4 (1.14–1.71) | 0.001 | 0.004 | 0.012 | 0.114 | 0.565 | |
| HR+ HER2− | 1.45 (1.16–1.81) | 0.001 | 0.005 | 0.015 | 0.141 | 0.623 | |
Notes:
Crude OR;
logistic regression analysis for genotype-frequency distributions.
Abbreviations: CI, confidence interval; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; OR, odds ratio.
Functional assay of rs217727 on H19 expression
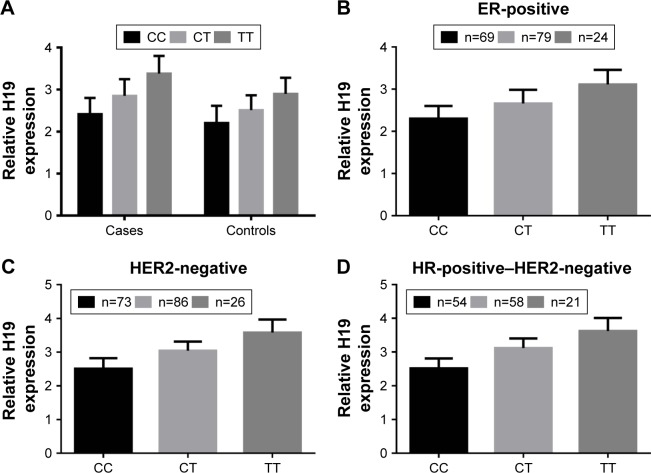
To explore the biological significance rs217727 further, we conducted a functional assay and examined associations between rs217727 genotypes and the expression level of H19 in 256 paired tissue samples of BC patients and corresponding normal tissue. As shown in Figure 2A, the expression level of H19 in BC tissue was significantly higher than in normal tissue (P=0.022). The rs217727 CT or TT genotype was also found to be significantly correlated with the elevated expression of H19 in BC patients compared with the CC genotype (P=0.013 and P<0.001, respectively). Moreover, for different molecular subtypes, relative H19 mRNA-expression levels were also consistently higher for the CT or TT genotype than the CC genotype in ER-positive (P=0.032 and P<0.001, respectively, Figure 2B), HER2-negative (both P<0.001, Figure 2C), and HR-positive–HER2-negative (both P<0.001, Figure 2D) subtypes.
Figure 2.
Functional assay of rs217727 on H19.
Notes: (A) Relative expression levels of H19 mRNA in cancerous and corresponding normal tissue; (B) relative H19 mRNA expression levels in ER-positive subtype; (C) relative H19 mRNA expression levels in HER2-negative subtype; (D) relative H19 mRNA expression levels in HR-positive–HER2-negative subtype.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hormone receptor.
Discussion
Further understanding of lncRNAs and their roles in tumor pathogenesis and metastasis could offer a great number of potential clues in developing novel therapeutic agents for BC. In the present case–control study, we focused on associations of two potential functional H19 polymorphisms (rs217727 and rs2839698) and BC susceptibility in a Chinese population. Results of the genotype distribution revealed that T variants of rs217727 (CT/TT genotype, CT + TT genotypes) were consistently significantly associated with increased risk of BC. With the disease risk in the Chinese population at 268.6 per 100,000, we had power (α=0.05) of 0.835 with an adjusted OR of 1.31 (95% CI 1.09–1.57) in this study, indicating that the mutant T allele of rs217727 may be a risk factor for BC.
H19 is an imprinted gene that can generate a 2.3-kb non-protein-coding molecule and play critical roles in embryonic development and growth control.22,23 H19 expression is decreased in maturing tissue after birth and is only found in cardiac and skeletal muscles. DMRs, located upstream of H19, have been reported to be essential in the regulation of H19 gene expression.23–26 H19 might promote carcinogenesis by acting in competitive endogenous RNA or precursors of microRNAs, and was confirmed to be highly upregulated in a variety of cancers, such as BC and hepatocellular, esophageal, and bladder cancers.28–31 Li et al revealed that the effect of H19 in gastric cancer was mediated by direct binding to ISM1, as well as indirect suppression of CALN1 expression via miR675,36 thus promoting proliferation, migration, invasion, and metastasis. As suggested by Luo et al,31 upregulated H19 is be able to increase bladder cancer-cell growth by regulating ID2 expression. In addition, H19 has also been demonstrated to promote pancreatic ductal adenocarcinoma-cell invasion and migration, partially by increasing HMGA2-mediated epithelial–mesenchymal transition through antagonizing Let7.37 The rs217727 C/T polymorphism is located in the exon 5 of the H19 gene and has been proved to contribute to the occurrence of many kinds of diseases. For instance, Gao et al38 identified that the T variant of rs217727 was associated with an increased risk of coronary artery disease (additive model, OR 2.05, 95% CI 1.35–3.12). A statistically significant increased risk of cervical cancer has also been observed for the T allele of rs217727 (OR 1.53, 95% CI 1.17–2.02).39 Another previous study by Yang et al32 found that the rs217727 CT + TT genotypes were correlated with significantly increased gastric cancer risk (OR 1.32, 95% CI 1.01–1.71), while in functional assay analysis, the rs217727 CT, TT, or CT + TT genotypes were found not to affect H19 mRNA expression levels compared with the CC genotype, suggesting the C/T mutation might potentially change translational efficiency and lead to a transformation in H19 structure, thereby influencing the function of H19.
In our study, we confirmed that the T variant of rs217727 was consistently associated with an increased risk of BC in the codominant model, as well as in the dominant model (OR 1.25, 95% CI 1.03–1.51, OR 1.56, 95% CI 1.15–2.09, and OR 1.31, 95% CI 1.09–1.57, respectively), with positive results all remaining significant after Bonferroni correction (P<0.025). Further stratified analysis among different subgroups of demographic characteristics and reproductive factors revealed that the elevated risks of rs217727 T variant were more evident in subgroups of younger patients (age <40 years), higher BMI individuals (BMI ≥23 kg/m2), premenopausal women, and subjects with later menarche, later menopause, earlier age at first live birth, and fewer pregnancies (two or fewer). These results were similar to previous studies by Hua et al34 and Yang et al,32 which identified that the variant genotypes of rs217727 were more pronounced in younger subjects in bladder cancer and gastric cancer. However, none of the subgroups passed the threshold for Bonferroni correction (P<0.0035), and thus were not involved in the subsequent FPRP calculation. It is well known that both genetic and environmental factors play important roles in the development of BC, so future larger studies with more subjects are still necessary to clarify better whether the H19 functional polymorphism rs217727 could have a gene–environment interaction in BC etiology. In stratified analysis of specific molecular subtypes, case-only studies according to ER, PR, and HER2 status were conducted. We noticed that the T variant of rs217727 was significantly correlated with ER-positive patients (OR 1.34, 95% CI 1.09–1.64), which was consistent with an investigation by Xia et al.33 This may partly have been due to H19 being a hormone-dependent gene and the overexpression of H19 being always accompanied by the presence of steroid receptors.40,41 A previous study also identified that H19 expression can present a positive relation with ERα expression in BC tumors and that the estrogen–ERα–H19 signaling axis might play an important role in regulating proliferation and differentiation potentials of normal luminal progenitors, as well as the development of ER-positive BC tumors.42 However, for the HER2 subgroup, positive associations were observed only in HER2-negative patients (OR 1.4, 95% CI 1.14–1.71) and not in HER2-positive patients (OR 1.07, 95% CI 0.81–1.41). In addition, the correlations were more profound in the HR-positive–HER2-negative molecular subtype (OR 1.45, 95% CI 1.16–1.81), but not in the triple-negative BC subtype (OR 1.25, 95% CI 0.89–1.75). All these results imply that the functional SNP rs217727 in H19 is highly likely to be involved in BC development in hormone-signaling pathways.
The C/T polymorphism rs2839689 is also located in the exon (3′-untranslated region) of the H19 gene. Yang et al32 reported that the T allele of rs2839689 was associated with elevated risks of gastric cancer, and the CT and TT genotypes were found to be correlated with increased serum mRNA-expression levels compared with the CC genotype. Li et al43 also revealed that the rs2839698 T allele had significantly an increased risk of colorectal cancer, with further bioinformatic analysis indicating that rs2839698 might be able to modulate promoter activity and change the folding structures of H19 by altering targeted microRNAs. It has been well demonstrated that the SNPs in lncRNAs can be directly regulated and modified by miRNAs.44,45 Also, SNPs might be a plausible cause of alterations in correlations between miRNAs and lncRNAs.46 Changes in target miRNAs could potentially affect the function and expression of lncRNA by genetic variants and ultimately modulate the risk of cancer and other diseases. In our study, we investigated the associations between the rs2839698 polymorphism and BC susceptibility for the first time. However, the distribution of rs2839698 genotypes between cases and cancer-free controls or within each subgroup indicated no significant differences. This may be explained by various environmental exposure and etiologies of diverse cancers, as well as differences in sampling in each investigation.
It has been well demonstrated that some genetic epidemiology studies tend to overestimate disease predisposition when conferred by a genetic polymorphism, and results are frequently for high probabilities of FP findings.47,48 Therefore, it is important to conduct FPRP analysis to verify if significant findings were chance findings or really noteworthy. In our study, the association between rs217727 CT + TT genotypes and BC risk remained noteworthy for all subjects (FPRP 0.197). As to stratified analysis, significantly increased risks were also observed in patients who were ER-positive, HER2-negative, and HR-positive–HER2-negative (FPRP 0.341, 0.114, and 0.141, respectively).
In conclusion, to the best of our knowledge, this is the second investigation on the association between H19 gene polymorphisms and BC susceptibility. In this hospital-based case–control study, we confirmed that the rs217727 C>T polymorphism was associated with an increased risk of BC. Further stratified analyses revealed that the association between BC risk and variant genotypes of rs217727 was more profound in patients who were ER-positive, HER2-negative, and HR-positive–HER2-negative. However, several limitations in the current study should also be mentioned. First, we included only two lncRNA H19 polymorphisms in the present study, while studies comprising more functional SNPs in H19 may be more capable of illuminating the precise role of genetic variants in BC carcinogenesis. Second, the sample size in this study was not large enough, which may have led to limited statistical power and impact on the precision and accuracy of results. Third, inherent selection and information bias may have been inevitable due to this form of hospital-based case–control design and the subjects enrolled in our study being restricted to a southeast China Han population. In spite of these limitations, the findings from our study are still informative for physicians and researchers in this field. Additional prospective population-based studies with larger samples involving different ethnicities, as well as further functional studies, are still needed to confirm our findings.
Acknowledgments
We would like to thank Minjun Lu, Jiantang Zhang, and Shuting Lin for their assistance in this study. The study was supported by grants from the National Nature Science Foundation (81302320), National Key Clinical Specialty Construction Program (201030404), and Sci-Tech Key Program of Fujian Province (2016J01549).
Footnotes
Author contributions
CW and FMF conceived and designed the experiments. YXL and YZC performed the experiments. MH analyzed the data. YXL wrote the manuscript. All authors contributed toward data analysis, drafting, and critically revising the paper; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 5.McPherson K, Steel C, Dixon J. Breast cancer: epidemiology, risk factors, and genetics. BMJ. 2000;321(7261):624–628. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133–140. doi: 10.1016/S1470-2045(00)00254-0. [DOI] [PubMed] [Google Scholar]
- 7.Kelsey JL, Gammon MD. The epidemiology of breast cancer. CA Cancer J Clin. 1991;41(3):146–165. doi: 10.3322/canjclin.41.3.146. [DOI] [PubMed] [Google Scholar]
- 8.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 11.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long J, Cai Q, Shu XO, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6(6):e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Chen GK, Stram DO, et al. A genome-wide association study of breast cancer in women of African ancestry. Hum Genet. 2013;132(1):39–48. doi: 10.1007/s00439-012-1214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fejerman L, Ahmadiyeh N, Hu D, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun. 2014;5:5260. doi: 10.1038/ncomms6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting CP, Oliver PL, Reik W. Evolution and functions of long non-coding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 23.Koukoura O, Sifakis S, Zaravinos A, et al. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32(1):51–57. doi: 10.1016/j.placenta.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Park JY, Lee JE, Park JB, Yoo H, Lee SH, Kim JH. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Res Treat. 2014;2(1):1–6. doi: 10.14791/btrt.2014.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10(23):2619–2626. doi: 10.1093/hmg/10.23.2619. [DOI] [PubMed] [Google Scholar]
- 26.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 27.Barrow TM, Barault L, Ellsworth RE, et al. Aberrant methylation of imprinted genes is associated with negative hormone receptor status in invasive breast cancer. Int J Cancer. 2015;137(3):537–547. doi: 10.1002/ijc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adriaenssens E, Dumont L, Lottin S, et al. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol. 1998;153(5):1597–1607. doi: 10.1016/S0002-9440(10)65748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Yang F, Yuan JH, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34(3):577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 30.Gao T, He B, Pan Y, et al. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clin Transl Oncol. 2014;16(4):410–417. doi: 10.1007/s12094-013-1098-x. [DOI] [PubMed] [Google Scholar]
- 31.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280(7):1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 32.Yang C, Tang R, Ma X, et al. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget. 2015;6(17):15311–15320. doi: 10.18632/oncotarget.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Z, Yan R, Duan F, Song C, Wang P, Wang K. Genetic polymorphisms in long noncoding RNA H19 are associated with susceptibility to breast cancer in Chinese population. Medicine (Baltimore) 2016;95(7):e2771. doi: 10.1097/MD.0000000000002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua Q, Lv X, Gu X, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. 2016;31(5):531–538. doi: 10.1093/mutage/gew018. [DOI] [PubMed] [Google Scholar]
- 35.Verhaegh GW, Verkleij L, Vermeulen SH, den Heijer M, Witjes JA, Kiemeney LA. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. 2008;54(5):1118–1126. doi: 10.1016/j.eururo.2008.01.060. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumor Biol. 2014;35(9):9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 38.Gao W, Zhu M, Wang H, et al. Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat Res. 2015;772:15–22. doi: 10.1016/j.mrfmmm.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Jin T, Wu X, Yang H, et al. Association of the miR-17-5p variants with susceptibility to cervical cancer in a Chinese population. Oncotarget. 2016;7(47):76647–76655. doi: 10.18632/oncotarget.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adriaenssens E, Lottin S, Dugimont T, et al. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18(31):4460–4473. doi: 10.1038/sj.onc.1202819. [DOI] [PubMed] [Google Scholar]
- 41.Zhang K, Luo Z, Zhang Y, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17(2):187–194. doi: 10.3233/CBM-160630. [DOI] [PubMed] [Google Scholar]
- 42.Basak P, Chatterjee S, Weger S, Bruce MC, Murphy LC, Raouf A. Estrogen regulates luminal progenitor cell differentiation through H19 gene expression. Endocr Relat Cancer. 2015;22(4):505–517. doi: 10.1530/ERC-15-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Hua Y, Jin J, et al. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. 2016;7(18):25470–25477. doi: 10.18632/oncotarget.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kallen AN, Zhou X-B, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis: a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 48.Colhoun HM, McKeigue PM, Smith GD. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361(9360):865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]