Abstract
Transforming growth factor-β (TGF-β) induces epithelial-mesenchymal transition (EMT) primarily via a Smad-dependent mechanism. However, there are few studies available on TGF-β-induced EMT through the activation of non-canonical pathways. In this study, the Cdc42-interacting protein-4 (CIP4)/partitioning-defective protein 6 (Par6) pathway was investigated in TGF-β1-stimulated NRK-52E cells. Rat NRK-52E cells were obtained and stimulated with TGF-β1. The expression levels of E-cadherin, α-smooth muscle actin (α-SMA) and CIP4 were then examined by western blot analyses. Rat NRK-52E cells were transfected with Par6 or CIP4 small interfering RNA (siRNA), and scrambled siRNA as controls. The cells were incubated with 20 ng/ml of TGF-β1 for 72 h in order to observe the effects of Par6 and CIP4 silencing. Confocal fluorescence microscopy was also applied to reveal the expression and distribution of E-cadherin, α-SMA, Par6 and CIP4. The results demonstrated that E-cadherin expression was decreased, and α-SMA expression was increased in the TGF-β1-stimulated cells. Simultaneously, the increased expression of CIP4 and p-Par6 was confirmed by western blot analyses. The results of confocal fluorescence microscopy revealed that rat CIP4 exhibited cluster formations located adjacent to the cell periphery; however, as for the protein expression and distribution of Par6, there was no obvious difference between the control cells and cells exposed to TGF-β1. siRNA molecules capable of CIP4 and Par6 knockdown were used to demonstrate reversed TGF-β1-induced EMT. Moreover, CIP4 loss of function reversed the increase in p-Par6 protein expression in the TGF-β1-stimulated NRK-52E cells. A similar result was observed with the decreased CIP4 protein expression due to Par6 loss of function. Our data thus suggest that the CIP4/Par6 complex plays an important role in the occurrence of EMT in TGF-β1-stimulated NRK-52E cells. The underlying mechanisms are mediated, at least in part, through the upregulation of CIP4, which occurrs due to stimulation with TGF-β1; subsequently, CIP4 increases the phosphorylation of Par6, which accelerates the process of EMT.
Keywords: Cdc42-interacting protein-4, partitioning-defective protein 6, epithelial-mesenchymal transition, transforming growth factor-β1
Introduction
Epithelial-mesenchymal transition (EMT) is a fundamental process driving morphogenesis in most metazoans conserved throughout evolution. Cells engaged in the EMT program will undergo complex changes in intercellular junctions, adhesive structures and apicobasal polarization (1,2). The process of EMT is known to be involved in various physiological and pathological states, including organ fibrosis (3,4), cancer metastasis (5) and resistance to chemotherapy (6). Recently, increasing evidence has shed light on the transforming growth factor-β1 (TGF-β1), which is a well-known inducer of EMT in epithelial cells (7,8). It binds to its receptors (TβRI, TβRII and TβRIII) and activates various transcription factors for cadherin isoform switching (9). TGF-β induces EMT primarily via a Smad-dependent mechanism. Smad2/3 is phosphorylated by Smad4 and then translocates to the nucleus, where it regulates the transcription of the target genes responsible for EMT (10,11). However, there are few studies available on TGF-β-induced EMT through the activation of non-canonical pathways (12). For example, it is still not well characterized that the Cdc42-interacting protein-4 (CIP4)/partitioning-defective protein 6 (Par6) pathway is responsible for TGF-β-induced EMT in NRK-52E cells.
Par6 is part of the Par polarity complex that localizes to the tight junction (TJ) and is comprised of 3 highly conserved proteins, Par6, Par3 and atypical protein kinase C (aPKC) (13). Previous studies have shown that Par6, a regulator of epithelial cell polarity and TJ assembly, is directly phosphorylated (at Ser345) and is activated by TβRII in response to TGF-β, which is essential for TGF-β-induced EMT and facilitates metastasis (13–15). CIP4 belongs to the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Members of this superfamily are noted for their involvement in membrane remodeling processes that occur in various cellular pathways, such as endocytosis, cytokinesis, T-tubule formation, cell migration and neuromorphogenesis (16). Earlier studies have demonstrated that CIP4 is capable of inducing extracellular matrix (ECM) deposition and exacerbating progressive fibrosis in chronic renal failure and is required for E-cadherin endocytosis (16–19). The interaction between Cdc42/Par6/aPKC and CIP4, as a downstream effector protein of Cdc42, may act as a link between Cdc42 signaling and the regulation of the actin cytoskeleton (17,20). However, the interaction between CIP4 and Par6 in NRK-52E cells undergoing TGF-β-induced EMT remains largely unknown.
In the present study, we demonstrated that the expression of CIP4 was significantly increased in NRK-52E cells stimulated with TGF-β1. Intriguingly, CIP4 lose-of-function with small interfering RNA (siRNA) reversed TGF-β1-induced EMT in NRK-52E cells. Moreover, we found that there was an interaction between Par6 and CIP4 in TGF-β1-stimulated NRK-52E cells undergoing EMT.
Materials and methods
Cell culture
Rat NRK-52E cells were obtained from the Chinese Academy of Sciences (Institute of Shanghai Cell Biology and Chinese Type Culture Collection, Shanghai, China), and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Life Technologies, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen Life Technologies) at 37°C in a humidified, 5% CO2, 95% air atmosphere. The medium was replenished every day. The cells were stimulated with TGF-β1 at 20 ng/ml.
Gene silencing by siRNA
For siRNA experiments, RNA primers complementary to rat Par6 and CIP4 were designed and synthesized by the Invitrogen Biotechnology Co., Ltd. (Shanghai, China). The NRK-52E cells were transfected with the annealed RNA primer pair using Lipofectamine 2000 (Invitrogen Life Technologies) in accordance with the instructions provided by the manufacturer. The cells transfected with scrambled siRNA served as controls. Five hours after transfection, the cells were incubated with 20 ng/ml of TGF-β1 for 72 h in order to observe the effects of Par6 and CIP4 silencing. The siRNA used had the following primers: Par6 forward, 5′-CUCACUGAGUGCGACAAGGUCUUCA-3′ and reverse, 5′-AGAAGAGGUUGUCGCUCACACACUG-3′; CIP4 forward, 5′-GCUAACAGUCGACUGUGCUAGUGU-3′ and reverse, 5′-UCGACAACGAGAGUGGACUCUAGC-3′; scramble forward, 5′-GACCAGUCGCUAUCACACAUGUCA-3′ and reverse, 5′-AGAUGACGAGUCUUACGCACUCGU-3′.
Reverse transcription-polymerase chain reaction (RT-PCR)
RNA extraction was performed using TRIzol reagent according to the manufacturer's instructions (Invitrogen Life Technologies). RNA integrity was verified by agarose gel electrophoresis. The synthesis of cDNA was performed by reverse transcription reactions with 2 μg of total RNA using moloney murine leukemia virus reverse transcriptase (Invitrogen Life Technologies) with oligo(dT) (15) primers (Fermentas, Pittsburgh, PA, USA) as described by the manufacturer. The first-strand cDNA served as the template for the regular PCR performed using a DNA engine (ABI 7300; Applied Biosystems, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control was used to normalize the data to determine the relative expression of the target genes. The reaction conditions were set according to the kit instructions. The PCR primers used in this study were as follows: Par6 forward, 5′-GCACGCAGAATGATGACGATT-3′ and reverse, 5′-GTCGTGCTACGATCGTAGTA-3′; CIP4 forward, 5′-ACACGGAGTTTGATGAGGAT-3′ and reverse, 5′-ATGGTGGAACGATGGTAGAA-3′; GAPDH forward, 5′-GGATTTGGTCGTATTGGG-3′ and reverse, 5′-GGAAGATGGTGATGGGATT-3′.
Immunoprecipitation (IP) and immunoblotting (IB)
Cell lysates were prepared as previously described (21). The lysates were then incubated with the indicated antibodies for 1 h at 4°C. The immune complexes were precipitated with protein A/G agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 1 h at 4°C, washed extensively with lysis buffer, resolved in 4–20% gradient SDS-PAGE, and analyzed by immunoblotting. All immunoblots were developed by enhanced chemiluminesence (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA).
Western blot analysis
The NRK-52E cells were homogenized and extracted in NP-40 buffer, followed by 5–10 min boiling and centrifugation to obtain the supernatant. Samples containing 50 μg of protein were separated on a 10% SDS-PAGE gel and transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). After saturation with 5% (w/v) non-fat dry milk in TBS and 0.1% (w/v) Tween-20 (TBST), the membranes were incubated with the following antibodies: E-cadherin (sc-71008; Santa Cruz Biotechnology, Inc.), α-SMA (SAB5500002; Sigma-Aldrich, St. Louis, MO, USA), rCIP4 (ab72220; Abcam, Cambridge, MA, USA), Par6 (sc-365323) and p-Par6 (both from Santa Cruz Biotechnoogy, Inc.), at dilutions ranging from 1:500 to 1:2,000 at 4°C overnight. Following 3 washes with TBST, the membranes were incubated with secondary immunoglobulins (Igs) conjugated to IRDye 800CW Infrared Dye (LI-COR Biotechnology, Lincoln, NE, USA), including donkey anti-goat IgG and donkey anti-mouse IgG at a dilution of 1:10,000–1:20,000. Following 1 h of incubation at 37°C, the membranes were washed 3 times with TBST. The blots were visualized by the Odyssey Infrared Imaging System (LI-COR Biotechnology). Signals were densitometrically assessed (Odyssey Application software version 3.0) and normalized to the GAPDH or β-actin signals to correct for unequal loading using the mouse monoclonal anti-GAPDH (MB001H) or anti-β-actin antibody (BS6007MH) (both from Bioworld Technology, Inc., St. Louis Park, MN, USA), respectively.
Confocal fluorescence microscopy
For immunocytochemical analysis, the NRK-52E cells were cultured on sterile glass cover-slips in 6-well plates. Thereafter, the cells were treated and fixed with iced acetone for 10 min, incubated with 0.1% Triton X-100 for 10 min in order to induce membrane rupture, and incubated with 1% BSA at 37°C for an additional 30 min for blocking. The cells were then incubated with anti-E-cadherin (sc-71008; Santa Cruz Biotechnology, Inc.), anti-α-SMA (55135-1-AP; Proteintech Group, Inc., St. Pearl, IL, USA), anti-Par6 (sc-365323; Santa Cruz Biotechnology, Inc.) and anti-CIP4 (ab72220; Abcam) antibodies (1:100) overnight at 4°C, followed by incubation with FITC-goat anti-rabbit IgG (SA00003-2; Proteintech Group, Inc.) at 37°C for 1 h. Thereafter, the nuclei were stained with DAPI (Sigma-Aldrich) for 5 min and analyzed using a laser confocal scanning microscope (Leica, Heidelberg, Germany). The imaging using the laser confocal scanning microscopy for the cell culture is better than that of total internal reflection fluorescence (TIRF) microscopy (22) that can only exploit a limited region adjacent to the substrate.
Statistical analysis
The data from these experiments are reported as the means ± standard error of the mean (SEM) for each group. All statistical analyses were performed by using GraphPad Prism version 5.0 software (GraphPad Software, La Jolla, CA, USA). Inter-group differences were analyzed by one-way analysis of variance (ANOVA), followed by Tukey's multiple comparison test as a post-test to compare the group means with an overall value of P<0.05. Differences with a P-value <0.05 were considered statistically significant.
Results
TGF-β1-induced EMT in NRK-52E cells
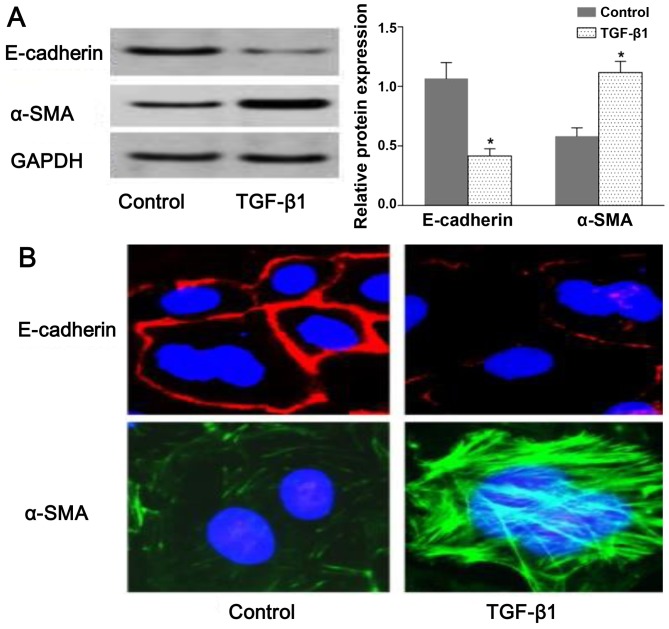
To assess TGF-β-induced EMT, we used rat NRK-52E cells as an in vitro model system for assessing EMT. The resutls of western blot analysis demonstrated that the expression of E-cadherin and α-SMA was decreased and increased, respectively, in the TGF-β1-stimulated group compared with the untreated group (Fig. 1A). Consistent with the results of western blot analysis, we observed a change in E-cadherin and α-SMA distribution in the NRK-52E cells following exposure to TGF-β by confocal fluorescence microscopy. The morphologic observations of the NRK-52E cells revealed a continuous distribution of E-cadherin near the perimeter of the control cells. On the contrary, a discontinuous distribution of E-cadherin was observed in the TGF-β1-stimulated cells (Fig. 1B). Furthermore, α-SMA was present exclusively in the cytosol of the TGF-β1-stimulated cells, and little endogenous expression in the control cells was observed (Fig. 1B). The results shown in Fig. 1 suggested that the NRK-52E cells underwent EMT following exposure to TGF-β1.
Figure 1.
Transforming growth factor-β1 (TGF-β1)-induced epithelial-mesenchymal transition (EMT) in NRK-52E cells. NRK-52E cells were untreated (control) or incubated with TGF-β1 (20 ng/ml) for 72 h. (A) The protein expression levels of E-cadherin and α-SMA were measured by western blot analyses and densitometric analyses. E-cadherin and α-SMA were visualized by confocal fluorescence microscopy. (B) Cell nuclei were enhanced by staining of the cell nuclei with DAPI for E-cadherin (×600 magnification). Values are expressed as the means ± SEM, n=3 in each group. *P<0.05 vs. control group.
TGF-β1 regulates the expression of CIP4 in rat NRK-52E cells
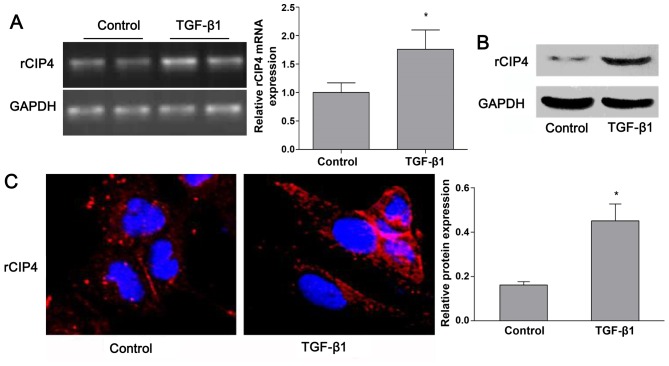
Based on the previous findings shown in the 5/6-nephrectomy rat model (23), the expression of rat CIP4 (rCIP4) in the NRK-52E cells stimulated with TGF-β1 in vitro was demonstrated. Following stimulation with TGF-β1 (20 ng/ml) for 72 h, the mRNA and protein levels of rCIP4 in NRK-52E cells were upregulated as compared to those of the control group (Fig. 2A and B). Confocal fluorescence microscopy revealed that rCIP4 exhibited a punctate localization throughout the cytosol, with the highest levels localized in the perinuclear region of the NRK-52E cells. Following exposure to TGF-β1, the rCIP4 levels increased, and rCIP4 was recruited into cluster formations located adjacent to the cell periphery. Gradually, these clusters were observed to be redistributed into the cytoplasm (Fig. 2C).
Figure 2.
Transforming growth factor-β1 (TGF-β1) regulates the expression of Cdc42-interacting protein-4 (CIP4) in rat NRK-52E cells. NRK-52E cells were untreated (control) or incubated with TGF-β1 (20 ng/ml) for 72 h, and (A) the mRNA and (B) protein expression levels of CIP4 were measured by RT-PCR and western blot analyses, respectively, and bar graphs represent the values are corrected for the loading control, GAPDH. (C) E-cadherin and α-SMA were visualized by confocal fluorescence microscopy (×600 magnification). Values are expressed as the means ± SEM, n=3 in each group. *P<0.05 vs. control group.
TGF-β1 upregulates the expression of p-Par6 in rat NRK-52E cells
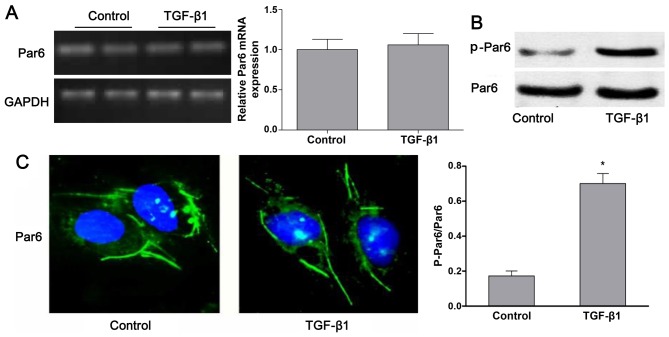
As shown in Fig. 3B, the protein expression of p-Par6 was increased by TGF-β1 stimulation as compared to the control group. There was no obvious difference in the mRNA and protein expression of Par6 between the control cells and the cells exposed to TGF-β1; this was confirmed by RT-PCR, western blot analyses and confocal fluorescence microscopy (Fig. 3A–C).
Figure 3.
Transforming growth factor-β1 (TGF-β1) upregulates the expression of p-Par6 in rat NRK-52E cells. NRK-52E cells were untreated (control) or incubated with TGF-β1 (20 ng/ml) for 72 h. (A) The mRNA expression of Par6 was measured by RT-PCR. (B) Representative western blots of Par6 and p-Par6 in control cells and TGF-β1-stimulated cells. The histogram shows the average volume density corrected for the loading control, GAPDH. (C) Par6 was visualized by confocal fluorescence microscopy (×600 magnification). Values are expressed as the means ± SEM, n=3 in each group. *P<0.05 vs. control group.
Stimulation with TGF-β1 enhances the interaction between Par6 and CIP4
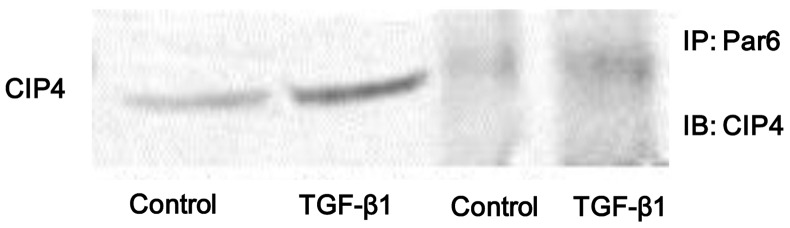
In the present study, we found that the protein expression of both p-Par6 and CIP4 was significantly increased in the NRK-52E cells stimulated with TGF-β1 in vitro. Thus, in order to examine whether the interaction between Par6 and CIP4 is dependent on TGF-β1, the cell lysates were prepared from TGF-β1-stimulated cells and immunoprecipitated with anti-Par6 antibody. The results indicated that the binding capacity between Par6 and CIP6 was weak in the control cells, but was increased in the NRK-52E cells stimulated with TGF-β1 (Fig. 4). Therefore, our data suggest that the interaction between Par6 and CIP6 may play a vital role in TGF-β1-induced EMT.
Figure 4.
Transforming growth factor-β1 (TGF-β1) stimulation enhances the interaction between partitioning-defective protein 6 (Par6) and Cdc42-interacting protein-4 (CIP4). NRK-52E cells were untreated (control) or incubated with TGF-β1 (20 ng/ml) for 72 h, and CIP4 interacted with Par6 in vitro. Cell lysates were prepared from control cells and TGF-β1-stimulated cells. Cell lysates were immunoprecipitated with anti-Par6 antibody. Immunopellets were analyzed by immunoblot analysis using anti-CIP4 antibody. IB, immunoblotting; IP, immunoprecipitation.
Silencing of Par6 inhibits TGF-β1-induced EMT
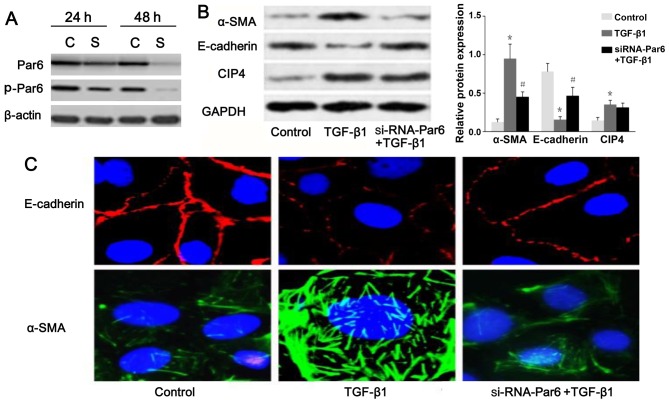
To inhibit the function of Par6, the NRK-52E cells were transfected with siRNA. The results of western blot analysis demonstrated that the expression of p-Par6 was markedly inhibited by transfection with siRNA-Par6. Moreover, the protein expression of p-Par6 was decreased with increasing treatment durations (Fig. 5A). Therefore, our data suggested that the siRNA experiments were successfully performed. As shown in Fig. 5B and C, as the E-cadherin protein levels increased, a marked decrease in α-SMA expression was clearly evident following transfection of the TGF-β1-stimulated cells with Par6-siRNA. The NRK-52E cells transfected with Par6-siRNA, however, demonstrated resistance to TGF-β1-induced EMT.
Figure 5.
Silencing of partitioning-defective protein 6 (Par6) inhibits transforming growth factor-β1 (TGF-β1)-induced epithelial-mesenchymal transition (EMT). (A) Small interfering RNA (siRNA) was transfected into NRK-52E cells for 24 and 48 h to suppress the expression of Par6 and p-Par6, which was measured by western blot analysis. C, si-RNA-control; S, siRNA-Par6. The NRK-52E cells were untreated (control), or incubated with TGF-β1 (20 ng/ml) for 71 h, or incubated with TGF-β1 (20 ng/ml) combined with Par6-siRNA for 72 h. (B) The protein expression levels of E-cadherin, α-SMA and Cdc42-interacting protein-4 (CIP4) were measured by western blot analysis and densitometric analyses. E-cadherin and α-SMA were visualized by confocal fluorescence microscopy. (C) Cell nuclei were enhanced by staining of the cell nuclei with DAPI for E-cadherin (×600 magnification). Values are expressed as the means ± SEM, n=3 in each group. *P<0.05 vs. control group; #P<0.05 vs. TGF-β1-stimulted group.
CIP4 is involved in TGF-β1-induced EMT and the regulation of Par6 expression
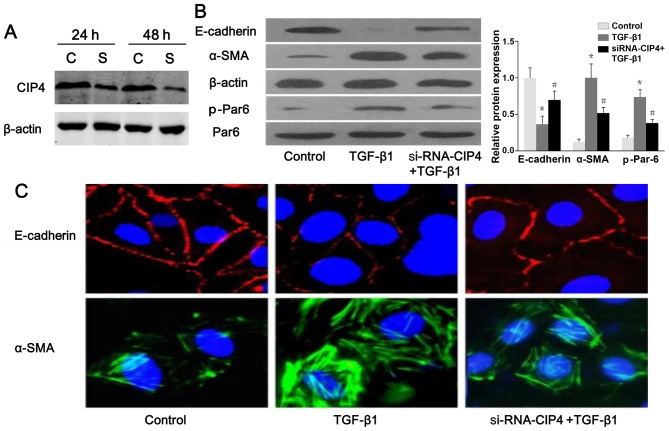
The results of western blot analysis and confocal fluorescence microscopy indicated a reduced protein expression and a discontinuous distribution of E-cadherin near the perimeter of TGF-β1-stimulated cells (Fig. 6A–C). Furthermore, α-SMA was present exclusively in the cytosol of TGF-β1-stimulated cells, and little endogenous expression was observed in the control cells (Fig. 6B and C), suggesting that these cells underwent EMT in response to TGF-β1. Following treatment with CIP4-siRNA, E-cadherin re-localized to regions surrounding the cellular junctions, and a reduction in α-SMA expression was observed compared with the TGF-β1-stimulated cells (Fig. 6B and C). Intriguingly, the protein level of p-Par6 in the TGF-β1-stimulated cells transfected with CIP4-siRNA was downregulated as compared to that of the TGF-β1-treated group (Fig. 6B). Therefore, our data suggest that CIP4 is involved in TGF-β1-induced EMT, and the underlying mechanisms are mediated, at least in part, through the upregulation of the expression of p-Par6.
Figure 6.
Cdc42-interacting protein-4 (CIP4) is involved in transforming growth factor-β1 (TGF-β1)-induced epithelial-mesenchymal transition (EMT) and regulation of partitioning-defective protein 6 (Par6) expression. (A) Small interfering RNA (siRNA) was transfected into NRK-52E cells for 24 and 48 h to suppress the expression of CIP4, which was measured by western blot analysis. C, si-RNA-control; S, siRNA-Par6. (B) Representative western blot of E-cadherin, α-SMA, Par6 and p-Par6 in control cells, TGF-β1-stimulated cells and TGF-β1-stimulated CIP4-siRNA cells. E-cadherin and α-SMA were visualized by confocal fluorescence microscopy. (C) Cell nuclei were enhanced by staining of the cell nuclei with DAPI for E-cadherin (×600 magnification). Values are expressed as the means ± SEM, n=3 in each group. *P<0.05 vs. control group; #P<0.05 vs. TGF-β1-stimulated group.
Discussion
TGF-β1 promotes the development of renal tubule interstitial fibrosis through EMT. In this study, the upregulation of rCIP4 and p-Par6 was demonstrated in NRK-52E cells stimulated with TGF-β1. Conversely, CIP4 or Par6 loss of function by siRNA reversed TGF-β1-induced EMT by increasing E-cadherin (an epithelial marker) and reducing α-SMA (a mesenchymal marker) expression. These findings indicated that CIP4/Par6 was involved in the TGF-β1-induced EMT processes, and subsequently in renal fibrosis.
In epithelial tissue, CIP4 has been identified to regulate the early E-cadherin trafficking/endocytosis (17), which is the key process of EMT in renal tubular epithelial cell (16,24). In 5/6-nephrectomized rats, the expression of rCIP4 has been shown to be increased in the renal tubules, and the overexpression of hCIP4 has been demonstrated to promote the development of EMT in HK-2 cells exposed to TGF-β1 (16). Moreover, Par6, a conserved polarity protein, regulates cell polarization processes. However, increasing evidence also suggests that Par6 is phosphorylated to facilitate EMT (25). Given the recent important roles reported for Par6 in the generation and progression of various types of cancer (26,27), Par6 phosphorylation may be central to various extrinsic cues that can lead to the EMT (28). It has also been revealed that Par6 can have different functions depending on the interacting partners (28). Furthermore, in cell-culture systems, E-cadherin endocytosis was identified as a model of EMT (25,26), and CIP4 acts as a link between Cdc42/Par6/aPKC and the early endocytic machinery to regulate E-cadherin endocytosis in epithelial cells. Taken together, our data suggested that CIP4/Par6 have a close interaction in the process of EMT induced by TGF-β1.
To more accurately determine the location of CIP4 and Par6, we provided superior assessment of the role of CIP4 and Par6 location in NRK-52E cells. Immunofluorescence laser scanning confocal microscopy showed that CIP4 exhibited punctate localization throughout the cytosol. Following stimulation with TGF-β1, CIP4 was increased and formed visible clusters adjacent to the cell periphery that gradually redistributed into the cytoplasm. These findings suggest that the upregulation of CIP4 plays an important role in the occurrence of EMT in NRK-52E cells. However, the cluster distribution of Par6 has no obvious difference in cell periphery. Interestingly, stimulation with TGF-β1 led to a closer interaction between CIP4 and Par6 in NRK-52E cells. Our results were measured by co-immunoprecipitation analysis, which showed that the binding capacity of CIP4 and Par6 was increased in TGF-β1-stimulated NRK-52E cells. CIP4 loss of function by siRNA reversed the increase in p-Par6 protein expression in TGF-β1-stimulated NRK-52E cells. A similar result was observed by decreasing of CIP4 protein expression due to Par6 loss of function by siRNA. These results suggested that there was an interaction between CIP4 and Par6, and that Par6 phosphorylation may be regulated by CIP4.
In conclusion, our data demonstrate that CIP4 and Par6 play an important role in the occurrence of EMT in TGF-β1-stimulated NRK-52E cells. The underlying mechanisms are mediated, at least in part, through the upregulation of the expression of CIP4 and p-Par6.
Acknowledgments
We would like to acknowledge the support by Natural Science Foundation of Shanghai (13ZR1407200).
References
- 1.Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Chua KN, Poon KL, Lim J, Sim WJ, Huang RY, Thiery JP. Target cell movement in tumor and cardiovascular diseases based on the epithelial-mesenchymal transition concept. Adv Drug Deliv Rev. 2011;63:558–567. doi: 10.1016/j.addr.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Simic P, Williams EO, Bell EL, Gong JJ, Bonkowski M, Guarente L. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Reports. 2013;3:1175–1186. doi: 10.1016/j.celrep.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarino M, Tosoni A, Nebuloni M. Direct contribution of epithelium to organ fibrosis: epithelial-mesenchymal transition. Hum Pathol. 2009;40:1365–1376. doi: 10.1016/j.humpath.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Zhao D, Besser AH, Wander SA, Sun J, Zhou W, Wang B, Ince T, Durante MA, Guo W, Mills G, et al. Cytoplasmic p27 promotes epithelial-mesenchymal transition and tumor metastasis via STAT3-mediated Twist1 upregulation. Oncogene. 2015;43:5447–5459. doi: 10.1038/onc.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng L, Shi Y, Wang H, Yin B, Xia J, et al. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–1749. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung H, Choi HS, Seo EK, Kang DH, Oh ES. Baicalin and baicalein inhibit transforming growth factor-β1-mediated epithelial-mesenchymal transition in human breast epithelial cells. Biochem Biophys Res Commun. 2015;458:707–713. doi: 10.1016/j.bbrc.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Liu N, Su X, Zhou G, Sun G, Du F, Bian X, Wang B. Epigallocatechin-3-gallate attenuates transforming growth factor-beta1 induced epithelial-mesenchymal transition via Nrf2 regulation in renal tubular epithelial cells. Biomed Pharmacother. 2015;70:260–267. doi: 10.1016/j.biopha.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci. 1999;112:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 10.Bae E, Kim SJ, Hong S, Liu F, Ooshima A. Smad3 linker phosphorylation attenuates Smad3 transcriptional activity and TGF-β1/Smad3-induced epithelial-mesenchymal transition in renal epithelial cells. Biochem Biophys Res Commun. 2012;427:593–599. doi: 10.1016/j.bbrc.2012.09.103. [DOI] [PubMed] [Google Scholar]
- 11.Meng XM, Huang XR, Chung AC, Qin W, Shao X, Igarashi P, Ju W, Bottinger EP, Lan HY. Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–1487. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avery-Cooper G, Doerr M, Gilbert RW, Youssef M, Richard A, Huether P, Viloria-Petit AM. Par6 is an essential mediator of apoptotic response to transforming growth factor beta in NMuMG immortalized mammary cells. Cancer Cell Int. 2014;14:19. doi: 10.1186/1475-2867-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 15.Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, Roncari L, Narimatsu M, Bose R, Moffat J, et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci USA. 2009;106:14028–14033. doi: 10.1073/pnas.0906796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai S, Zeng R, Zhou Q, Liao W, Zhang Y, Xu C, Han M, Pei G, Liu L, Liu X, et al. Cdc42-interacting protein-4 promotes TGF-B1-induced epithelial-mesenchymal transition and extracellular matrix deposition in renal proximal tubular epithelial cells. Int J Biol Sci. 2012;8:859–869. doi: 10.7150/ijbs.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–1648. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 18.Wirtz-Peitz F, Zallen JA. Junctional trafficking and epithelial morphogenesis. Curr Opin Genet Dev. 2009;19:350–356. doi: 10.1016/j.gde.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 20.Aspenström P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr Biol. 1997;7:479–487. doi: 10.1016/S0960-9822(06)00219-3. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Kimura A, Baumann CA, Saltiel AR. APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol Cell Biol. 2002;22:3599–3609. doi: 10.1128/MCB.22.11.3599-3609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, He C, Stoykovich MP, Schwartz DK. Nanoscale topography influences polymer surface diffusion. ACS Nano. 2015;9:1656–1664. doi: 10.1021/nn506376n. [DOI] [PubMed] [Google Scholar]
- 23.Xu C, Zhou Q, Liu L, Liu P, Pei G, Zeng R, Han M, Xu G. Cdc42-interacting protein 4 represses E-Cadherin expression by promoting β-Catenin translocation to the nucleus in murine renal tubular epithelial cells. Int J Mol Sci. 2015;16:19170–19183. doi: 10.3390/ijms160819170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng G, Lyons JG, Tan TK, Wang Y, Hsu TT, Min D, Succar L, Rangan GK, Hu M, Henderson BR, et al. Disruption of E-cadherin by matrix metalloproteinase directly mediates epithelial-mesenchymal transition downstream of transforming growth factor-beta1 in renal tubular epithelial cells. Am J Pathol. 2009;175:580–591. doi: 10.2353/ajpath.2009.080983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunaratne A, Di Guglielmo GM. Par6 is phosphorylated by aPKC to facilitate EMT. Cell Adhes Migr. 2013;7:357–361. doi: 10.4161/cam.25651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, Muthuswamy SK. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan L, Shen Y, Lu Z, Shang D, Zhao Z, Lu Y, Wu Y, Zhang Y, Tu Z, Liu H. Roles of partitioning-defective protein 6 (Par6) and its complexes in the proliferation, migration and invasion of cancer cells. Clin Exp Pharmacol Physiol. 2017;44:909–913. doi: 10.1111/1440-1681.12794. [DOI] [PubMed] [Google Scholar]
- 28.Gunaratne A, Thai BL, Di Guglielmo GM. Atypical protein kinase C phosphorylates Par6 and facilitates transforming growth factor β-induced epithelial-to-mesenchymal transition. Mol Cell Biol. 2013;33:874–886. doi: 10.1128/MCB.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]