Figure 4. The effect of changing the p85 hinge region located between iSH2 and cSH2.
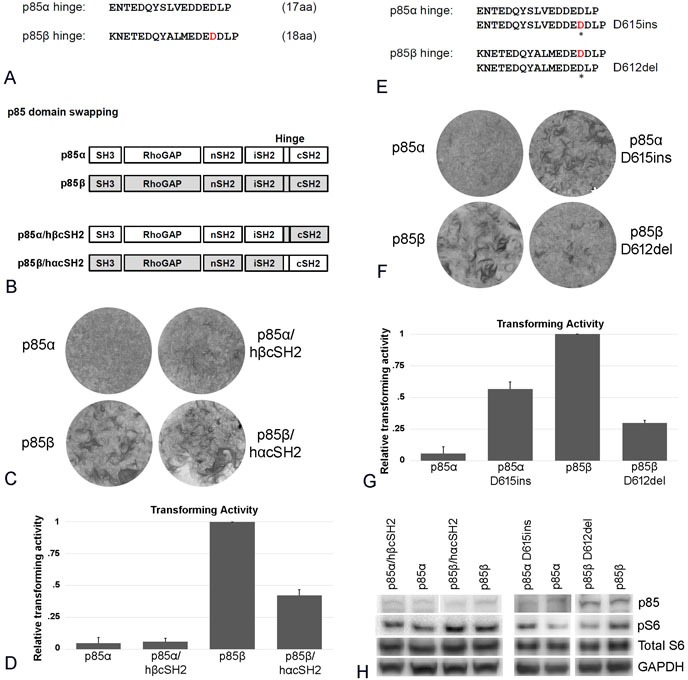
A. Comparison of the amino acid sequences of the hinge regions of p85α and p85β. B. Schematic representation of the p85 hinge region constructs. C. Representative focus assays of p85α and p85β wild type and of the hinge region-cSH2 exchange constructs depicted in B. D. Efficiencies of cellular transforming activity of p85 hinge region constructs relative to the transforming activity of wild type p85β. CEF were transfected with RCAS(A) constructs expressing p85β, p85α, p85β/hαcSH2, p85α/hβcSH2). Transforming activity was standardized to 0.5 μg transfected DNA. E. Sequence of single amino acid insertions and deletion in the p85 hinge regions (p85α D615ins and p85β D612del). F. Representative focus assays of single amino acid insertions or deletions in the hinge region of p85. G. Efficiencies of cellular transforming activity of p85 single amino acid insertion or deletion mutant constructs relative to the transforming activity of wild type p85β. CEF were transfected with RCAS(A) constructs expressing p85α, p85β, p85αD615ins, and p85βD612del. Transforming activity was standardized to 0.5 μg transfected DNA. H. PI3K signaling in cells expressing p85 hinge region constructs and the single-amino acid insertion or deletion mutants as reflected by Western blots of S6 phosphorylated at Ser235/236. The data in Figure 4H are from a representative experiment that was repeated three times with consistent results (for details, see the Materials and Methods section).
