Abstract
Renal cell carcinoma (RCC) is considered a cytokine-responsive tumor. However, with the lack of diagnostic screening biomarkers, early diagnosis of RCC is challenging. Our study was investigated the association of IL1 gene polymorphisms and RCC risk. We conducted a case-control study of 291 RCC cases and 463 controls to evaluation the IL1RN of single nucleotide polymorphisms (SNPs) on RCC risk. We selection of 16 SNPs in IL1RN, IL1A, IL1B genes were analyzed. Using the chi-squared (χ2) test and genetic model analysis, we found an association with RCC risk for five SNPs [rs3783550 (IL1A), rs3783546 (IL1A), rs1609682 (IL1A), rs3783521 (IL1A), and rs1143623 (IL1B)] and increased the risk of RCC. Stratified analyses show that smoking, not drinking and age>55 populations relative to nonsmoking, drinking and age<55 more susceptible. Our study suggested that IL1B and IL1A may involve in the development of RCC in Chinese Han population.
Keywords: renal cell carcinoma (RCC), case-control studies, IL1, single-nucleotide polymorphism (SNP)
INTRODUCTION
Kidney cancer accounting for 2% of all malignancies, it has become one of the most important health problems worldwide, cases are increasing year by year [1]. Renal cell carcinoma (RCC) is the major cancer type in kidney, and accounts for about 3% of all human malignancies, with a male-to-female ratio of approximately 2:1 [1, 2]. RCC risk factors included smoking tobacco [3], obesity [4, 5], hypertension [6], and a history of chronic kidney disease [7]. Besides the environmental risk factors, inherited risk has been considered crucial risk factors of RCC. A recent study has demonstrated that genetic factors contribute greatly to the occurrence of RCC. [8, 9]. Genome-wide association studies (GWAS) have shown EPAS1, HIF-2α, CCND1, ITPR2 were significantly associated with RCC susceptibility in western populations [10–13]. However, these GWAS identified RCC risk SNPs identified in western populations was rarely replicated in Chinese populations. The potential role of RCC risk in immune response has been demonstrated [14, 15]. RCC tissue is often infiltrated by lymphocytes, macrophages and dendritic cells, reflecting the immunogenicity of RCC [16–18]. The present pathogenic mechanism of RCC, because of these two inflammatory conditions, is characterized by the presence of a wide range of proinflammatory cytokines. It has also been shown a variety of cytokines such as tumor necrosis factor alpha (TNF- alpha), interleukin -6 (IL-6), type 1 interferons (IFNs) and interferon gamma in vitro [19–22]. The effects of cytokines observed in these studies were either growth promoting or antiproliferative [21]. In addition, polymorphisms in the immune response may explain human diversity because they lead to differences in the ability of individuals to produce cytokines, resulting in a wide variety of biological consequences. Although many studies investigating the frequency of cytokine gene polymorphisms appear in chronic kidney disease patients, there is still a lack of IL1 regarding how renal cancer can be related. Therefore, the purpose of this study was to investigate the IL-1 cytokine gene polymorphism and cytokine receptor gene polymorphism in RCC patients.
RESULTS
Demographic and clinical features of the RCC and the control group are shown in Table 1. The cases were well matched with the controls in terms of age (P = 0.273) and gender (P = 0.017), smoking status (P = 0.193). However, drinking status exist significant difference between two groups (P< 0.05). In Table 2, we listed the primers in the study. All SNPs were in Hardy–Weinberg equilibrium among control subjects, in addition to rs928940. The distributions of the allele frequencies for all selected polymorphisms are presented in Table 3. Our results indicated that polymorphism rs3783550, rs3783546, rs1609682 and rs3783521 in IL1A gene are associated with susceptibility to RCC. We hypothesized that harboring minor allele per SNP is a risk factor, compared with owning wild-type alleles. The results of various genetic models are shown in Table 4. Five SNPs were significantly associated with RCC risk in Chinese han population, and were used to establish the genetic risk models. Our results showed that the rs3783550 T allele carriers (dominant, Log-additive model) was associated with an increased risk of RCC (dominant, OR=1.39, 95%CI = 1.03-1.88, P= 0.031; Log-additive, OR=1.32, 95%CI = 1.05-1.64, P= 0.015). The genotype “C/C” and “G/C-C/C” of rs3783546 in IL1A gene was associated with of RCC risk in Codominant(OR = 1.77, 95% CI=1.09-2.87, p = 0.04), dominant model (OR=1.41, 95%CI = 1.04-1.91, P= 0.025). For rs1609682, carriers “T” allel individual, increased RCC risk (OR=1.32, 95%CI = 1.06-1.65, P= 0.013). The genotype “G/G” and “G/A-G/G” of rs3783521 was associated with an increased risk of RCC in Codominant, dominant, and Log-additive model (OR = 1.33, 95% CI= 1.06-1.66, p = 0.012). Meanwhile, we adjustment for age, sex, we found rs1143623(G) were associated with RCC risk in recessive model(OR = 1.54, 95% CI= 1.02-2.32, p = 0.04).
Table 1. Characteristics of RCC patients and control participants.
| Variable | Case | Control | P |
|---|---|---|---|
| Total | 291 | 463 | |
| Age(Mean±SD) | 56.88±11.66 | 50.65±11.79 | 0.273 |
| BMI(Mean±SD) | 23.96±2.90 | 23.79±3.77 | 0.098 |
| Gender | |||
| Female | 99 | 198 | 0.017 |
| Male | 192 | 265 | |
| Smoking status | |||
| Smoking | 120 | 169 | 0.193 |
| Nosmoking | 171 | 294 | |
| Drinking status | |||
| Drinking | 53 | 174 | <0.050 |
| Nodrinking | 238 | 289 |
Table 2. PCR primers.
| SNP | 1st-PCR primer sequences | 2st-PCR primer sequences | UEP sequences |
|---|---|---|---|
| rs3783550 | ACGTTGGATGTGAAGGCCAAATGCTAAGGG | ACGTTGGATGCTCAGGCATCTCCTATGAAG | cGAATTCTGTTAGAGAACAAGATG |
| rs3783546 | ACGTTGGATGACACTGCTGTTGGCACTATG | ACGTTGGATGGTAGGGCAGTAGCTTCATTC | ccTTATTCACTGAGAGCCTT |
| rs2856838 | ACGTTGGATGCTGGTGTCAGAGAAGACAAC | ACGTTGGATGGTTAGTTATGCCATCCTGAG | CCAGGTGTCTGTCTCCTAA |
| rs1609682 | ACGTTGGATGCATGGGACTGCTATTCTTAC | ACGTTGGATGACCTCTAGTGAGGGTAAAAC | gAGTGAGGGTAAAACAAAAGTATT |
| rs3783521 | ACGTTGGATGCCAACTGGCTACATTTCTGC | ACGTTGGATGTCAGGAGGAGAGGGTTAATC | ccTGTTGCCTAAAGAGGAA |
| rs2853550 | ACGTTGGATGCGAAGACTATCCTCCTCACC | ACGTTGGATGTGCAGTGCTTCAGCTGATCC | CAGCTGATCCTGTTCCA |
| rs1143643 | ACGTTGGATGCCTCAGCATTTGGCACTAAG | ACGTTGGATGACTCCTGAGTTGTAACTGGG | GGGCCCCCAACTTTC |
| rs3136558 | ACGTTGGATGAAGGGCTTGAAAGAATCCCG | ACGTTGGATGGATTCATCCACCTCGGCTTC | aaccCGCCTGGCCCAGAGAGGGATGA |
| rs1143630 | ACGTTGGATGTCTTGAGTCTGCCTCTAACC | ACGTTGGATGAGATTATCCCTCTCTGAAGC | AGCTCAAGGAGGTTAAG |
| rs1143627 | ACGTTGGATGTCTCAGCCTCCTACTTCTGC | ACGTTGGATGTTGTGCCTCGAAGAGGTTTG | gtTCCCTCGCTGTTTTTAT |
| rs16944 | ACGTTGGATGCTGTCTGTATTGAGGGTGTG | ACGTTGGATGAGAGGCTCCTGCAATTGACA | AATTGACAGAGAGCTCC |
| rs1143623 | ACGTTGGATGACCTATTTCCCTCGTGTCTC | ACGTTGGATGATGTGCCAGGTATCGTGCTC | tttaGTGCTCGCTCTGCATTAT |
| rs17042888 | ACGTTGGATGTGGAGTTGGAGTCTTGTTGG | ACGTTGGATGCTACTTGCTCAGCACCATAC | agcGGTGTTGAAATCCCAAAA |
| rs928940 | ACGTTGGATGACATGGTTCCATCTCTATCC | ACGTTGGATGAGAAGAGAAAAGTTGACGGG | aTGACGGGGTGCATACTC |
| rs3181052 | ACGTTGGATGCTTTATGTTTGTCTGGGCCG | ACGTTGGATGACAGTCCCCATATCTGGAAG | cttaACTCATACACCCACAGAGCC |
| rs452204 | ACGTTGGATGAAAAGAGCCTCAACATGCAG | ACGTTGGATGTAGACTTAGCCACGTGACTG | gcccATAGGATGATGCAAGCAGAAGT |
Table 3. Basic information of candidate SNPs in this study.
| SNP | Chr | Gene | allel | MAF(case) | MAF(control) | HWE | OR | 95%CI | P | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs3783550 | 2q13 | IL1A | T/G | 0.38 | 0.33 | 0.399 | 1.28 | 1.04 | 1.58 | 0.018* |
| rs3783546 | 2q13 | IL1A | C/G | 0.38 | 0.33 | 0.459 | 1.30 | 1.05 | 1.60 | 0.014* |
| rs2856838 | 2q13 | IL1A | A/G | 0.27 | 0.25 | 0.320 | 1.12 | 0.89 | 1.40 | 0.338 |
| rs1609682 | 2q13 | IL1A | T/G | 0.38 | 0.33 | 0.398 | 1.29 | 1.05 | 1.59 | 0.016* |
| rs3783521 | 2q13 | IL1A | G/A | 0.38 | 0.33 | 0.399 | 1.29 | 1.05 | 1.59 | 0.015* |
| rs2853550 | 2q13 | IL1B | A/G | 0.11 | 0.09 | 0.067 | 1.29 | 0.92 | 1.79 | 0.137 |
| rs1143643 | 2q13 | IL1B | C/T | 0.48 | 0.47 | 0.191 | 1.01 | 0.83 | 1.23 | 0.926 |
| rs3136558 | 2q13 | IL1B | G/A | 0.41 | 0.37 | 0.617 | 1.21 | 0.99 | 1.48 | 0.066 |
| rs1143630 | 2q13 | IL1B | T/G | 0.18 | 0.16 | 1.000 | 1.16 | 0.89 | 1.51 | 0.262 |
| rs1143627 | 2q13 | IL1B | G/A | 0.49 | 0.48 | 0.160 | 1.05 | 0.86 | 1.28 | 0.618 |
| rs16944 | 2q13 | IL1B | A/G | 0.49 | 0.48 | 0.162 | 1.06 | 0.87 | 1.29 | 0.556 |
| rs1143623 | 2q13 | IL1B | G/C | 0.41 | 0.40 | 0.286 | 1.04 | 0.85 | 1.28 | 0.675 |
| rs17042888 | 2q13 | IL1RN | A/G | 0.27 | 0.26 | 0.470 | 1.05 | 0.84 | 1.31 | 0.697 |
| rs928940 | 2q13 | IL1RN | T/G | 0.41 | 0.39 | 0.000 | 1.07 | 0.87 | 1.31 | 0.510 |
| rs3181052 | 2q13 | IL1RN | G/A | 0.43 | 0.41 | 0.248 | 1.09 | 0.89 | 1.34 | 0.384 |
| rs452204 | 2q13 | IL1RN | G/A | 0.36 | 0.35 | 1.000 | 1.06 | 0.86 | 1.30 | 0.594 |
*p value ≤ 0.05 indicates statistical significance; A/B stands for minor/major alleles on the control sample frequencies. HWE: Hardy–Weinberg equilibrium; MAF: minor allele frequency; OR: odds ratio; SNP: single-nucleotide polymorphisms.
Table 4. Association between IL1 polymorphisms genotypes and RCC risk under different genotypic models.
| Model | Genotype | control | case | OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| rs3783550 | Codominant | G/G | 206 (44.5%) | 106 (36.5%) | 1 | |
| G/T | 212 (45.8%) | 144 (49.7%) | 1.32 (0.96-1.81) | 0.053 | ||
| T/T | 45 (9.7%) | 40 (13.8%) | 1.73 (1.06-2.81) | |||
| Dominant | G/G | 206 (44.5%) | 106 (36.5%) | 1 | 0.031* | |
| G/T-T/T | 257 (55.5%) | 184 (63.5%) | 1.39 (1.03-1.88) | |||
| Recessive | G/G-G/T | 418 (90.3%) | 250 (86.2%) | 1 | 0.089 | |
| T/T | 45 (9.7%) | 40 (13.8%) | 1.49 (0.94-2.34) | |||
| Log-additive | --- | --- | --- | 1.32 (1.05-1.64) | 0.015* | |
| rs3783546 | Codominant | G/G | 206 (44.7%) | 106 (36.4%) | 1 | 0.040* |
| G/C | 210 (45.5%) | 144 (49.5%) | 1.33 (0.97-1.83) | |||
| C/C | 45 (9.8%) | 41 (14.1%) | 1.77 (1.09-2.87) | |||
| Dominant | G/G | 206 (44.7%) | 106 (36.4%) | 1 | 0.025* | |
| G/C-C/C | 255 (55.3%) | 185 (63.6%) | 1.41 (1.04-1.91) | |||
| Recessive | G/G-G/C | 416 (90.2%) | 250 (85.9%) | 1 | 0.072 | |
| C/C | 45 (9.8%) | 41 (14.1%) | 1.52 (0.97-2.38) | |||
| Log-additive | --- | --- | --- | 1.33 (1.07-1.66) | 0.011* | |
| rs1609682 | CoDominant | G/G | 205 (44.4%) | 106 (36.4%) | 1 | 0.046* |
| G/T | 212 (45.9%) | 144 (49.5%) | 1.31 (0.96-1.80) | |||
| T/T | 45 (9.7%) | 41 (14.1%) | 1.76 (1.09-2.86) | |||
| Dominant | G/G | 205 (44.4%) | 106 (36.4%) | 1 | 0.031* | |
| G/T-T/T | 257 (55.6%) | 185 (63.6%) | 1.39 (1.03-1.88) | |||
| Recessive | G/G-G/T | 417 (90.3%) | 250 (85.9%) | 1 | 0.070 | |
| T/T | 45 (9.7%) | 41 (14.1%) | 1.52 (0.97-2.39) | |||
| Log-additive | --- | --- | --- | 1.32 (1.06-1.65) | 0.013* | |
| rs3783521 | CoDominant | A/A | 206 (44.5%) | 106 (36.4%) | 1 | 0.043* |
| G/A | 212 (45.8%) | 144 (49.5%) | 1.32 (0.96-1.81) | |||
| G/G | 45 (9.7%) | 41 (14.1%) | 1.77 (1.09-2.87) | |||
| Dominant | A/A | 206 (44.5%) | 106 (36.4%) | 1 | 0.028* | |
| G/A-G/G | 257 (55.5%) | 185 (63.6%) | 1.40 (1.04-1.89) | |||
| Recessive | A/A-G/A | 418 (90.3%) | 250 (85.9%) | 1 | 0.069 | |
| G/G | 45 (9.7%) | 41 (14.1%) | 1.52 (0.97-2.39) | |||
| Log-additive | --- | --- | --- | 1.33 (1.06-1.66) | 0.012* | |
| rs1143623 | CoDominant | C/C | 159 (34.6%) | 110 (37.9%) | 1 | 0.065 |
| C/G | 233 (50.6%) | 124 (42.8%) | 0.82 (0.59-1.16) | |||
| G/G | 68 (14.8%) | 56 (19.3%) | 1.38 (0.88-2.17) | |||
| Dominant | C/C | 159 (34.6%) | 110 (37.9%) | 1 | 0.720 | |
| C/G-G/G | 301 (65.4%) | 180 (62.1%) | 0.94 (0.69-1.30) | |||
| Recessive | C/C-C/G | 392 (85.2%) | 234 (80.7%) | 1 | 0.040* | |
| G/G | 68 (14.8%) | 56 (19.3%) | 1.54 (1.02-2.32) | |||
| Log-additive | --- | --- | --- | 1.10 (0.88-1.37) | 0.400 |
The LD and haplotype analysis were investigated. We found one block in studied IL1A SNPs (Figure 1). In Table 5 we listed the association between IL1A haplotype and the risk of RCC. Haplotype estimation analysis showed that the haplotype of rs3783550T/ rs3783546C/ rs2856838G/ rs1609682T/rs3783521G(TCGTG) It could be a potential risk factor for RCC (OR = 1.68, 95% CI= 1.19 - 2.37, p = 0.0032).
Figure 1. Haplotype block map for the IL1 SNPs genotyped in this study.
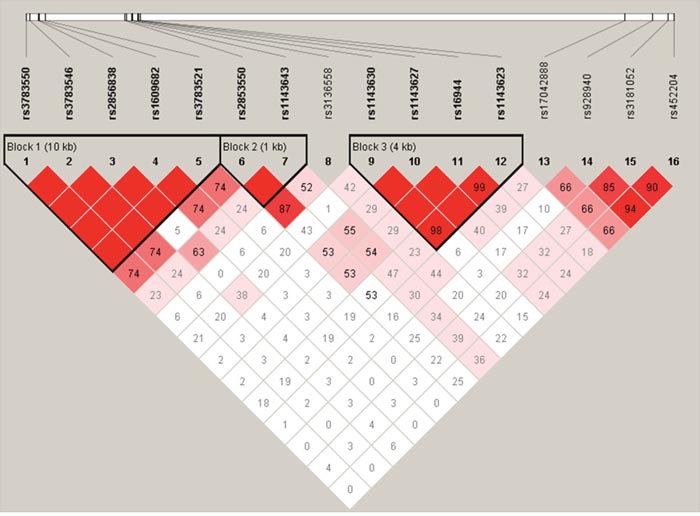
Table 5. Haplotype frequencies and their associations with RCC risk.
| Block | rs3783550 | rs3783546 | rs2856838 | rs1609682 | rs3783521 | Freq | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|---|---|
| 1 | G | G | G | G | A | 0.6499 | 1 | --- |
| 2 | T | C | A | T | G | 0.2555 | 1.20 (0.93 - 1.54) | 0.160 |
| 3 | T | C | G | T | G | 0.0946 | 1.68 (1.19 - 2.37) | 0.003* |
In addition we also carried out smoking, drinking, age stratification analysis, we found in smoking population there have six SNPs rs3783550 (P=0.029), rs3783546 (P =0.019), rs1609682 (P=0.023), rs3783521 (P=0.021), rs1143630 (P=0.045), rs3136558 (P=0.020) polymorphism were associated with RCC, however, in nosmoking population we did not found any SNPs with RCC. Meanwhile, in nodrinking there was also a significant relationship between the risk of RCC of six SNPs except rs1143630 and rs3136558. For age > 55, five SNPs polymorphism (rs3783550 (P=0.003), rs3783546 (P =0.003), rs2856838 (P=0.020), rs1609682 (P=0.003), rs3783521 (P=0.003) were associated with RCC risk (Table 6).
Table 6. The association between SNPs and smoking, drinking status, age analysis of RCC patients.
| SNP | Allel | Smoking status | Drinking status | Age | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Smoking | Nosmoking | Drinking | Nodrinking | >55 | ≤55 | ||||||||
| OR(95%CI) | P | OR(95%CI) | P | OR(95%CI) | P | OR(95%CI) | P | OR(95%CI) | P | OR(95%CI) | P | ||
| rs3783550 | T/G | 1.37(1.03-1.81) | 0.029 | 1.20(0.85-1.68) | 0.308 | 0.95(0.60-1.50) | 0.823 | 1.43(1.11-1.85) | 0.006 | 1.43(1.11-1.85) | 0.003 | 1.01(0.74-1.38) | 0.959 |
| rs3783546 | C/G | 1.39(1.05-1.84) | 0.019 | 1.20(0.85-1.68) | 0.308 | 0.95(0.60-1.51) | 0.836 | 1.45(1.13-1.87) | 0.004 | 1.45(1.13-1.87) | 0.003 | 1.03(0.76-1.41) | 0.831 |
| rs2856838 | A/G | 1.18(0.87-1.61) | 0.284 | 0.99(0.68-1.43) | 0.941 | 0.93(0.57-1.53) | 0.774 | 0.83(0.63-1.10) | 0.198 | 0.83(0.63-1.10) | 0.020 | 0.81(0.57-1.14) | 0.227 |
| rs1609682 | T/G | 1.38(1.04-1.82) | 0.023 | 1.20(0.85-1.68) | 0.308 | 0.95(0.60-1.50) | 0.823 | 1.44(1.12-1.85) | 0.005 | 1.44(1.12-1.85) | 0.003 | 1.03(0.75-1.41) | 0.845 |
| rs3783521 | G/A | 1.39(1.05-1.83) | 0.021 | 1.20(0.85-1.68) | 0.308 | 0.95(0.60-1.50) | 0.823 | 1.45(1.12-1.86) | 0.004 | 1.45(1.12-1.86) | 0.003 | 1.03(0.75-1.41) | 0.845 |
| rs2853550 | A/G | 1.43(0.93-2.21) | 0.106 | 1.17(0.66-2.06) | 0.591 | 0.80(0.36-1.80) | 0.589 | 1.49(0.99-2.23) | 0.052 | 1.49(0.99-2.23) | 0.142 | 1.26(0.78-2.03) | 0.340 |
| rs1143643 | C/T | 1.10(0.84-1.43) | 0.495 | 1.30(0.93-1.82) | 0.121 | 0.95(0.62-1.48) | 0.836 | 1.01(0.79-1.29) | 0.939 | 1.01(0.79-1.29) | 0.453 | 0.98(0.73-1.31) | 0.869 |
| rs3136558 | G/A | 1.38(1.05-1.82) | 0.020 | 0.94(0.67-1.32) | 0.708 | 1.11(0.71-1.74) | 0.644 | 1.20(0.93-1.53) | 0.157 | 1.20(0.93-1.53) | 0.817 | 1.36(1.01-1.84) | 0.044 |
| rs1143630 | T/G | 1.43(1.01-2.04) | 0.045 | 0.82(0.53-1.28) | 0.381 | 0.79(0.43-1.46) | 0.453 | 0.77(0.56-1.07) | 0.114 | 0.77(0.56-1.07) | 0.162 | 1.06(0.72-1.56) | 0.764 |
| rs1143627 | G/A | 1.03(0.78-1.34) | 0.851 | 1.10(0.79-1.54) | 0.568 | 0.95(0.61-1.47) | 0.814 | 1.03(0.81-1.32) | 0.803 | 1.03(0.81-1.32) | 0.414 | 0.98(0.73-1.32) | 0.915 |
| rs16944 | A/G | 1.03(0.79-1.35) | 0.803 | 1.11(0.79-1.54) | 0.548 | 0.96(0.62-1.48) | 0.847 | 1.04(0.82-1.33) | 0.738 | 1.04(0.82-1.33) | 0.291 | 0.97(0.72-1.30) | 0.834 |
| rs1143623 | G/C | 0.97(0.74-1.27) | 0.801 | 1.16(0.83-1.64) | 0.378 | 1.02(0.65-1.60) | 0.933 | 1.01(0.79-1.29) | 0.944 | 1.01(0.79-1.29) | 0.592 | 1.01(0.75-1.36) | 0.965 |
| rs17042888 | A/G | 0.82(0.60-1.11) | 0.194 | 1.34(0.92-1.95) | 0.129 | 0.89(0.53-1.49) | 0.663 | 1.02(0.77-1.34) | 0.915 | 1.02(0.77-1.34) | 0.974 | 1.00(0.72-1.40) | 0.996 |
| rs928940 | T/G | 0.89(0.68-1.18) | 0.429 | 1.20(0.86-1.69) | 0.287 | 0.95(0.61-1.49) | 0.824 | 1.03(0.80-1.32) | 0.831 | 1.03(0.80-1.32) | 0.528 | 1.14(0.84-1.54) | 0.406 |
| rs3181052 | G/A | 1.00(0.77-1.31) | 0.983 | 1.13(0.80-1.57) | 0.491 | 0.89(0.57-1.39) | 0.620 | 1.10(0.86-1.41) | 0.451 | 1.10(0.86-1.41) | 0.226 | 1.33(0.99-1.79) | 0.061 |
| rs452204 | G/A | 0.93(0.70-1.23) | 0.605 | 1.22(0.86-1.72) | 0.268 | 1.06(0.67-1.67) | 0.811 | 1.02(0.79-1.32) | 0.876 | 1.02(0.79-1.32) | 0.398 | 1.25(0.92-1.70) | 0.159 |
DISCUSSION
In our case-control study of RCC, we elaborated the associations of 16 SNPs in IL1A, IL1B and IL1RN with the risk of RCC in Chinese han population. Our study revealed that IL1A and IL1B SNPs were associated with susceptibility to RCC, but there was no significant association between SNP and the risk of RCC in IL1RN.
We selected locus 2q13 in IL1A, IL1B, IL1RN three candidate genes were analyzed. The proteins encoded by these genes belong to interleukin -1 (IL-1). The three molecules in the IL-1 cluster, namely, IL-1α, IL-1β, and interleukin-1 receptor antagonist (IL-1Ra), are encoded by the IL1A, IL1B, and IL1RN genes, respectively [23–25]. IL-1 cytokines are a major inflammatory cytokine and have been implicated in the mediation of acute and chronic inflammatory diseases. It is a key mediator of inflammation, with pleiotropic effects on several cells and signaling pathways. IL-1 is a pleiotropic cytokine that participates in stimulating immune responses, such as inflammation, and regulates cellular growth, differentiation, and movement of cells. It is secreted by viruses, bacteria, and fungi that react to various antigens [26]. IL-1 cytokines are known to contribute to the development and progression of a number of renal diseases [27, 28], and polymorphisms of the IL-1 family cytokine genes have been studied for associations with IgAN susceptibility, progression, and clinical manifestations, such as hematuria and proteinuria. Several associations have been reported between IL-1 cluster gene polymorphisms and IgAN. However, IL-1cluster gene polymorphisms did not reported in RCC. The IL gene reported more focused on asthma, liver cancer, breast cancer, oral cancer, cervical cancer, leukemia and other research, at about rs3783550, rs3783546, rs1609682, rs3783521 polymorphisms were renal cell carcinoma and the related reports.
Smoking has been regarded as one of the risk factors of RCC. The risk of RCC caused by polymorphism is particularly significant in the smoking population, which may be caused by cigarette smoking induced immune suppression, so that renal cell carcinoma escape immune surveillance, thereby promoting the occurrence and development of renal cell carcinoma [29]. Studies have reported that between alcohol consumption and risk of renal cell carcinoma has a dose-dependent relationship, namely alcohol consumption to achieve a certain degree of risk exists only a small amount of alcohol, but can play the role of is beneficial to the body [30], this can be a very good explanation to our other hierarchical analysis results, the relative risk of drinkers for non-drinkers carrying the variant genotype and higher risk of RCC. These risk factors and genetic factors interact to affect the susceptibility of renal cancer. However, due to the limitations of data collection, the study of environmental factors are limited to smoking, drinking and other factors, so it is not able to completely analyze the interaction of gene environment. Therefore, it is necessary to carry out a complete analysis of the gene environment interaction, and to verify the results of the study.
Our results need to be validated in a larger sample and in other races with functional analysis to clarify the potential mechanisms underlying the links between SNPs of IL-1 and susceptibility to RCC.
MATERIALS AND METHODS
Study subjects
The case-control study included 291 histopathologically confirmed RCC patients and 463 controls. None of the patients had a family history of carcinoma. A total of 291 patients with RCC were prospectively recruited from People's Hospital of Hainan Province, Haikou people's Hospital Affiliated to Xiangya Medical School of Central South University, and First Affiliated Hospital of Sun Yat-sen University, and 463 healthy controls from the same region were enrolled in this study. Detailed characteristics of the RCC patients and controls are shown in Table 1. The control group had no history of other tumors and was unrelated to the genes in RCC patients. General examinations, laboratory tests, and medical histories confirmed their health. Everyone personally asked the trained interviewer to collect information about demographic data and related factors using a pre-test questionnaire (smoking, drinking, BMI et al). This study was approved by People's Hospital of Hainan Province, Haikou people's Hospital Affiliated to Xiangya Medical School of Central South University, and First Affiliated Hospital of Sun Yat-sen University, and all individuals involved in this study have signed informed consent. After informed consent was issued by all individuals, 5-ml samples of venous blood were collected for genomic DNA extraction.
Selection of single nucleotide polymorphisms and genotyping
Candidate SNPs were selected according to the following restrictions: (1) The SNPs located at IL1; (2) each SNP had minor allele frequency (MAF) of > 5% in Chinese Han population; (3) each SNP had an r2 of > 0.80. Thus, for this study, we have selected the 16 SNPs described (Table 2). Whole blood were extracted using GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Co. Ltd. Xi’an City, China). We measured DNA concentration using NanoDrop 2000 spectrophotometer. Sequenom MassARRAY Assay Design 3.0 Software was used to design a Multiplexed SNP MassEXTEND assay [31]. Sequenom MassARRAY RS1000 was used for genotyping, and the related data were managed using Sequenom Typer 4.0 Software [31, 32].
Laboratory personnel were blinded to the genotyping results of all samples.
Statistical analysis
All SNPs performed Hardy Weinberg balance (HWE) performed by comparing the observed and expected frequencies of genotypes using χ2 analysis [33]. OR and 95%CI were calculated using the logistic regression model. The association between SNPs and RCC risk were analyzed by logistic regression (Four genetic models dominant, recessive, and additive model). The haplotype frequencies were determined using the SHEsis program.
Finally, we used statistical packages (4.2 Edition) to perform linkage disequilibrium (LD), haplotype genetic association construction, and polymorphism at loci [34, 35]. All p values were two-sided, and p <0.05 was indicated statistical significance.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Number: 81560250 and 81460450) and the Hainan Province Scientific and Technological Cooperation Special Foundation (Grant Number: KJHZ2015-25) and the Hainan Provincial Natural Science Foundation (Grant Number: 812146 and 813256 and 817379 and 20168312) and the Hainan Province Natural Science Foundation of Innovation Research Team Project (2017CXTD010). We thank all of the participants for their involvement in this study.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Ferlay J, Shin H, Bray F, Forman D, Mathers C, Parkin D. Estimates of worldwide burden of cancer in 2012: GLOBOCAN 2012. Int J Cancer. 2012 doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Ervik M. Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. Int J Cancer. 2012 [Google Scholar]
- 3.Theis RP, Dolwick Grieb SM, Burr D, Siddiqui T, Asal NR. Smoking, environmental tobacco smoke, and risk of renal cell cancer: a population-based case-control study. Bmc Cancer. 2008;8:1–11. doi: 10.1186/1471-2407-8-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergström A, Hsieh CC, Lindblad P, Lu CM, Cook NR, Wolk A. Obesity and renal cell cancer--a quantitative review. Journal of Urology. 2002;168:877. doi: 10.1054/bjoc.2001.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gati A, Kouidhi S, Marrakchi R, El Gaaied A, Kourda N, Derouiche A, Chebil M, Caignard A, Perier A. Obesity and renal cancer: Role of adipokines in the tumor-immune system conflict. Oncoimmunology. 2014;3:e27810. doi: 10.4161/onci.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deckers IA, van den Brandt PA, van Engeland M, van Schooten FJ, Godschalk RW, Keszei AP, Schouten LJ. Polymorphisms in genes of the renin-angiotensin-aldosterone system and renal cell cancer risk: interplay with hypertension and intakes of sodium, potassium and fluid. Int J Cancer. 2014;136:1104–1116. doi: 10.1002/ijc.29060. [DOI] [PubMed] [Google Scholar]
- 7.Linehan WM, Walther MM, Zbar B. The Genetic Basis of Cancer of the Kidney. Journal Durologie Médicale Et Chirurgicale. 2003;170:2163–2172. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- 8.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nature Reviews Urology. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clague J, Lin J, Cassidy A, Matin S, Tannir NM, Tamboli P, Wood CG, Wu XF. Family history and risk of renal cell carcinoma: results from a case-control study and systematic meta-analysis. Cancer Epidemiology Biomarkers & Prevention. 2009;18:801–807. doi: 10.1158/1055-9965.EPI-08-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Petursdottir V, Hardarson S, Gudjonsson SA, Johannsdottir H, Helgadottir HT, Stacey SN. A common variant at 8q24.21 is associated with renal cell cancer. Nature Communications. 2013;4:2776. doi: 10.1038/ncomms3776. [DOI] [PubMed] [Google Scholar]
- 11.Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, Prokhortchouk E, Wu X, Kiemeney LA, Gaborieau V. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nature Genetics. 2011;43:60. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schödel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, Tomlinson IP, Ratcliffe PJ, Mole DR. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nature genetics. 2012;44:1–2. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X. A genome-wide association study identifies a novel susceptibility locus for renal cell carcinoma on 12p11.23. Human Molecular Genetics. 2012;21:456–462. doi: 10.1093/hmg/ddr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and Interleukin-13 Signaling Connections Maps. Science. 2003;300:1527–1528. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura E, Megumi Y, Kobayashi T, Kamoto T, Ishitoya S, Terachi T, Tachibana M, Matsushiro H, Habuchi T, Kakehi Y. Genetic Polymorphisms of the Interleukin-4 Receptor α Gene Are Associated with an Increasing Risk and a Poor Prognosis of Sporadic Renal Cell Carcinoma in a Japanese Population. Clinical Cancer Research. 2002;8:2620–2625. [PubMed] [Google Scholar]
- 16.Thurnher M, Radmayr C, Hobisch A, Böck G, Romani N, Bartcch G, Klocker H. Tumor-infiltrating T lymphocytes from renal-cell carcinoma express B7-1 (CD80): T-Cell expansion by T-T cell co-stimulation. International Journal of Cancer. 1995;62:559–564. doi: 10.1002/ijc.2910620512. [DOI] [PubMed] [Google Scholar]
- 17.Ikemoto S, Yoshida N, Narita K, Wada S, Kishimoto T, Sugimura K, Nakatani T. Role of tumor-associated macrophages in renal cell carcinoma. Oncology Reports. 2003;10:1843. [PubMed] [Google Scholar]
- 18.Thurnher M, Radmayr C, Ramoner R, Ebner S, Böck G, Klocker H, Romani N, Bartsch G. Human renal - cell carcinoma tissue contains dendritic cells. International Journal of Cancer. 1996;68:1–7. doi: 10.1002/(SICI)1097-0215(19960927)68:1<1::AID-IJC1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 19.Alberti L, Thomachot MC, Bachelot T, Menetrier-Caux C, Puisieux I, Blay JY. IL-6 as an intracrine growth factor for renal carcinoma cell lines. International Journal of Cancer. 2004;111:653–661. doi: 10.1002/ijc.20287. [DOI] [PubMed] [Google Scholar]
- 20.Falkensammer C, Jöhrer K, Gander H, Ramoner R, Putz T, Rahm A, Greil R, Bartsch G, Thurnher M. IL-4 inhibits the TNF-α induced proliferation of renal cell carcinoma (RCC) and cooperates with TNF-α to induce apoptotic and cytokine responses by RCC: implications for antitumor immune responses. Cancer Immunology & Immunotherapy. 2006;55:1228. doi: 10.1007/s00262-006-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obiri NI, Husain SR, Debinski W, Puri RK. Interleukin 13 inhibits growth of human renal cell carcinoma cells independently of the p140 interleukin 4 receptor chain. Clinical Cancer Research. 1996;2:1743. [PubMed] [Google Scholar]
- 22.Yu SJ, Kim HS, Cho SW, Sohn J. IL-4 inhibits proliferation of renal carcinoma cells by increasing the expression of p21WAF1 and IRF-1. Experimental & Molecular Medicine. 2004;36:372–379. doi: 10.1038/emm.2004.49. [DOI] [PubMed] [Google Scholar]
- 23.Dinarello CA. The many worlds of reducing interleukin-1. Arthritis & Rheumatism. 2005;52:1960–1967. doi: 10.1002/art.21107. [DOI] [PubMed] [Google Scholar]
- 24.Dinarello CA. Blocking IL-1 in systemic inflammation. Journal of Experimental Medicine. 2005;201:1355–1359. doi: 10.1084/jem.20050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald AA, Leclercq SA, Yan A, Homik JE, Dinarello CA. Rapid responses to anakinra in patients with refractory adult-onset Still's disease. Arthritis & Rheumatism. 2005;52:1794–1803. doi: 10.1002/art.21061. [DOI] [PubMed] [Google Scholar]
- 26.Barksby H, Lea S, Preshaw P, Taylor J. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clinical & Experimental Immunology. 2007;149:217–225. doi: 10.1111/j.1365-2249.2007.03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensen JT, Langefeld CD, Li L, Mccall CE, Cousart SL, Dryman BN, Freedman BI, Bowden DW. Association of an IL-1A 3′UTR polymorphism with end-stage renal disease and IL-1 alpha expression. Kidney International. 2003;63:1211–1219. doi: 10.1046/j.1523-1755.2003.00856.x. [DOI] [PubMed] [Google Scholar]
- 28.Freedman BI, Yu H, Spray BJ, Rich SS, Rothschild CB, Bowden DW. Genetic linkage analysis of growth factor loci and end-stage renal disease in African Americans. Kidney International. 1997;51:819–825. doi: 10.1038/ki.1997.115. [DOI] [PubMed] [Google Scholar]
- 29.Germano G, Allavena P, Mantovani A. Cytokines as a key component of cancer-related inflammation. Cytokine. 2008;43:374–379. doi: 10.1016/j.cyto.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Lee JE, Hunter DJ, Spiegelman D, Adami HO, Albanes D, Bernstein L, van den Brandt PA, Buring JE, Cho E, Folsom AR, Freudenheim JL, Giovannucci E, Graham S, et al. Alcohol intake and renal cell cancer in a pooled analysis of 12 prospective studies. Journal of the National Cancer Institute. 2007;99:801–810. doi: 10.1093/jnci/djk181. [DOI] [PubMed] [Google Scholar]
- 31.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2: Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 32.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 33.Adamec C. [Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples]. [Article in Czech] Cesk Zdrav. 1964;12:613–619. [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. The odds ratio. Bmj. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]


