Abstract
Accumulating evidence suggested that long non-coding RNAs (lncRNAs) play essential roles in various biological processes, including tumorigenesis. Aberrant expression of LINC00161 has been reported in some cancer types, however, the association of LINC00161 and hepatocellular carcinoma (HCC) has not been evaluated. Here, we measured the expression of LINC00161 in HCC tissues and corresponding normal liver tissues using real-time PCR. The result showed that the expression level of LINC00161 was significantly higher in HCC tissues. Further analysis indicated that HCC patients with higher LINC00161 expression have shorter survival. Multivariate Cox regression analysis showed that LINC00161 expression was an independent prognostic factor for the overall survival. Furthermore, our result indicated that knock-down of LINC00161 can significantly inhibit liver cancer cell migration and invasion. The present work indicated that LINC00161 might serve as an oncogenic gene and play a pivotal role in promoting tumor migration and invasion in HCC. Our work implicates the promising effect of LINC00161 on the prognosis of HCC.
Keywords: LINC00161, hepatocellular carcinoma, prognosis, tumor cell migration, invasion
INTRODUCTION
Liver cancer has become the fifth most frequently diagnosed cancer and third leading cause of cancer death worldwide [1, 2]. Hepatocellular carcinoma (HCC) is the most common histological subtype of liver cancers, and accounts for 75% of all cases. The risk factors of HCC patients are associated with geographic region and ethnic background [3]. Until now, the most effective therapy method for HCC is still complete resection. It is still difficult for the early diagnosis of HCC. So, exploring the pathogenesis and biological features of HCC is crucial for early detection and treatment.
Long non-coding RNAs (lncRNAs) are a class of transcripts with the size longer than 200 nucleotides. Recent works have shown that lncRNAs play important roles in various biological processes by modulating gene expression through chromatin organization [4, 5]. Many cancer-associated lncRNAs have been reported to be novel independent biomarkers for cancer diagnosis and prognosis in different types of cancers, such as breast cancer [6, 7], oesophageal squamous cell carcinoma [8], colorectal cancer [9], lung cancer [10] and ovarian cancer [11]. For example, lncRNA-p21 has been documented to be involved in cell apoptosis and regulation of Warburg effect [12]. Moreover, some lncRNAs have been identified to regulate chemoresistance in many cancers, such as MEG3 [13].
Previous works have identified several lncRNAs to be associated with the progression of HCC, such as FTX and PCAT-1 [14–16]. In the present work, we examine the influence of LINC00161 expression on HCC tumorigenesis. LINC00161 is a novel lncRNA, which has been reported to play important role in osteosarcoma [17]. Here, we examined the expression difference of LINC00161 between HCC tissues and corresponding normal liver tissues, and evaluated the clinical significance of LINC00161 in HCC patients. Moreover, we examined the effects of LINC00161 on HCC cells migration and invasion, which suggests the potential effects of LINC00161 on HCC prognosis.
RESULTS
The expression of LINC00161 in HCC tissues and cell lines
Here, we examined the expression level of LINC00161 in 104 pairs of HCC tissues and adjacent liver tissues using quantitative RT-PCR. The result demonstrated that LINC00161 expression level was significantly up-regulated in HCC tissues compared with matched normal liver tissues (P-value < 0.001, Figure 1A). Next, we analyzed the expression level of LINC00161 in different cell lines, and the result showed that LINC00161 expressions in HCC cell lines (HepG2, Hep3B, Huh7 and SMMC7721) were significantly higher compared with normal liver cell line LO2 (Figure 1B). This result suggests that LINC00161 might play an oncogenic role in liver cancer tumorigenesis.
Figure 1. The relative expression levels of LINC00161 in HCC tissues and cell lines.
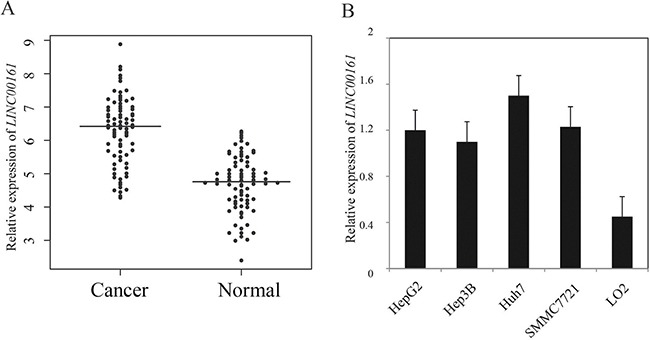
(A) The relative expression of LINC00161 in HCC tissues and adjacent non-tumor liver tissues. The bars represent the means of the relative expression of LINC00161. (B) the relative expression of LINC00161 in HCC cell lines.
Clinical significance of LINC00161 expression in HCC
Next, we evaluated the association between LINC00161 expression and clinicopathological characteristics. The enrolled 104 HCC patients were divided into two equal groups according to the median value of LINC00161 expression level (high-expression group and low-expression group). Then, we examined the correlation between LINC00161 expression level and clinicopathological factors of HCC patients. The result showed that higher expression value of LINC00161 was tightly associated with tumor grade (P-value = 0.0012, Table 1), and no significant differences of other characteristics were found in HCC patients. Kaplan-Meier survival analysis revealed that HCC patients with higher LINC00161 expression level correlated with shorter overall survival (P-value = 0.001, Figure 2). Univariate proportional hazard model indicated that the tumor grade and LINC00161 expression level was prognostic predictors. Those characteristics of HCC patients which are associated with overall survival in the univariate Cox analysis were further evaluated in a multivariate Cox model. The result showed that LINC00161 expression level (P-value = 0.001, Table 2) was an independent prognostic factor for predicting the 5-year overall survival of HCC patients.
Table 1. Clinicopathological associations of LINC00161 expression in HCC patients.
| LINC00161 expression | |||
|---|---|---|---|
| Variable | High (n = 52) | Low (n = 52) | P-value |
| Ages (years) | 0.88 | ||
| < 50 | 20 | 23 | |
| ≥ 50 | 32 | 29 | |
| Gender | 0.96 | ||
| Male | 35 | 33 | |
| Female | 17 | 19 | |
| Tumor size | 0.22 | ||
| < 5 cm | 31 | 38 | |
| ≥ 5 cm | 21 | 14 | |
| Serum AFP (ng/l) | 0.74 | ||
| < 400 | 19 | 17 | |
| ≥ 400 | 33 | 35 | |
| Lymph node metastasis | 0.24 | ||
| No | 21 | 24 | |
| Yes | 31 | 28 | |
| Alcohol abuse | 0.99 | ||
| No | 26 | 25 | |
| Yes | 26 | 27 | |
| Tumor grade | 0.0012 | ||
| G1 | 13 | 26 | |
| G2 | 18 | 16 | |
| G3 | 21 | 10 | |
Figure 2. Kaplan-Meier survival curves of patients with HCC based on LINC00161 expression status.
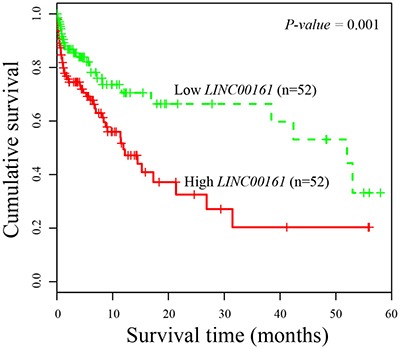
Patients with higher expression group have significantly poorer prognosis than those in lower expression group.
Table 2. Univariate and multivariate analyses of prognostic factors in HCC patients.
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| age (Years) | 1.44 | 0.96–1.65 | 0.28 | 1.41 | 0.72–1.52 | 0.36 |
| gender | 1.43 | 0.92–2.14 | 0.33 | 1.34 | 0.91–2.11 | 0.48 |
| Alcohol abuse | 1.68 | 1.01–2.11 | 0.41 | 1.58 | 1.02–1.98 | 0.55 |
| Lymph node metastasis | 1.58 | 0.89–1.88 | 0.91 | 1.44 | 0.77–1.67 | 0.66 |
| tumor size | 1.48 | 1.06–1.72 | 0.08 | 1.36 | 0.91–1.68 | 0.18 |
| tumor grade | 2.06 | 1.05–3.46 | 0.0015 | 1.57 | 1.21–3.33 | 0.092 |
| LINC00161 | 1.44 | 0.83–1.89 | < 0.001 | 1.37 | 0.85–1.86 | 0.001 |
In vitro effect of LINC00161 on HCC cell migration and invasion
To investigate whether LINC00161 can influence HCC cell migration and invasion, we performed in vitro Transwell assay in Huh7 cell line. The result showed that knock-down of LINC00161 significantly inhibit HCC cell migration and invasion (Figure 3), suggesting that LINC00161 might play an important role in the tumorigenesis of HCC.
Figure 3. Effects of LINC00161 on migration and invasion of Huh7 cell line.
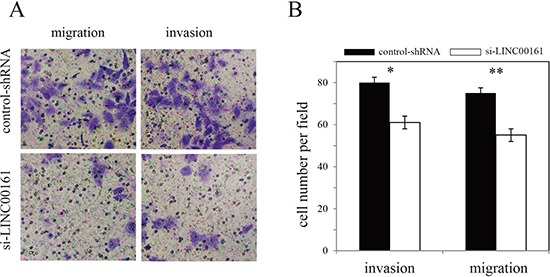
(A) Transwell assay of Huh7 cells and representative fields of invasive cells. (B) Average number of invasive cells per field from three independent experiments.
DISCUSSION
HCC is one of the most common malignancy tumors with lower survival rate. Prognostic factor detection in HCC is necessary to predict the overall survival rate and select optimal therapeutic strategy. Although many efforts have been devoted to detect potential markers in HCC [18–21], the underlying molecular mechanism is still limited. Increasing evidence has suggested that lncRNAs are widely existed in mammalian genomes and play crucial role in cell biology [22]. Until now, lncRNAs has been implemented in diverse pathological processes in cancers.
Dysregulation of LINC00161 has been documented to play an important function in osteosarcoma [17]. Increasing LINC00161 can accelerate cisplatin-induced apoptosis and reduce chemoresistance. Furthermore, LINC00161 can sponge endogenous miR-645 and inhibit its expression leading to the induced expression of IFIT2. However, no study has been performed on the association between LINC00161 and HCC. In the present work, we explored the relationship between LINC00161 expression and the clinicopathogical features of HCC patients for the first time. We found that LINC00161 was significantly over-expressed in HCC tissues and cell lines compared with adjacent normal liver tissues and LO2 cell line, respectively. Multivariate survival analysis showed that LINC00161 could be used as a potential prognostic biomarker for HCC. Further functional analysis indicated that know-down of LINC00161 could significantly inhibit cell migration and invasion, which indicates that LINC00161 might function as an oncogenic gene in HCC.
In summary, our work showed that LINC00161 expression was up-regulated in HCC and is significantly associated with cancer cell migration and invasion. Our findings implicated for the first time that LINC00161 expression was an independent prognostic factor and molecular therapeutic target in HCC.
MATERIALS AND METHODS
Patients and tissue samples
Liver tumor tissues and paired normal adjacent tissues were obtained from patients with a diagnosis of HCC who underwent surgery at the Yi Ji-shan Hospital (Anhui Province, China) between April 2008 and Nov 2014. Informed written consents were obtained from all enrolled HCC patients, and all patients had complete 5-year follow-up. These HCC patients had never received any radiotherapy before surgery excision. All liver tissues were immediately frozen in liquid nitrogen after surgery. This study was approved by the Ethics Committee of Shanghai Cancer Center, Fudan University.
RNA extraction and RT-PCR
The total RNA from tissue samples and cells were isolated using the TRIzol solution (Invitrogen, USA) according to the manufacturer's protocol. Quantitative RT-PCR was performed using BioRad Chromo4 real-time PCR system. GAPDH was used as an internal control, and the expression level of LINC00161 was measured using the following primer sequences: F:5-ACTTGAGTGAGGTGGGTTTC-3 and R:5-TTGGTGTTCCTTGGCTTGTA-3. Relative quantification of LINC00161 was calculated by using the 2−ΔΔCT method.
Cell culture
Four human HCC cell lines (HepG2, Hep3B, Huh7 and SMMC7721) and normal human liver cells (LO2) were purchased from Shanghai Institutes for Biological Sciences, China. The cells were cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS), 100 μ/ml penicillin and 100 mg/ml streptomycin. All these cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
LINC00161 knockdown by lentiviruses
To generate lentiviruses expressing LINC00161 shRNA and control shRNAs, Huh7 cells were transfected with 5μg of shRNAs. After transfection, Huh7 cells were cultured with DMEM medium containing 10% FBS for 36 hours. The medium containing lentivirus particles was centrifuged at 10000×g for 2 min and used for infection.
Cell migration and invasion assays
The cell invasion assay was performed using a 24-well Transwell chamber (Costar, USA) without Matrigel coating. For migration assay, a total of 2 × 105 transfected cells were added to the upper chamber of Transwell assay inserts. Then, the inserts were added to the bottom chamber wells filled with conditioned medium. After the incubation for 24 hours and stained with hematoxylin for 10 minutes, we calculated the number of cell in five random fields for each chamber. For invasion assay, transfected cells were plated in the top chamber with a Matrigel-coated membrane in the bottom chambers.
Statistical analysis
Statistical significance between groups was measured using student's t-test. Kaplan-Meier method and log-rank test were used to measure the overall survival rate. The Cox proportional hazards model was applied for the multivariate analysis. All statistical analyses were carried out using SPSS version 16.0, and results were considered statistically significant at P-value < 0.05.
Footnotes
CONFLICTS OF INTEREST
The authors have declared no competing interests exist.
Authors' contributions
Conceived and designed the experiments: YW, LW, QP; Performed the experiments: LX, QNC, XL, XW and QC; Wrote the paper: YW.
Ethical statements
Patients were identified from a prospective institutional database and the study was approved by the local Research Ethics Committee.
REFERENCES
- 1.Center MM, Jemal A. International trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2011;20:2362–68. doi: 10.1158/1055-9965.EPI-11-0643. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Yang F, Gao B, Yu Z, Liu X, Xie F, Zhang J. Hepatitis B virus infection in hepatocellular carcinoma tissues upregulates expression of DNA methyltransferases. Int J Clin Exp Med. 2015;8:4175–85. [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013;14:18790–808. doi: 10.3390/ijms140918790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Y, Katsaros D, Loo LW, Hernandez BY, Chong C, Canuto EM, Biglia N, Lu L, Risch H, Chu WM, Yu H. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget. 2015;6:8579–8592. doi: 10.18632/oncotarget.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Chen X, Wang Z, Guo M, Shi H, Wang X, Cheng L, Zhou M. A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep. 2015;5:16553. doi: 10.1038/srep16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, Sun N, Liu Z, Skogerboe G, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–10. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230–2242. doi: 10.18632/oncotarget.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao H, Zhong Z, Sun J. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget. 2016;7:32433–32448. doi: 10.18632/oncotarget.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–90. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Y, He Z, Liu B, Wang P, Chen Y. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/β-catenin signaling pathway. Mol Med Rep. 2015;12:4530–37. doi: 10.3892/mmr.2015.3897. [DOI] [PubMed] [Google Scholar]
- 14.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, Zhou WP, Sun SH. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–34. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 15.Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481–86. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 16.Qiu X, Hong Y, Yang D, Xia M, Zhu H, Li Q, Xie H, Wu Q, Liu C, Zuo C. ISG15 as a novel prognostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Int J Clin Exp Med. 2015;8:17140–50. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang L, Zheng X, Zhong W, Tian X, Yin B, Tian K, Zhang W. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382:137–46. doi: 10.1016/j.canlet.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhayat SA, Hüsing A, Senninger N, Schmidt HH, Haier J, Wolters H, Kabar I. Circulating microRNA-200 Family as Diagnostic Marker in Hepatocellular Carcinoma. PLoS One. 2015;10:e0140066. doi: 10.1371/journal.pone.0140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitai S, Kudo M, Minami Y, Haji S, Osaki Y, Oka H, Seki T, Kasugai H, Sasaki Y, Matsunaga T. Validation of a new prognostic staging system for hepatocellular carcinoma: a comparison of the biomarker-combined Japan Integrated Staging Score, the conventional Japan Integrated Staging Score and the BALAD Score. Oncology. 2008;75:83–90. doi: 10.1159/000173428. [DOI] [PubMed] [Google Scholar]
- 21.Bagi CM, Andresen CJ. Models of hepatocellular carcinoma and biomarker strategy. Cancers (Basel) 2010;2:1441–52. doi: 10.3390/cancers2031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–27. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]


