Abstract
The CASP8 -652 6N insertion/deletion (I/D) polymorphism reduces expression of caspase 8. We conducted a meta-analysis to clarify the relationship between this polymorphism and cancer risk. Eligible articles were retrieved from PubMed, EMBASE, CNKI, and WANFANG databases through February 2017. A total of 33 articles with 49 studies, including 33,494 cases and 36,397 controls, were analyzed. We found that the CASP8 -652 6N ins/del polymorphism was associated with decreased overall cancer risk in five genetic models [DD vs. II: odds ratio (OR)=0.76, 95% confidence interval (CI)=0.69–0.84, ID vs. II: OR=0.87, 95% CI=0.83–0.92, DD vs. ID/II: OR=0.82, 95% CI=0.75–0.89, ID/DD vs. II: OR=0.85, 95% CI=0.80–0.90, and D vs. I: OR=0.87, 95% CI=0.83–0.91]. Stratified analyses showed that the polymorphism was associated with decreased risk of colorectal, breast, esophageal, renal cell, lung, cervical, bladder, gastric, and other cancers. Overall cancer risk was reduced in Asian and Caucasian patients, both hospital- and population-based studies, and both high and low quality studies. Our results highlight the role of the CASP8 -652 6N ins/del polymorphism in decreasing cancer risk. Further studies with large-cohort populations, especially for specific cancer types and ethnic groups, are needed to confirm our findings.
Keywords: CASP8, -652 6N insertion/deletion, polymorphism, cancer risk, meta-analysis
INTRODUCTION
Cancer is a substantial public health burden worldwide and is the second leading cause of death in the United States. An estimated 1,688,780 new cancer cases and 600,920 cancer deaths will occur in the United States this year [1]. Approximately 14 million new cancer cases occurred worldwide in 2012, and by 2025, global cancer incidence is predicted to rise to 20 million new cases annually [2]. Although there are many cancer risk factors, genetic abnormalities play crucial roles in carcinogenesis [3–6].
Apoptosis is a control mechanism to prevent over-proliferation in normal cells [7], and apoptosis pathway aberrations are implicated in cancer development [8]. Caspases are the main regulatory enzymes in the apoptosis pathway [9]. Caspase 8 mediates the extrinsic apoptosis pathway [10, 11]. Human CASP8 is located on chromosome 2q33∼q34, has 11 exons [12], and is highly polymorphic with more than 474 single nucleotide polymorphisms (SNPs) according to the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP). The CASP8 -652 6N ins/del polymorphism (rs3834129) is a six-nucleotide insertion/deletion variant located in the CASP8 promoter region [13], and leads to decreased CASP8 expression. Impaired caspase 8 function reduces T lymphocyte “activation-induced cell death” (AICD) activity, which is important in immune surveillance of cancer cells [13].
Extensive epidemiological studies have assessed the association between the CASP8 -652 6N ins/del polymorphism and cancer risk. However, these studies have not produced conclusive results. The most recent previous meta-analysis of this association, conducted in 2014, assessed a relatively small number of studies. We performed this meta-analysis with a larger sample size to more precisely describe the association of interest.
RESULTS
Study characteristics
Our study selection workflow is shown in Figure 1. Our systematic computer-based search initially identified 108 potentially relevant articles. After scanning titles and abstracts, 67 articles about unrelated topics were excluded. We further excluded 12 articles: eight were meta-analyses [14–21], three were case only studies [22–24], and one deviated from HWE [25]. Articles incorporating several ethnic groups or cancer types were separated into corresponding independent studies. In total, our analysis included datasets from 33 articles with 49 studies [13, 26–57].
Figure 1. Flow diagram of the study selection process.
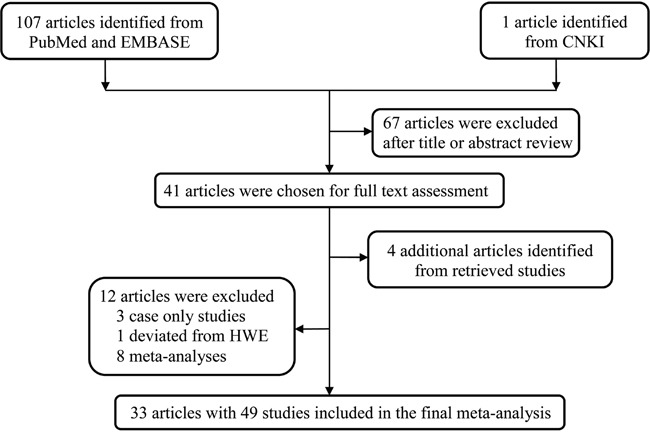
Characteristics for 33,494 cases and 36,397 controls are summarized in Table 1. Of the included studies, 12 were conducted on colorectal cancer, nine on other cancers, eight on breast cancer, three on esophageal cancer, three on renal cell carcinoma, and two on lung cancer, cervical cancer, prostate cancer, bladder cancer, lymphoma cancer, and gastric cancer, respectively. Twenty-seven studies were conducted in Asians, 20 in Caucasians, one in Africans, and one in mixed populations. Twenty-four studies were of population-based design, 22 studies were of hospital-based design, and three did not mention study design in the original data. We also classified the studies as either low quality (25 studies) or high quality (24 studies) by quality score.
Table 1. Characteristics of studies included in the meta-analysis.
| Author last name | Year | Cancer type | Country | Ethnicity | Design | Genotype method | Case | Control | MAF | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II | ID | DD | All | II | ID | DD | All | ||||||||||
| Sun | 2007 | Lung | China | Asian | PB | PCR-RFLP | 756 | 348 | 45 | 1149 | 640 | 407 | 64 | 1111 | 0.24 | 0.947 | 11 |
| Sun | 2007 | Esophagus | China | Asian | PB | PCR-RFLP | 652 | 328 | 38 | 1018 | 543 | 338 | 56 | 937 | 0.24 | 0.724 | 11 |
| Sun | 2007 | Gastric | China | Asian | PB | PCR-RFLP | 262 | 142 | 16 | 420 | 233 | 152 | 25 | 410 | 0.25 | 0.975 | 11 |
| Sun | 2007 | Colorectal | China | Asian | PB | PCR-RFLP | 605 | 280 | 33 | 918 | 528 | 304 | 58 | 890 | 0.24 | 0.116 | 11 |
| Sun | 2007 | Breast | China | Asian | PB | PCR-RFLP | 699 | 371 | 49 | 1119 | 513 | 419 | 72 | 1004 | 0.28 | 0.279 | 11 |
| Sun | 2007 | Cervical | China | Asian | PB | PCR-RFLP | 199 | 102 | 13 | 314 | 314 | 211 | 42 | 567 | 0.26 | 0.428 | 10 |
| Yang | 2008 | Pancreatic | China | Asian | PB | PCR-RFLP | 268 | 111 | 18 | 397 | 521 | 323 | 63 | 907 | 0.25 | 0.185 | 13 |
| Pittman | 2008 | Colorectal | England | Caucasian | PB | AS-PCR | 995 | 1897 | 987 | 3879 | 892 | 1872 | 897 | 3661 | 0.50 | 0.170 | 9 |
| Frank | 2008 | Breast | Germany | Caucasian | HB | Fluorescent | 298 | 535 | 221 | 1054 | 270 | 506 | 263 | 1039 | 0.50 | 0.403 | 7 |
| Frank | 2008 | Breast | England | Caucasian | PB | Fluorescent | 235 | 541 | 251 | 1027 | 245 | 608 | 321 | 1174 | 0.53 | 0.169 | 10 |
| Frank | 2008 | Breast | Germany | Caucasian | PB | Fluorescent | 280 | 509 | 222 | 1011 | 285 | 492 | 229 | 1006 | 0.47 | 0.550 | 9 |
| Frank | 2008 | Breast | England | Caucasian | PB | Fluorescent | 1133 | 2115 | 1050 | 4298 | 1149 | 2263 | 1062 | 4474 | 0.49 | 0.422 | 8 |
| Cybulski | 2008 | Breast | Poland | Caucasian | PB | AS-PCR | 178 | 314 | 126 | 618 | 274 | 499 | 192 | 965 | 0.46 | 0.195 | 6 |
| Cybulski | 2008 | Prostate | Poland | Caucasian | PB | AS-PCR | 139 | 236 | 110 | 485 | 274 | 499 | 192 | 965 | 0.46 | 0.195 | 6 |
| Li | 2008 | Melanoma | USA | Caucasian | HB | PCR | 243 | 385 | 177 | 805 | 207 | 440 | 188 | 835 | 0.49 | 0.116 | 11 |
| Wang | 2009 | Bladder | China | Asian | HB | PCR-RFLP | 238 | 115 | 12 | 365 | 205 | 138 | 25 | 368 | 0.26 | 0.786 | 10 |
| Gangwar | 2009 | Bladder | India | Asian | HB | PCR-RFLP | 121 | 84 | 7 | 212 | 133 | 101 | 16 | 250 | 0.27 | 0.584 | 9 |
| De Vecchi | 2009 | Breast | Italy | Caucasian | PB | PCR-RFLP | 162 | 301 | 117 | 580 | 106 | 206 | 94 | 406 | 0.49 | 0.752 | 7 |
| Zhu | 2010 | RCC | China | Asian | HB | PCR-RFLP | 226 | 119 | 8 | 353 | 205 | 139 | 21 | 365 | 0.25 | 0.686 | 11 |
| Srivastava | 2010 | Gallbladder | India | Asian | PB | PCR-RFLP | 147 | 69 | 12 | 228 | 122 | 84 | 24 | 230 | 0.29 | 0.103 | 11 |
| Liu | 2010 | Colorectal | China | Asian | PB | PCR-RFLP | 233 | 116 | 21 | 370 | 528 | 278 | 32 | 838 | 0.20 | 0.538 | 13 |
| Li | 2010 | HNSCC | USA | Caucasian | HB | PCR–RFLP | 311 | 456 | 256 | 1023 | 257 | 542 | 253 | 1052 | 0.50 | 0.324 | 10 |
| Xiao | 2011 | Lymphoma | China | Asian | NM | PCR-PAGE | 43 | 17 | 4 | 64 | 89 | 38 | 6 | 133 | 0.19 | 0.460 | 3 |
| Xiao | 2011 | Lymphoma | China | Asian | NM | PCR-PAGE | 49 | 23 | 3 | 75 | 63 | 40 | 4 | 107 | 0.22 | 0.442 | 3 |
| Umar | 2011 | Esophageal | India | Asian | PB | PCR | 139 | 103 | 17 | 259 | 138 | 93 | 28 | 259 | 0.29 | 0.046 | 11 |
| Theodoropoulos | 2011 | Colorectal | Greece | Caucasian | HB | RFLP-PCR | 103 | 201 | 98 | 402 | 120 | 254 | 106 | 480 | 0.49 | 0.194 | 9 |
| Malik | 2011 | Esophageal | India | Asian | HB | RFLP-PCR | 68 | 59 | 8 | 135 | 96 | 75 | 24 | 195 | 0.32 | 0.127 | 8 |
| Malik | 2011 | Gastric | India | Asian | HB | RFLP-PCR | 59 | 44 | 5 | 108 | 96 | 75 | 24 | 195 | 0.32 | 0.127 | 8 |
| Ma | 2011 | Ovarian | China | Asian | HB | MassARRAY | 128 | 87 | 3 | 218 | 138 | 122 | 25 | 285 | 0.30 | 0.789 | 8 |
| Liamarkopoulos | 2011 | Gastric | Greece | Caucasian | HB | PCR-RFLP | 35 | 42 | 11 | 88 | 120 | 254 | 106 | 480 | 0.49 | 0.194 | 7 |
| Hart | 2011 | Lung | Norway | Caucasian | PB | TaqMan | 125 | 210 | 101 | 436 | 106 | 209 | 118 | 433 | 0.51 | 0.481 | 10 |
| Chatterjee | 2011 | Cervical | South Africa | African | HB | PCR-RFLP | 18 | 63 | 25 | 106 | 43 | 129 | 85 | 257 | 0.58 | 0.614 | 6 |
| Fu | 2011 | Prostate | China | Asian | HB | PCR-RFLP | 257 | 132 | 17 | 406 | 211 | 159 | 38 | 408 | 0.29 | 0.315 | 10 |
| Wang | 2012 | RCC | China | Asian | HB | PCR-RFLP | 192 | 101 | 7 | 300 | 168 | 114 | 18 | 300 | 0.25 | 0.817 | 10 |
| Wang | 2012 | PTC | China | Asian | HB | PCR–RFLP | 65 | 45 | 8 | 118 | 106 | 92 | 15 | 213 | 0.29 | 0.408 | 7 |
| Tong | 2012 | ALL | China | Asian | HB | PCR-RFLP | 217 | 113 | 31 | 361 | 338 | 153 | 28 | 519 | 0.20 | 0.057 | 10 |
| Hashemi | 2012 | Breast | Iran | Asian | HB | AS-PCR | 113 | 107 | 16 | 236 | 79 | 91 | 33 | 203 | 0.39 | 0.434 | 6 |
| George | 2012 | Prostate | India | Asian | HB | PCR-RFLP | 84 | 69 | 12 | 165 | 116 | 83 | 6 | 205 | 0.23 | 0.050 | 9 |
| Xiao | 2013 | Colorectal | China | Asian | HB | PCR-PAGE | 187 | 107 | 11 | 305 | 212 | 115 | 15 | 342 | 0.21 | 0.905 | 7 |
| Wu | 2013 | Colorectal | China | Asian | HB | PCR-SSCP | 284 | 152 | 15 | 451 | 358 | 244 | 29 | 631 | 0.24 | 0.119 | 11 |
| De Martino | 2013 | RCC | Austria | Caucasian | HB | PCR-RFLP | 72 | 138 | 40 | 250 | 53 | 129 | 68 | 250 | 0.53 | 0.572 | 9 |
| Pardini | 2014 | Colorectal | Spain | Caucasian | PB | Taqman | 500 | 996 | 482 | 1978 | 425 | 802 | 420 | 1647 | 0.50 | 0.290 | 11 |
| Pardini | 2014 | Colorectal | Italy | Caucasian | PB | Taqman | 195 | 285 | 137 | 617 | 783 | 1230 | 538 | 2551 | 0.45 | 0.178 | 9 |
| Pardini | 2014 | Colorectal | USA | Caucasian | PB | Taqman | 237 | 514 | 259 | 1010 | 383 | 794 | 403 | 1580 | 0.51 | 0.835 | 9 |
| Pardini | 2014 | Colorectal | England | Caucasian | PB | Taqman | 410 | 825 | 341 | 1576 | 165 | 393 | 209 | 767 | 0.53 | 0.436 | 11 |
| Pardini | 2014 | Colorectal | Czech | Caucasian | PB | Taqman | 239 | 479 | 249 | 967 | 169 | 326 | 177 | 672 | 0.51 | 0.443 | 10 |
| Pardini | 2014 | Colorectal | Netherlands | Caucasian | PB | Taqman | 169 | 282 | 134 | 585 | 106 | 177 | 76 | 359 | 0.46 | 0.895 | 8 |
| Tang | 2015 | OSCC | China | Asian | HB | PCR-RFLP | 328 | 159 | 18 | 505 | 276 | 197 | 34 | 507 | 0.26 | 0.885 | 10 |
| Carvalho | 2015 | ALL | Brazil | Mixed | NM | PCR | 23 | 81 | 26 | 130 | 47 | 53 | 25 | 125 | 0.41 | 0.163 | 4 |
MAF: minor allele frequency; HWE: Hardy-Weinberg equilibrium; OSCC: oral squamous cell carcinoma; PTC: papillary thyroid carcinoma; RCC: renal cell carcinoma; HNSCC: head and neck squamous cell carcinoma; ALL: acute lymphocytic leukemia; PB: population based; HB: hospital based; NM: not mentioned; PCR-PAGE: polymerase chain reaction-polyacrylamide gel electrophoresis; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; AS-PCR: allele-specific polymerase chain reaction.
Quantitative analysis
Overall meta-analysis information is shown in Table 2 and Figure 2. In the pooled analysis, the CASP8 -652 6N ins/del polymorphism was associated with reduced overall cancer risk in all five genetic models (homozygous: DD vs. II: odds ratio (OR)=0.76, 95% confidence interval (CI)=0.69–0.84; heterozygous: ID vs. II: OR=0.87, 95% CI=0.83–0.92; recessive: DD vs. ID/II: OR=0.82, 95% CI=0.75–0.89; dominant: ID/DD vs. II: OR=0.85, 95% CI=0.80–0.90; and allele: D vs. I: OR=0.87, 95% CI=0.83–0.91.
Table 2. Meta-analysis of the association between the CASP8 -652 6N ins/del polymorphism and overall cancer risk.
| Variables | No. of studies | Sample size | Homozygous | Heterozygous | Recessive | Dominant | Allele | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DD vs. II | ID vs. II | DD vs. ID/II | ID/DD vs. II | D vs. I | ||||||||
| OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | |||
| All | 49 | 33494/36397 | 0.76 (0.69-0.84) | <0.001 | 0.87 (0.83-0.92) | <0.001 | 0.82 (0.75-0.89) | <0.001 | 0.85 (0.80-0.90) | <0.001 | 0.87 (0.83-0.91) | <0.001 |
| Cancer type | ||||||||||||
| Colorectal | 12 | 13058/14418 | 0.93 (0.82-1.05) | 0.018 | 0.94 (0.88-0.99) | 0.529 | 0.96 (0.87-1.06) | 0.019 | 0.93 (0.87-1.00) | 0.190 | 0.96 (0.90-1.01) | 0.012 |
| Breast | 8 | 9943/10271 | 0.80 (0.67-0.96) | 0.001 | 0.90 (0.81-1.01) | 0.018 | 0.85 (0.74-0.99) | 0.002 | 0.87 (0.77-0.99) | 0.002 | 0.89 (0.80-0.98) | <0.001 |
| Esophageal | 3 | 1412/1196 | 0.56 (0.40-0.78) | 0.901 | 0.93 (0.74-1.17) | 0.206 | 0.58 (0.42-0.79) | 0.812 | 0.83 (0.71-0.97) | 0.385 | 0.81 (0.72-0.92) | 0.712 |
| RCC | 3 | 903/915 | 0.39 (0.26-0.59) | 0.852 | 0.78 (0.64-0.95) | 0.998 | 0.46 (0.32-0.66) | 0.732 | 0.71 (0.58-0.86) | 0.949 | 0.70 (0.61-0.82) | 0.966 |
| Lung | 2 | 1585/1544 | 0.66 (0.51-0.87) | 0.473 | 0.75 (0.64-0.88) | 0.385 | 0.75 (0.59-0.95) | 0.458 | 0.73 (0.63-0.85) | 0.453 | 0.78 (0.69-0.87) | 0.273 |
| Cervical | 2 | 420/824 | 0.58 (0.36-0.93) | 0.456 | 0.86 (0.59-1.25) | 0.230 | 0.59 (0.39-0.88) | 0.728 | 0.76 (0.59-0.98) | 0.355 | 0.76 (0.63-0.92) | 0.556 |
| Prostate | 2 | 650/205 | 1.54 (0.67-3.55) | 0.100 | 0.99 (0.79-1.23) | 0.411 | 1.50 (0.74-3.07) | 0.135 | 1.05 (0.85-1.29) | 0.321 | 1.11 (0.93-1.33) | 0.255 |
| Bladder | 2 | 577/618 | 0.44 (0.25-0.77) | 0.799 | 0.79 (0.62-1.01) | 0.334 | 0.48 (0.27-0.84) | 0.907 | 0.74 (0.59-0.93) | 0.317 | 0.74 (0.61-0.90) | 0.338 |
| Lymphoma | 2 | 139/240 | 1.19 (0.44-3.23) | 0.729 | 0.82 (0.52-1.31) | 0.635 | 1.26 (0.47-3.39) | 0.789 | 0.86 (0.56-1.34) | 0.559 | 0.93 (0.64-1.35) | 0.535 |
| Gastric | 2 | 196/675 | 0.35 (0.19-0.63) | 0.939 | 0.74 (0.44-1.23) | 0.145 | 0.45 (0.26-0.78) | 0.538 | 0.64 (0.40-1.01) | 0.171 | 0.66 (0.51-0.84) | 0.487 |
| ALL | 2 | 491/644 | 1.85 (1.20-2.87) | 0.655 | 1.83 (0.69-4.85) | 0.004 | 1.32 (0.81-2.14) | 0.228 | 1.79 (0.81-3.97) | 0.014 | 1.33 (1.10-1.61) | 0.443 |
| Others | 9 | 4120/4847 | 0.57 (0.43-0.75) | 0.009 | 0.72 (0.65-0.79) | 0.976 | 0.65 (0.49-0.88) | 0.001 | 0.70 (0.64-0.77) | 0.855 | 0.75 (0.68-0.84) | 0.013 |
| Ethnicity | ||||||||||||
| Asian | 27 | 10569/11219 | 0.58 (0.48-0.70) | <0.001 | 0.80 (0.75-0.85) | 0.231 | 0.62 (0.52-0.74) | 0.002 | 0.77 (0.72-0.83) | 0.016 | 0.79 (0.73-0.84) | <0.001 |
| Caucasian | 20 | 22689/24796 | 0.90 (0.83-0.98) | 0.006 | 0.92 (0.88-0.97) | 0.225 | 0.95 (0.89-1.02) | 0.007 | 0.92 (0.87-0.97) | 0.079 | 0.95 (0.91-0.99) | 0.008 |
| African | 1 | 106/257 | 0.70 (0.35-1.43) | / | 1.17 (0.62-2.19) | / | 0.63 (0.37-1.05) | / | 0.98 (0.54-1.80) | / | 0.82 (0.60-1.13) | / |
| Mixed | 1 | 130/125 | 2.13 (1.01-4.46) | / | 3.12 (1.70-5.73) | / | 1.00 (0.54-1.85) | / | 2.80 (1.57-5.00) | / | 1.50 (1.05-2.12) | / |
| Source of control | ||||||||||||
| PB | 24 | 25259/26848 | 0.83 (0.75-0.92) | <0.001 | 0.89 (0.84-0.94) | 0.008 | 0.89 (0.82-0.96) | <0.001 | 0.87 (0.81-0.93) | <0.001 | 0.89 (0.85-0.95) | <0.001 |
| HB | 22 | 7966/9184 | 0.61 (0.49-0.75) | <0.001 | 0.83 (0.77-0.89) | 0.213 | 0.67 (0.55-0.82) | <0.001 | 0.79 (0.73-0.87) | 0.024 | 0.81 (0.75-0.88) | <0.001 |
| NM | 3 | 269/365 | 1.73 (0.95-3.14) | 0.619 | 1.30 (0.53-3.20) | 0.003 | 1.07 (0.63-1.80) | 0.896 | 1.29 (0.58-2.88) | 0.005 | 1.14 (0.79-1.64) | 0.156 |
| Quality score | ||||||||||||
| >9 | 24 | 16745/16831 | 0.67 (0.58-0.77) | <0.001 | 0.81 (0.76-0.87) | 0.008 | 0.75 (0.66-0.85) | <0.001 | 0.78 (0.73-0.84) | <0.001 | 0.81 (0.76-0.87) | <0.001 |
| ≤9 | 25 | 16749/19566 | 0.87 (0.77-0.99) | <0.001 | 0.95 (0.90-1.01) | 0.289 | 0.90 (0.81-1.00) | <0.001 | 0.94 (0.88-1.00) | 0.048 | 0.94 (0.90-0.99) | <0.001 |
Values were in bold, if the 95% CI excluded 1 or P<0.05.
Het: heterogeneity; RCC: renal cell carcinoma; ALL: acute lymphocytic leukemia; HB: hospital based; PB: population based; NM: not mentioned.
Figure 2. Forest plot of the association between the CASP8 -652 6N ins/del polymorphism and cancer risk via the homozygous model.

The OR and 95% CI for each study are plotted as a box and horizontal line. ◊, pooled ORs and the corresponding 95% CIs.
In cancer type stratification analysis, the CASP8 -652 6N ins/del polymorphism decreased risk for colorectal cancer, breast cancer, esophageal cancer, renal cell carcinoma, lung cancer, cervical cancer, bladder cancer, gastric cancer, and other cancers. However, acute lymphocytic leukemia risk was increased (DD vs. II: OR=1.85, 95% CI=1.20–2.87; and D vs. I: OR=1.33, 95% CI=1.10–1.61). We observed no correlations between the CASP8 -652 6N ins/del polymorphism and prostate cancer or lymphoma.
Stratification analysis by ethnicity revealed a decreased cancer risk for Asians (DD vs. II: OR=0.58, 95% CI=0.48–0.70) and Caucasians (DD vs. II: OR=0.90, 95% CI=0.83–0.98), and an increased risk in mixed populations (DD vs. II: OR=2.13, 95% CI=1.01–4.46). We also found that the CASP8 -652 6N ins/del polymorphism decreased cancer risk in population-based (DD vs. II: OR=0.83, 95% CI=0.75–0.92) and hospital-based groups (DD vs. II: OR=0.61, 95% CI=0.49–0.75). Similarly, the CASP8 -652 6N ins/del polymorphism was associated with decreased cancer risk in both the high quality (DD vs. II: OR=0.67, 95% CI=0.58–0.77) and low quality study groups (DD vs. II: OR=0.87, 95% CI=0.77–0.99).
Heterogeneity and sensitivity analysis
Heterogeneity was observed in all five genetic models (P<0.001, Q test). Therefore, the random-effect model was adopted to generate ORs and 95% CIs. We also conducted a sequential leave-one-out sensitivity analysis to evaluate the impact of a single study on the pooled estimates. Omission of no single study influenced the pooled ORs, indicating the statistical robustness of this meta-analysis (data not shown).
Publication bias
Begg's funnel plot shapes did not suggest any obvious asymmetry (Figure 3). Egger's test results (DD vs. II: t=-4.17, P<0.001; ID vs. II: t=-0.12, P=0.905; DD vs. ID/II: t=-1.15, P=0.257; ID/DD vs. II: t=-1.09, P=0.281; and D vs. I: t=-3.33, P=0.002) suggested that publication bias existed in the homozygote and allele models.
Figure 3. Funnel plot analysis to detect publication bias for the CASP8 -652 6N ins/del polymorphism via the homozygous model.

Each point represents a separate study for the indicated association.
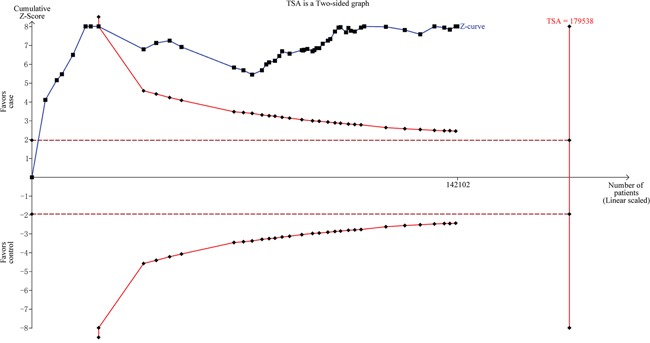
Trial sequential analysis
To minimize random errors and strengthen the robustness of our conclusions, we performed trial sequential analysis (TSA) (Figure 4). The cumulative Z-curve crossed the trial sequential monitoring boundary before the required information size was reached, suggesting that our study conclusion was convincing and no additional evidence was needed to verify said conclusion.
Figure 4. Trial sequential analysis for the CASP8 -652 6N ins/del polymorphism via the allele contrast model.
DISCUSSION
The present meta-analysis comprehensively evaluated the relationship between the CASP8 -652 6N ins/del polymorphism and cancer risk across 49 studies (33,494 cases and 36,397 controls). The CASP8 -652 6N ins/del polymorphism was associated with decreased cancer risk in all five genetic models, and in the following subgroups: colorectal cancer, breast cancer, esophageal cancer, renal cell carcinoma, lung cancer, cervical cancer, bladder cancer, gastric cancer, other cancers, Asian, Caucasian, mixed population, population-based controls, hospital-based controls, high quality score, and low quality score.
Human immune cells play critical roles in eliminating potentially malignant cells [58]. Caspase 8 protein (encoded by CASP8) maintains immune cells by mediating the activation-apoptosis balance [59]. Low caspase 8 expression or functional aberrations may decrease T lymphocyte apoptotic reactivity [13]. The CASP8 -652 6N del variant inactivates the transcription factor stimulatory protein 1 binding site, decreasing CASP8 transcription [13]. Thus, this variant may affect cancer susceptibility by influencing immune surveillance.
The first case-control study of the CASP8 -652 6N del variant-cancer association, with 4,995 cases and 4,972 controls, was conducted by Sun, et al. in 2007 [13]. The authors found that the CASP8 -652 6N deletion allele decreased susceptibility to lung, colorectal, esophageal, breast, cervical, and gastric cancers. Biochemical assays illustrated that this variant might decrease apoptotic reactivity in cancer cell-stimulated T lymphocytes. However, Umar, et al. did not detect any association between the CASP8 -652 6N polymorphism and esophageal squamous cell carcinoma (ESCC) risk in 259 patients and 259 healthy controls in an Indian population [45]. Several meta-analyses have attempted to address these contradictory conclusions. A 2012 meta-analysis by Chen, et al., including 19 case-control studies with 23,172 cases and 26,532 controls, associated the del allele, ins/del genotype, and del allele carriers with reduced overall cancer risk [16]. Similarly, in a meta-analysis incorporating 11 reports with 27,459 cases and 31,614 controls, Yin, et al. associated the CASP8 -652 5N del polymorphism with reduced overall cancer risk via homozygous, dominant, and recessive models [15]. In 2014, breast cancer- and colorectal cancer-specific meta-analyses [19, 20] concluded that the CASP8 -652 6N del polymorphism reduced cancer risk. However, no association was observed between this polymorphism and prostate cancer susceptibility in a meta-analysis by Zhang, et al. [21].
To provide a more robust clarification, our meta-analysis included all eligible studies published in either the English or Chinese language. In agreement with the four previously published meta-analyses, we found that the CASP8 -652 6N ins/del polymorphism was associated with reduced overall cancer risk. In subgroup analyses, the polymorphism was associated with reduced risk of colorectal cancer, breast cancer, esophageal cancer, renal cell carcinoma, lung cancer, cervical cancer, bladder cancer, gastric cancer, and other cancers, but not prostate cancer or lymphoma. A prostate cancer-specific meta-analysis also failed to detect a significant association. This may be attributed to cancer-specific inherent heterogeneity [60, 61]. Additionally, we observed an association with decreased cancer risk among Asians and Caucasians, but not Africans or mixed ethnicity populations. However, the limited number of studies in Africans and mixed ethnicity population may account for this finding, and CASP8 -652 6N ins/del polymorphism allelic distributions might vary geographically and ethnically.
Our meta-analysis of the association between the CASP8 -652 6N ins/del polymorphism and cancer risk is by far the largest such meta-analysis with the greatest statistical power published thus far. We conducted subgroup analyses to provide a more precise, cancer type-specific conclusion, and we assessed studies in both Chinese and English to minimize selection bias. However, our study had certain limitations. First, for some types of cancers, the calculated association was not robust enough due to limited numbers of original studies. Second, only one CASP8 genetic variant was considered, and confounding factors, such as other genetic mutations and environmental exposures, also influence cancer susceptibility. Third, the observed between-study heterogeneity may reduce the validity of our conclusions. Finally, publication bias, language bias, or selection bias might lead to false positive or negative findings.
The present work robustly concludes that the CASP8 -652 6N ins/del polymorphism is associated with reduced overall cancer risk. Refined studies with larger sample sizes, especially for certain cancer types and ethnic groups, are needed to fully validate this relationship.
MATERIALS AND METHODS
Search strategy
We conducted a literature search in PubMed and EMBASE using the following combined terms: ‘Caspase 8’ or ‘CASP8’ and ‘polymorphism’ or ‘polymorphisms’ or ‘single nucleotide polymorphism’ or ‘SNP’ or ‘variant’ and ‘cancer’ or ‘tumor’ or ‘carcinoma’ or ‘carcinogenesis’ or ‘neoplasm’. We also searched studies written in Chinese from two databases, WANFANG and CNKI. We searched for articles published through February 2017 without imposing language limitations. Relevant references were also collected from retrieved articles. Only the largest or the most recent study was retained if studies contained overlapping data.
Inclusion/exclusion criteria
Studies included in our analysis met the following criteria: (1) evaluated CASP8 -652 6N ins/del polymorphism with respect to cancer risk; (2) case-control design; (3) sufficient information to extract genotype frequencies for all subjects; (4) genotype frequency of controls consistent with Hardy-Weinberg equilibrium (HWE); (5) publication language was English or Chinese. Criteria for exclusion included: (1) abstract only, review, or meta-analysis; (2) case only studies; (3) no detailed genotyping data provided; (4) repeated publication.
Data extraction
Two authors (Jiarong Cai and Qingjian Ye) independently identified all eligible studies, and extracted data was included in the meta-analysis following consensus. The following items were recorded from each study: first author's name, year of publication, country, patient ethnicity, cancer type, source of controls, genotyping method, and genotype distributions of cases and controls. If reports contained more than one ethnic group or cancer type, we separated them into different studies.
Trial sequential analysis
After adopting a risk of 5% for type I errors and 30% for type II errors, the required information size (sample sizes from all included trials) was calculated. TSA monitoring boundaries were built based on required information size and risk for type I and type II errors. If the cumulative Z-curve crossed the TSA monitoring boundary before the required information size was reached (i.e. if a sufficiently small P-value was achieved), further trials were unnecessary.
Statistical analyses
We used the Chi-square test to ensure that all control genotype frequencies were in agreement with HWE. Odds ratios (ORs) with corresponding 95% confidence intervals (CIs) obtained from case and control genotype frequencies were used to assess the strength of association between the CASP8 -652 6N ins/del polymorphism and cancer risk. Pooled ORs were calculated for the following five genetic models: homozygote model (DD vs. II), heterozygote model (ID vs. II), recessive model (DD vs. ID/II), dominant model (ID/DD vs. II), and allele model (D vs. I). The Cochran's Chi-square-based Q-test and the inconsistency index (I2 statistics) were adopted to assess heterogeneity between study results. I2<50% or P>0.10 indicates heterogeneity. The fixed-effects model (Mantel-Haenszel method) was used to estimate the pooled OR if no heterogeneity existed (I2<50% or P>0.10). Otherwise, the random-effects model (DerSimonian and Laird method) was applied. Quality assessment for each study was performed using the quality assessment criteria described previously (Supplementary Table 1) [62–65]. To decrease heterogeneity among studies, we conducted stratification analyses by ethnicity, cancer type, control source, and quality score. By adopting one-way sensitivity analysis, we recalculated the pooled ORs to assess the robustness of the results. We also conducted Begg's funnel plot and Egger's regression asymmetry test to examine potential publication bias [66–69]. STATA software v. 11.0 (Stata Corporation, College Station, TX) was used for statistical analyses [70]. P<0.05 (two-sided) was considered statistically significant.
SUPPLEMENTARY MATERIALS TABLES
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 31172044).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008;359:2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Wu XS, He J, Ma T, Lei W, Shen ZY. A novel TP53 variant (rs78378222 A > C) in the polyadenylation signal is associated with increased cancer susceptibility: evidence from a meta-analysis. Oncotarget. 2016;7:32854–32865. doi: 10.18632/oncotarget.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue W, Zhu M, Wang Y, He J, Zheng L. Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta-analysis. Sci Rep. 2015;5:7986. doi: 10.1038/srep07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 8.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 9.Nunez G, Benedict MA, Hu Y, Inohara N. Caspases: the proteases of the apoptotic pathway. Oncogene. 1998;17:3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 10.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 11.Feltham R, Vince JE, Lawlor KE. Caspase-8: not so silently deadly. Clin Transl Immunology. 2017;6:e124. doi: 10.1038/cti.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenet J, Teitz T, Wei T, Valentine V, Kidd VJ. Structure and chromosome localization of the human CASP8 gene. Gene. 1999;226:225–232. doi: 10.1016/s0378-1119(98)00565-4. [DOI] [PubMed] [Google Scholar]
- 13.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, Guo Y, Yang M, Zhang X, Zhang Q, Zeng C, Lin D. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 14.Sergentanis TN, Economopoulos KP. Association of two CASP8 polymorphisms with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;120:229–234. doi: 10.1007/s10549-009-0471-5. [DOI] [PubMed] [Google Scholar]
- 15.Yin M, Yan J, Wei S, Wei Q. CASP8 polymorphisms contribute to cancer susceptibility: evidence from a meta-analysis of 23 publications with 55 individual studies. Carcinogenesis. 2010;31:850–857. doi: 10.1093/carcin/bgq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D, Ma T, Liu XW, Liu Z. CASP-8 -652 6N ins/del polymorphism and cancer risk: a literature-based systematic HuGE review and meta-analysis. Exp Ther Med. 2012;4:762–770. doi: 10.3892/etm.2012.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Yang Y, Guo C, Wang Y. CASP8 -652 6N del polymorphism and cancer risk: a meta-analysis of 30 case-control studies in 50,112 subjects. Mutagenesis. 2012;27:559–566. doi: 10.1093/mutage/ges019. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YJ, Zhong XP, Chen Y, Liu SR, Wu G, Liu YF. Association between CASP-8 gene polymorphisms and cancer risk in some Asian population based on a HuGE review and meta-analysis. Genet Mol Res. 2013;12:6466–6476. doi: 10.4238/2013.February.28.3. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Wang J, Wang F, Ma Z, Yu Z. CAS P8 -652 6N del polymorphism and breast cancer risk: a systematic review and meta-analysis. Neth J Med. 2014;72:10–16. [PubMed] [Google Scholar]
- 20.Peng Q, Lao X, Tang W, Chen Z, Li R, Wang J, Deng Y, Li T, Qin X, Li S. CASP8 -652 6N del polymorphism contributes to colorectal cancer susceptibility: evidence from a meta-analysis. PLoS One. 2014;9:e87925. doi: 10.1371/journal.pone.0087925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang CD, Li HT, Liu K, Lin ZD, Peng QL, Qin X, He M, Wu H, Mo ZN, Yang XL. Impact of caspase-8 (CASP8) -652 6N del and D302H polymorphisms on prostate cancer in different ethnic groups. Asian Pac J Cancer Prev. 2014;15:7713–7718. doi: 10.7314/apjcp.2014.15.18.7713. [DOI] [PubMed] [Google Scholar]
- 22.Yoo SS, Choi JE, Lee WK, Choi YY, Kam S, Kim MJ, Jeon HS, Lee EB, Kim DS, Lee MH, Kim IS, Jheon S, Park JY. Polymorphisms in the CASPASE genes and survival in patients with early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:5823–5829. doi: 10.1200/JCO.2009.23.1738. [DOI] [PubMed] [Google Scholar]
- 23.Choi JY, Kim JG, Lee YJ, Chae YS, Sohn SK, Moon JH, Kang BW, Jung MK, Jeon SW, Park JS, Choi GS. Prognostic impact of polymorphisms in the CASPASE genes on survival of patients with colorectal cancer. Cancer Res Treat. 2012;44:32–36. doi: 10.4143/crt.2012.44.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhlmann JD, Bankfalvi A, Schmid KW, Callies R, Kimmig R, Wimberger P, Siffert W, Bachmann HS. Prognostic relevance of caspase 8 -652 6N InsDel and Asp302His polymorphisms for breast cancer. BMC Cancer. 2016;16:618. doi: 10.1186/s12885-016-2662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kesarwani P, Mandal RK, Maheshwari R, Mittal RD. Influence of caspases 8 and 9 gene promoter polymorphism on prostate cancer susceptibility and early development of hormone refractory prostate cancer. BJU Int. 2011;107:471–476. doi: 10.1111/j.1464-410X.2010.09533.x. [DOI] [PubMed] [Google Scholar]
- 26.Cybulski C, Wokolorczyk D, Gliniewicz B, Sikorski A, Gorski B, Jakubowska A, Huzarski T, Byrski T, Debniak T, Gronwald J, Lubinski J, Narod SA. A six-nucleotide deletion in the CASP8 promoter is not associated with a susceptibility to breast and prostate cancers in the Polish population. Breast Cancer Res Treat. 2008;112:367–368. doi: 10.1007/s10549-007-9864-5. [DOI] [PubMed] [Google Scholar]
- 27.Frank B, Rigas SH, Bermejo JL, Wiestler M, Wagner K, Hemminki K, Reed MW, Sutter C, Wappenschmidt B, Balasubramanian SP, Meindl A, Kiechle M, Bugert P, et al. The CASP8 -652 6N del promoter polymorphism and breast cancer risk: a multicenter study. Breast Cancer Res Treat. 2008;111:139–144. doi: 10.1007/s10549-007-9752-z. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Zhao H, Hu Z, Liu Z, Wang LE, Gershenwald JE, Prieto VG, Lee JE, Duvic M, Grimm EA, Wei Q. Genetic variants and haplotypes of the caspase-8 and caspase-10 genes contribute to susceptibility to cutaneous melanoma. Hum Mutat. 2008;29:1443–1451. doi: 10.1002/humu.20803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, Penegar S, Tomlinson I, Houlston RS. CASP8 variants D302H and -652 6N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer. 2008;98:1434–1436. doi: 10.1038/sj.bjc.6604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M, Sun T, Wang L, Yu D, Zhang X, Miao X, Liu J, Zhao D, Li H, Tan W, Lin D. Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res. 2008;14:3230–3236. doi: 10.1158/1078-0432.CCR-08-0177. [DOI] [PubMed] [Google Scholar]
- 31.De Vecchi G, Verderio P, Pizzamiglio S, Manoukian S, Barile M, Fortuzzi S, Ravagnani F, Pierotti MA, Radice P, Peterlongo P. Evidences for association of the CASP8 -652 6N del promoter polymorphism with age at diagnosis in familial breast cancer cases. Breast Cancer Res Treat. 2009;113:607–608. doi: 10.1007/s10549-008-9963-y. [DOI] [PubMed] [Google Scholar]
- 32.Gangwar R, Mandhani A, Mittal RD. Caspase 9 and caspase 8 gene polymorphisms and susceptibility to bladder cancer in north Indian population. Ann Surg Oncol. 2009;16:2028–2034. doi: 10.1245/s10434-009-0488-3. [DOI] [PubMed] [Google Scholar]
- 33.Wang M, Zhang Z, Tian Y, Shao J, Zhang Z. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter associated with risk and progression of bladder cancer. Clin Cancer Res. 2009;15:2567–2572. doi: 10.1158/1078-0432.CCR-08-2829. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Lu J, Liu Z, Wang LE, Zhao H, El-Naggar AK, Sturgis EM, Wei Q. The six-nucleotide deletion/insertion variant in the CASP8 promoter region is inversely associated with risk of squamous cell carcinoma of the head and neck. Cancer Prev Res (Phila) 2010;3:246–253. doi: 10.1158/1940-6207.CAPR-08-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Zhang Y, Jin M, Ni Q, Liang X, Ma X, Yao K, Li Q, Chen K. Association of selected polymorphisms of CCND1, p21, and caspase8 with colorectal cancer risk. Mol Carcinog. 2010;49:75–84. doi: 10.1002/mc.20579. [DOI] [PubMed] [Google Scholar]
- 36.Srivastava K, Srivastava A, Mittal B. Caspase-8 polymorphisms and risk of gallbladder cancer in a northern Indian population. Mol Carcinog. 2010;49:684–692. doi: 10.1002/mc.20641. [DOI] [PubMed] [Google Scholar]
- 37.Zhu J, Qin C, Wang M, Yan F, Ju X, Meng X, Ding Q, Li P, Yang J, Cao Q, Zhang Z, Yin C. Functional polymorphisms in cell death pathway genes and risk of renal cell carcinoma. Mol Carcinog. 2010;49:810–817. doi: 10.1002/mc.20656. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee K, Williamson AL, Hoffman M, Dandara C. CASP8 promoter polymorphism is associated with high-risk HPV types and abnormal cytology but not with cervical cancer. J Med Virol. 2011;83:630–636. doi: 10.1002/jmv.22009. [DOI] [PubMed] [Google Scholar]
- 39.Fu G, Tang J, Wang M, Qin C, Yan F, Ding Q, Yin C, Wang X, Zhang Z. CASP8 promoter polymorphism, mRNA expression and risk of prostate cancer among Chinese men. J Biomed Res. 2011;25:128–134. doi: 10.1016/S1674-8301(11)60016-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart K, Landvik NE, Lind H, Skaug V, Haugen A, Zienolddiny S. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 2011;71:123–129. doi: 10.1016/j.lungcan.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Liamarkopoulos E, Gazouli M, Aravantinos G, Tzanakis N, Theodoropoulos G, Rizos S, Nikiteas N. Caspase 8 and caspase 9 gene polymorphisms and susceptibility to gastric cancer. Gastric Cancer. 2011;14:317–321. doi: 10.1007/s10120-011-0045-1. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Zhang J, Liu S, Huang Y, Chen B, Wang D. Polymorphisms in the CASP8 gene and the risk of epithelial ovarian cancer. Gynecol Oncol. 2011;122:554–559. doi: 10.1016/j.ygyno.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 43.Malik MA, Zargar SA, Mittal B. A six-nucleotide deletion polymorphism in the casp8 promoter is associated with reduced risk of esophageal and gastric cancers in Kashmir valley. Indian J Hum Genet. 2011;17:152–156. doi: 10.4103/0971-6866.92090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theodoropoulos GE, Gazouli M, Vaiopoulou A, Leandrou M, Nikouli S, Vassou E, Kouraklis G, Nikiteas N. Polymorphisms of caspase 8 and caspase 9 gene and colorectal cancer susceptibility and prognosis. Int J Colorectal Dis. 2011;26:1113–1118. doi: 10.1007/s00384-011-1217-5. [DOI] [PubMed] [Google Scholar]
- 45.Umar M, Upadhyay R, Kumar S, Ghoshal UC, Mittal B. CASP8 -652 6N del and CASP8 IVS12-19G>A gene polymorphisms and susceptibility/prognosis of ESCC: a case control study in northern Indian population. J Surg Oncol. 2011;103:716–723. doi: 10.1002/jso.21881. [DOI] [PubMed] [Google Scholar]
- 46.Xiao MS, Zhang DF, Zeng Y, Cheng YF, Yao YG. Polymorphisms in the promoter region of the CASP8 gene are not associated with non-Hodgkin's lymphoma in Chinese patients. Ann Hematol. 2011;90:1137–1144. doi: 10.1007/s00277-011-1265-5. [DOI] [PubMed] [Google Scholar]
- 47.George GP, Mandal RK, Kesarwani P, Sankhwar SN, Mandhani A, Mittal RD. Polymorphisms and haplotypes in caspases 8 and 9 genes and risk for prostate cancer: a case-control study in cohort of North India. Urol Oncol. 2012;30:781–789. doi: 10.1016/j.urolonc.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 48.Hashemi M, Eskandari-Nasab E, Fazaeli A, Rezaei H, Mashhadi MA, Arbabi F, Taheri M. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 -652 6N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505:176–179. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 49.Tong N, Zhang L, Sheng X, Wang M, Zhang Z, Fang Y, Xue Y, Li J, Zhang Z. Functional polymorphisms in FAS, FASL and CASP8 genes and risk of childhood acute lymphoblastic leukemia: a case-control study. Leuk Lymphoma. 2012;53:1360–1366. doi: 10.3109/10428194.2011.654117. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Yuan Q, Yin C, Zhu J, Qin C, Zhang W. Association of caspases 8-652 6N ins/del polymorphism and risk of renal cell carcmoma in Chinese Han population. Chin J Exp Surg. 2012;29:2409–2412. [Google Scholar]
- 51.Wang YX, Zhao L, Wang XY, Liu CM, Yu SG. Role of Caspase 8, Caspase 9 and Bcl-2 polymorphisms in papillary thyroid carcinoma risk in Han Chinese population. Med Oncol. 2012;29:2445–2451. doi: 10.1007/s12032-011-0121-8. [DOI] [PubMed] [Google Scholar]
- 52.de Martino M, Haitel A, Schatzl G, Klingler HC, Klatte T. The CASP8 -652 6N insertion/deletion promoter polymorphism is associated with renal cell carcinoma risk and metastasis. J Urol. 2013;190:717–722. doi: 10.1016/j.juro.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Li Y, Li S, Zhu L, Li G, Yu Z, Zhao X, Ge J, Cui B, Dong X, Tian S, Hu F, Zhao Y. Association between main Caspase gene polymorphisms and the susceptibility and prognosis of colorectal cancer. Med Oncol. 2013;30:565. doi: 10.1007/s12032-013-0565-0. [DOI] [PubMed] [Google Scholar]
- 54.Xiao MS, Chang L, Li WL, Du YS, Pan Y, Zhang DF, Wen Y, Luo J, Li XY, Yao YG. Genetic polymorphisms of the CASP8 gene promoter may not be associated with colorectal cancer in Han Chinese from southwest China. PLoS One. 2013;8:e67577. doi: 10.1371/journal.pone.0067577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardini B, Verderio P, Pizzamiglio S, Nici C, Maiorana MV, Naccarati A, Vodickova L, et al. Veneroni S, Daidone MG, Ravagnani F, Bianchi T, Bujanda L, et al. Association between CASP8 -652 6N del polymorphism (rs3834129) and colorectal cancer risk: results from a multi-centric study. PLoS One. 2014;9:e85538. doi: 10.1371/journal.pone.0085538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvalho DC, Wanderley AV, Amador MA, Fernandes MR, Cavalcante GC, Pantoja KB, Mello FA, de Assumpcao PP, Khayat AS, Ribeiro-Dos-Santos A, Santos S, Dos Santos NP. Amerindian genetic ancestry and INDEL polymorphisms associated with susceptibility of childhood B-cell leukemia in an admixed population from the Brazilian Amazon. Leuk Res. 2015 doi: 10.1016/j.leukres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Tang YI, Liu Y, Zhao W, Yu T, Yu H. Caspase-8 polymorphisms and risk of oral squamous cell carcinoma. Exp Ther Med. 2015;10:2267–2276. doi: 10.3892/etm.2015.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 59.Ho PK, Hawkins CJ. Mammalian initiator apoptotic caspases. FEBS J. 2005;272:5436–5453. doi: 10.1111/j.1742-4658.2005.04966.x. [DOI] [PubMed] [Google Scholar]
- 60.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan H, Bao J, Zhou X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci Rep. 2015;5:12566. doi: 10.1038/srep12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu C, Zhu J, Fu W, Liang Z, Song S, Zhao Y, Lyu L, Zhang A, He J, Duan P. MDM4 rs4245739 A > C polymorphism correlates with reduced overall cancer risk in a meta-analysis of 69477 subjects. Oncotarget. 2016;7:71718–71726. doi: 10.18632/oncotarget.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu W, Zhuo ZJ, Chen YC, Zhu J, Zhao Z, Jia W, Hu JH, Fu K, Zhu SB, He J, Liu GC. NFKB1 -94insertion/deletion ATTG polymorphism and cancer risk: evidence from 50 case-control studies. Oncotarget. 2017;8:9806–9822. doi: 10.18632/oncotarget.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue WQ, He YQ, Zhu JH, Ma JQ, He J, Jia WH. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci Rep. 2014;4:6791. doi: 10.1038/srep06791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer. 2013;133:1765–1775. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 67.He J, Xi B, Ruiter R, Shi TY, Zhu ML, Wang MY, Li QX, Zhou XY, Qiu LX, Wei QY. Association of LEP G2548A and LEPR Q223R polymorphisms with cancer susceptibility: evidence from a meta-analysis. PLoS One. 2013;8:e75135. doi: 10.1371/journal.pone.0075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu J, Hua RX, Jiang J, Zhao LQ, Sun X, Luan J, Lang Y, Sun Y, Shang K, Peng S, Ma J. Association studies of ERCC1 polymorphisms with lung cancer susceptibility: a systematic review and meta-analysis. PLoS One. 2014;9:e97616. doi: 10.1371/journal.pone.0097616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He J, Wang F, Zhu JH, Chen W, Cui Z, Jia WH. No association between MTR rs1805087 A > G polymorphism and non-Hodgkin lymphoma susceptibility: evidence from 11 486 subjects. Leuk Lymphoma. 2015;56:763–767. doi: 10.3109/10428194.2014.935370. [DOI] [PubMed] [Google Scholar]
- 70.Niu Y, Li F, Tang B, Shi Y, Yu P. Association of hOGG1 Ser326Cys polymorphism with gastric cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:6563–6568. doi: 10.1007/s11033-012-1485-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


