Abstract
Pituitary adenomas are among the most frequent intracranial neoplasms and treatment depends on tumor subtype and clinical features. Unfortunately, non responder cases occur, then new molecular targets are needed.
Notch system component expression and activation data are scarce in pituitary tumorigenesis, we therefore aimed to characterize Notch system in pituitary tumors of different histotype. In human pituitary adenomas we showed NOTCH1-4 receptors, JAGGED1 ligand and HES1 target gene expression with positive correlations between NOTCH1,2,4 and HES1, and NOTCH3 and JAGGED1 denoting Notch system activation in a subset of tumors. Importantly, NOTCH3 positive cells were higher in corticotropinomas and somatotropinomas compared to non functioning adenomas. In accordance, Notch activation was evidenced in AtT20 tumor corticotropes, with higher levels of NOTCH1-3 active domains, Jagged1 and Hes1 compared to normal pituitary.
In the prolactinoma cell lines GH3 and MMQ, in vivo GH3 tumors and normal glands, Notch system activation was lower than in corticotropes. In MMQ cells only the NOTCH2 active domain was increased, whereas NOTCH1 active domain was higher in GH3 tumors. High levels of Jagged1 and Dll1 were found solely in GH3 cells, and Hes1, Hey1 and Hey2 were expressed in a model dependent pattern.
Prolactinomas harbored by lacDrd2KO mice expressed high levels of NOTCH1 active domain and reduced Hes1.
We show a differential expression of Notch system components in tumoral and normal pituitaries and specific Notch system involvement depending on adenoma histotype, with higher activation in corticotropinomas. These data suggest that targeting Notch pathway may benefit non responder pituitary adenomas.
Keywords: Notch, pituitary, corticotropinoma, prolactinoma, Jagged1
INTRODUCTION
Pituitary adenomas are among the most frequent intracranial neoplasms, with a prevalence estimated at 22.5% by autopsy and magnetic resonance imaging studies [1, 2]. They are classified as functional or non functioning tumors, the former with clinical specific symptoms due to increased hormonal secretion and activity. Non functioning pituitary adenomas, with no symptoms or signs secondary to hormone over-secretion, are identified instead by mass effect or incidentally found in autopsy [1, 3, 4]. Treatment depends on tumor subtype, size and mass compression on cerebral neighboring tissue. Surgery is the first line of treatment for non functioning adenomas. Instead, effective medical therapy has been demonstrated in functioning adenomas, in particular in prolactin and growth hormone secreting pituitary tumors [5]. Unfortunately, non responder cases occur in all tumor subtypes [6]. Therefore, new molecular targets are needed to improve treatments.
The Notch system is conserved across species [7, 8]. It has been extensively characterized as a regulator of cell fate decisions in a variety of organisms and tissues, and participates in tumor generation so that its presence and involvement in pituitary tumor development merits a comprehensive study.
There are four Notch receptors (Notch1–4) in mammals. They are single pass type I transmembrane molecules coded by a single precursor that is activated by post-transductional proteolytic cleavage resulting in a non-covalently linked heterodimer composed of an extracellular fragment and a transmembrane-intracellular subunit [9].
There are five mammalian Notch ligands, three of which belong to the Delta family (Dll1, Dll3 and Dll4) and two to the Jagged family (Jagged1 and Jagged2). Notch ligands are also transmembrane proteins so that Notch pathway activation involves cell to cell contact. Receptor–ligand interaction triggers crucial signaling cleavages within the Notch receptor [10]. The clipping of the extracellular domain creates a membrane–tethered intermediate which is a substrate for an intramembrane cleaving protease, the γ-secretase [11]. γ-secretase-induced cleavage finally releases the Notch intracellular domain (NICD) from the membrane, and the released NICD translocates directly to the nucleus, where it forms a transcriptional complex with the DNA binding protein CSL, Mastermind and transcriptional coactivators to drive the expression of Notch target genes [7, 8]. Target genes regulated by Notch are highly dependent on cell type and can include genes whose products are involved in fundamental aspects of cell biology, such as cell cycle regulation [12, 13], cellular differentiation, and metabolism [14]. Common targets of the pathway include the Hes and Hey families of transcription repressors [15–17] as well as Myc transcription factor [14, 18, 19]. The binding and function of Notch on DNA involves a rapid and dynamic process controlled by phosphorylation and ubiquitination and subsequent proteasomal degradation [20, 21], which shuts off the pathway. In the absence of NICD, CSL forms complexes with a variety of corepressors to suppress the transcription of Notch target genes [7, 8].
Given the important and widespread roles of Notch signaling across a range of tissues, it is not surprising that abnormal Notch signaling is involved in several inherited human diseases [22, 23]. Furthermore, Notch malfunction has been associated with diseases linked to changes in cell fate and cell proliferation including cancer [22]. These syndromes present a broad range of clinical symptoms, emphasizing the highly pleiotropic nature of Notch. Moreover, both solid tumors and leukemias have been associated with deregulation of the Notch system in which, depending on the context, it can act both as an oncogene or a tumor suppressor. Notch signaling is overexpressed or constitutively activated in colorectal cancer partly because of mutations in regulators of Notch signaling [24–26]. In particular, Jagged1 expressed in tumor cells or produced by endothelial cells, is thought to be a key ligand for Notch activation in colorectal cancer cells [27, 28]. Notch signaling plays a crucial role in the early stages of colorectal cancer development by controlling the fate of stem cells and cancer stem cells [29]. In mammals, adult stem cell subpopulation sustains tissue homeostasis granting cell replacement during aging or damage [30]. Similarly, different cell populations were described in tumors in which has been identified a small fraction of tumor-initiating cells, representing the so-called cancer stem cells [31]. Notch system also participates at the later stages of tumor invasion and metastasis [29]. A number of studies have confirmed that activation of Notch signaling also plays an oncogenic role in breast cancer [32, 33]. In contrast, more recent studies indicate that hyperactivation of Notch3 may be detrimental to breast cancer cells by inducing senescence [34].
Various components of the Notch pathway are expressed during pituitary development, including Notch2 and Notch3 receptors, the ligand Jagged 1 and the downstream effector Hes1 [35–37]. Interestingly, in the anterior pituitary of adult mice, Notch receptor genes were found to be expressed in side population cells (SP), a cellular fraction which is enriched in stem/ progenitor cells [38–40]. In the adult rat pituitary, Tando and colleagues determined that the Notch system is involved in follicle stellate- S-100 positive cell proliferation, and that Notch1, Notch2, and Jagged1 colocalized with S-100 protein, while Jagged2 expression was described in melanotropes [41]. Recently, the same group also described that Notch2 activation in the pituitary gland needed E-cadherin mediated cell attachment [42].
In pituitary tumorigenesis, published data related to Notch system expression and functions are scarce. GeneChip microarrays and proteomics analyses demonstrated increased expression of NOTCH3 in non functioning and prolactin secreting adenomas in humans while in somatotropinomas a significantly reduced expression of NOTCH3 was found [43, 44]. Furthermore, microarray analysis performed in the fractioned SP and main population from human GH and non functioning pituitary adenomas cells showed more than 1.5 fold increased expression of components of the Notch system in the SP, including HES1, JAG1 and NOTCH paralogs [40]. It is known that Notch and Wnt pathway genes as other key markers, represent not only stem cell signaling systems but also regulatory circuits known for their critical role in pituitary embryonic development [45].
Notch3 and Jagged1 were also overexpressed in human clinically non functioning pituitary adenomas compared to normal pituitary gland [46, 47], while no significant differences were determined for prolactin or growth hormone secreting adenomas in that study [46].
Evidence points to an association of increased activation of the Notch system with more aggressive pituitary adenomas. However, there is no complete description of all Notch receptors, ligands and downstream effectors in the normal and pathological pituitary gland. Indeed, there are only few studies evaluating Notch system in human ACTH secreting adenomas [48] or prolactinoma models. There is definitely a potential therapeutic benefit for targeting Notch in tumorigenesis, as evidence in pituitary adenomas is lacking. But, because Notch function and system components can substantially differ and be dependent on cell type and tissue, and specific for each type of cancer, it is vital to characterize gene expression and activation in each adenoma type.
Therefore, in the present study we decided to evaluate the expression of the different Notch receptors and other components of the system in a comparative manner in tumoral and normal pituitaries in human and rodent samples. In this way, we aimed to elucidate Notch system significance in pituitary tumor development in search of new targets for the treatment of adenomas with resistance or intolerance to pharmacological therapy in which no alternatives exist other than pituitary surgery.
RESULTS
Notch signaling component expression in human pituitary adenomas
In human pituitary adenomas NOTCH1-4 mRNA expression was detected in all tumors analyzed (Table 1), with variable levels among the same adenoma type (Figure 1A–1D). The expression of JAGGED1 and HES1 was also determined in samples in which sufficient RNA was available and variable levels of expression were quantified independently of tumor type (Figure 1E–1F).
Table 1. Clinical and tumor features from human samples used in qRT-PCR experiments.
| Histotype | Age | Sex | Micro / macroadenoma | Recurrence | Ki67 |
|---|---|---|---|---|---|
| NF1 | 47 | M | macroadenoma | no | 3, 0 |
| NF2 | 62 | F | macroadenoma | no | > 3 |
| NF3 | 42 | F | macroadenoma | no | 4, 5 |
| NF4 | 45 | M | macroadenoma | no | < 1 |
| NF5 | M | - | no | 5 | |
| NF6 | 67 | M | macroadenoma | - | - |
| NF7 | M | - | no | - | |
| NF8 | 41 | F | macroadenoma | no | 1, 8 |
| NF9 | 42 | F | macroadenoma | no | 4, 5 |
| NF10 | 46 | F | - | no | < 1 |
| NF11 | 68 | F | macroadenoma | yes | 4 |
| NF12 | 58 | F | macroadenoma | no | 3 |
| ACTH1 | 28 | F | microadenoma | no | < 1 |
| ACTH2 | 56 | M | macroadenoma | no | 3, 4 |
| ACTH3 | 25 | F | microadenoma | no | < 1 |
| ACTH4 | 67 | F | macroadenoma | yes | 9 |
| ACTH5 | 38 | M | macroadenoma | no | 1, 5 |
| ACTH6 | 65 | F | macroadenoma | yes | 11, 2 |
| GH1 | 36 | F | - | - | - |
| GH2 | 60 | F | macroadenoma | no | 1 |
| GH3 | 35 | M | macroadenoma | yes | 8, 0 |
Age, sex, recurrence and tumor size, histotype and Ki67 proliferation index from human samples used in qRT-PCR experiments. N = 21.
Figure 1. Notch system components are expressed in human pituitary adenomas.

mRNA expression of NOTCH1-4 receptors, JAGGED1 ligand and HES1 target gene were determined by qRT-PCR. Gene levels normalized to the housekeeping gene Gadph are shown as percentage of change of NF average (which was considered 100%) (NF = non functioning adenoma; ACTH = corticotropinoma; GH = somatotropinoma).
Remarkably, positive correlations between the expression of NOTCH1-2,4 and the HES1 target gene, and between NOTCH3 with the ligand JAGGED1 were found in the cohort of samples used when all adenomas were considered, independently of tumor histotype (Figure 2). These significant correlations clearly denote activation of the Notch system in a subset of pituitary adenomas.
Figure 2. NOTCH-JAGGED1 and NOTCH-HES1 correlations.

Relation between mRNA levels of NOTCH1-4 and JAGGED1 (n = 13, 8, 13, 6 (A–D)) or HES1 (n = 11, 10, 11, 12 (E–H)) was determined in all adenomas tested. The equation of linear regression, R2 coefficient of determination and coefficient of Spearman are shown in each graph. p ≤ 0.05 denotes a significant correlation.
Instead, no correlation between Ki-67 and any of the Notch system components analyzed was found (data not shown). Correlations of gene expression and tumor recurrence or extrasellar extension could not be performed as only 4 were recurrent tumors, and most were macroadenomas (Table 1).
To evaluate Notch protein expression, distribution and cellular localization in human pituitary adenomas (see Table 2) we performed NOTCH3 immunohistochemistry and found that non functioning adenomas, prolactinomas, somatotropinomas and corticotropinomas expressed NOTCH3 protein in the cytoplasm and membrane of tumor cells (Figure 3A–3D). In prolactinomas and non functioning adenomas NOTCH3 cells were scattered and isolated (Figure 3A, 3B). Conversely, in somatotropinomas and corticotropinomas, NOTCH3-positive cells were observed organized in clusters with particularly intense staining in ACTH tumors (Figure 3C, 3D). Quantitation of percentage of NOTCH3 positive cells per total cells showed higher levels in corticotropinomas and somatotropinomas compared to non functioning adenomas (Figure 3E).
Table 2. Clinical and tumor features from human samples used in immunohistochemistry experiment.
| Histotype | Age | Sex | Micro / macroadenoma | Recurrence | Ki67 |
|---|---|---|---|---|---|
| ACTH-a | F | microadenoma | no | − | |
| ACTH-b | 46 | F | microadenoma | no | < 1 |
| ACTH-c | F | microadenoma | no | − | |
| GH-a | 49 | F | macroadenoma | no | 3 |
| GH-b | 58 | F | microadenoma | no | < 1 |
| GH-c | 47 | F | – | no | 3.0 |
| GH-d | 26 | M | macroadenoma | yes | 3, 5 |
| PRL-a | 50 | F | macroadenoma | no | 12 |
| PRL-b | 50 | M | macroadenoma | no | – |
| NF-a | 37 | F | microadenoma | no | – |
| NF-b | F | macroadenoma | no | – | |
| NF-c | 67 | M | macroadenoma | no | – |
Age, sex, recurrence and tumor size, histotype and Ki67 proliferation index from human samples used in NOTCH3 immunohistochemistry experiment. N = 12.
Figure 3. NOTCH3 protein expression determined by immunohistochemistry.

Percentage of positive cells, distribution and cellular localization in human pituitary adenomas. Representative images of NOTCH3 stained cells, (A–B) NOTCH3+ cells were isolated within prolactinomas and NF pituitary tumors, and in the GH and ACTH tumors (C–D) specific stained groups of positive cells were evidenced. NOTCH staining was visualized in membranes and cytoplasms of human adenoma cells. Arrows indicate staining in cytoplasm, and arrowheads in cell membranes. (E) Average of the percentage of NOTCH3+ cells in relation to total cells for Non Functioning adenomas (NF), prolactinomas (PRLoma), corticotropinomas (ACTHoma) and somatotropinomas (GHoma). N = 3,2,3,4 patients, respectively. *P ≤ 0.05 vs. NF.
Notch system is active in the corticotropinoma cell line AtT20
mRNA expression of the components of the Notch system was determined in the AtT20 tumoral corticotrope cell line by qRT-PCR and compared with normal mouse pituitaries. Notch1, Notch2 and Notch4 mRNA expression levels were similar in AtT20 cells and normal adult pituitaries whereas Notch3 mRNA expression was reduced in corticotrope tumor cells in relation to normal mouse pituitary cells (Figure 4A).
Figure 4. Notch system component expression in mouse corticotrope tumor cells and normal pituitaries.

(A–C) Gene expression normalized to Gapdh in AtT20 cells and mouse pituitaries determined by qRT-PCR and expressed as percentage of change of the normal pituitaries which were considered 100%. *p = 0.0003 for Notch3, Notch1,2,4 NS, n = 3–5; 3–8 for AtT20 and pituitary respectively (A), p = 0.0111 and p = 0.0105 for the Notch ligands Dll1 and Jagged1, respectively vs pituitary and (B) p = 0.0016, 0.0184 and 0.0032 for the Notch target genes Hes1, Hes5 and Hey2, respectively, vs. pituitary n = 2–3; 3 for AtT20 and pituitary respectively (C). (D, E) Active and membrane domains of NOTCH1-3 receptor levels determined by Western Blot: Bars show the mean of the receptor normalized to β actin levels, expressed as the percentage of normal mouse pituitary. *p = 0.026 for NOTCH1, p < 0.0001 for NOTCH2 and p = 0.0146 for NOTCH3 vs. pituitary, for the active domains (80KDa) n = 3–6; 5–7 for AtT20 and pituitary respectively (D). No significant differences were found for the membrane domain (110 KDa) n = 3–6; 3–7 for AtT20 and pituitary respectively (E). Representative Western blots showing the active and membrane domains of NOTCH receptors in normal pituitaries (Pit) and ATt20 cells (F).
The Notch ligands Dll1 and Jagged1 were also analyzed and significantly increased Jagged1 and decreased Dll1 mRNA levels in AtT20 cells compared to normal pituitary cells were found (Figure 4B). On the other hand, Notch target genes Hes1, Hes5 and Hey2 were expressed with a particular pattern for each gene, with higher Hes1 expression in tumor corticotropes and reduced Hes5 and Hey2 mRNA levels compared with normal pituitary glands, Hey2 being almost undetectable in ATt20 cells. These results could be related with the specific function they might have in normal and tumoral cells (Figure 4C).
At the protein level, expression of the active NOTCH intracellular domains of NOTCH1-3 receptors were significantly increased in the tumoral cells suggesting these receptors are activated and involved in the tumorigenic process of the corticotropinoma cell line (Figure 4D, 4F). Instead, the membrane domain of the Notch receptor showed no differences between the tumor cell line and the normal pituitary (Figure 4E, 4F).
The Notch system is expressed in GH3 and MMQ cell lines and in experimental prolactinomas
In order to determine the relative expression of each Notch receptor in different tumoral lactotrope cell lines, we first performed qRT-PCR of the four Notch receptors in the rat prolactinoma MMQ cells (prolactin secreting cells) and GH3 (growth hormone and prolactin secreting cells). We also analyzed the in vivo model of tumor xenografts generated by subcutaneous injection of GH3 cells in Nude mice, and compared gene expression to that of normal rat pituitary glands.
GH3 and MMQ cells, and also GH3 in vivo tumors expressed Notch RNA messengers, and with the exception of Notch2 in which no differences with normal pituitaries were found, the rest of Notch paralogs showed lower levels of mRNA in tumoral cell lines. GH3 xenografts instead, showed only reduced Notch3 levels compared to control pituitaries (Figure 5A).
Figure 5. Expression pattern of Notch pathway genes and proteins in prolactinoma cell lines, tumor xenografts and normal pituitaries.

(A–C) Differential gene expression normalized to Gapdh and expressed as percentage of change of the normal pituitaries considered 100% determined by qRT-PCR. For the figures in this panel, different letters indicate significant differences (p ≤ 0.05) between groups. (A) Notch1-4 receptors n = 4–5, 5, 4, 4–6 for MMQ, GH3, GH3 in vivo tumors and rat pituitaries. (B) Notch ligands Dll1 and Jagged1 n = 3, 3, 3, 2 and (C) Notch target genes Hes1, Hey1 and Hey2 n = 3, 3, 3, 2 for MMQ, GH3, GH3 in vivo tumors and rat pituitaries, respectively. (D–E) Active and membrane domains of NOTCH1-3 receptors were determined by Western blot. Bars show the mean of the relation of the receptor levels to β actin expression, showed as the percentage of normal rat pituitary. (D) The active domains (80KDa): n = 5–6; 5, 3, 6. (E) The membrane domains (110KDa): n = 3–6, 2–5, 0–3, 3–6 for MMQ, GH3, GH3 tumors and normal rat pituitaries. (F) Representative Western blots showing the active and membrane domains of NOTCH receptors in MMQ, GH3, GH3 in vivo tumors and normal pituitaries.
Nevertheless, we found that the intracellular active domain of NOTCH2 was significantly increased in the MMQ prolactinoma cell line. Instead, NOTCH1 active domain expression was higher in GH3 in vivo tumors compared to prolactin secreting cell lines while NOTCH3 active domain showed no differences (Figure 5D, 5F). The membrane domains of the NOTCH receptors showed no differences, with the exception of NOTCH3 which was lower in prolactin secreting cell lines and tumor xenografts compared to normal pituitaries (Figure 5E, 5F), in correlation with its mRNA expression.
The expression of the ligands Jagged1 and Dll1 was higher in GH3 cells than in control cells, (Figure 5B). Therefore, in the light of the results presented herein showing high levels of active Notch receptors and the Notch ligands Jagged1 and Dll1 in GH3 cells or Jagged1 in AtT20 cells, an activation of the Notch system in tumoral pituitary cell lines can be inferred.
Additionally, analysis of the expression of the target genes Hes1, Hey1 and Hey2 suggested variable activation which differed substantially between clonal cell lines, tumor xenografts and normal pituitaries. This variability may reflect different effectors and actions of the Notch system in normal and tumor cells. In particular, Hes1 expression predominated in normal rat pituitary when compared to prolactin secreting cell lines and xenografts, Hey1 showed higher levels in the GH3 tumor relative to MMQ cells and normal gland, and the target Hey2 was detected in the GH3 xenograft model and in the normal pituitary but was almost absent in the tumor MMQ and GH3 cell lines, similarly to results found in AtT20 cells for this transcription factor (Figure 5C).
Differences in GH3 in vivo tumors and GH3 cells were also noteworthy. Higher levels of NICD1 and lower levels of NICD2 were found in GH3 xenografts compared to the cell line, as well as reduced expression of Dll1 and an increment of Hey2.
Comparing results between cell lines established from adenomas of different histotypes, it became evident that the Notch system was more activated in ATt20 compared to GH3 and MMQ cells. First, the active intracellular domains of Notch1-3 were increased in this corticotrope cell line, while no increase in the activated paralogs was found in GH3 cells, and only NICD2 but not NICD1 or NICD3 was increased in MMQ cells. Secondly, the canonical target Hes1 was increased in ATt20, but decreased in GH3 cells. And finally, Notch mRNA expression (Notch1, 3 and 4) was consistently decreased in GH3 and MMQ cells while only Notch3 was decreased in ATt20. Consistent among the three cell lines a dramatic decrease in Hey2 mRNA in comparison to normal pituitaries was determined.
Notch system is upregulated in prolactinomas harbored by lacDrd2KO mice in vivo
Clear upregulation of the Notch system was observed in prolactinoma-bearing pituitaries of female mice lacking the dopamine D2 receptor exclusively in lactotropes, compared to their counterparts, pituitaries from Drd2loxP/loxP mice. Notch1 and Notch3 receptor mRNA levels were significantly increased in the lacDrd2KO pituitaries (Figure 6A). Moreover, significantly increased NOTCH1 active domain and NOTCH2 and NOTCH3 membrane domains were found in the tumoral pituitaries by Western blot assessment (Figure 6D–6F).
Figure 6. Prolactinomas of lacDrd2KO female mice have an active Notch system.
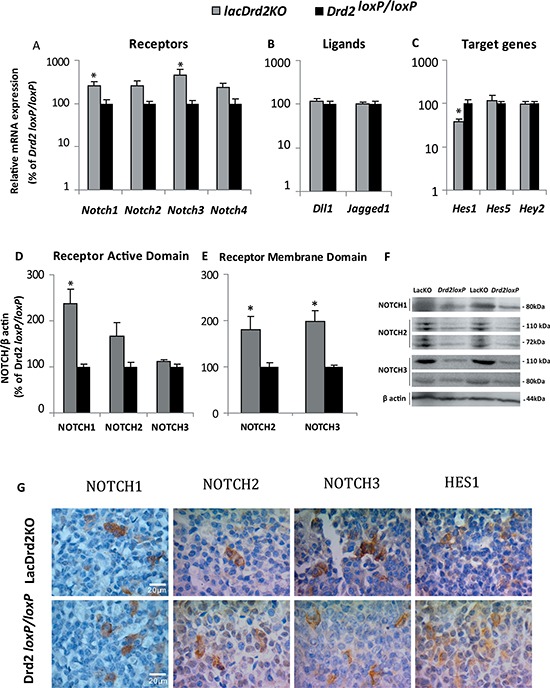
(A–C) Gene expression determined by qRT-PCR in pituitaries of lacDrd2KO and Drd2loxP/loxP counterparts normalized to Gapdh and expressed as percentage of change of the Drd2loxP/loxP pituitaries considered 100%. Bars show mean values. Notch1 *p = 0.0431, Notch3 *p = 0.0089 vs. Drd2loxp/loxp; Notch2, 4 NS, n = 7, 5 for lacDrd2KO and Drd2loxP/loxP mice, respectively (A). Notch ligands Dll1 and Jagged1 NS (B), and Notch target genes Hes1 (*p = 0.0106 lacDrd2KO vs. Drd2loxP/loxP), Hes5 and Hey2, NS (C); n = 7, 5 for lacDrd2KO and Drd2loxP/loxP mice, respectively. (D–E) The expression of the active and membrane domains of NOTCH1-3 receptors was determined by Western blot, related to the correspondent β actin levels, and expressed as the percentage of Drd2loxP/loxP pituitary average. *p = 0.01 for NOTCH1, NOTCH2, 3 NS active domain, p =0.0604 and p = 0.0042 for NOTCH2 and NOTCH3 membrane domains, respectively for lacDrd2KO vs Drd2loxP/loxP mice. n = 4, 3–4 for lacDrd2KO and Drd2loxP/loxP. Representative Western blots showing the active and membrane domains of NOTCH receptors in lacDrd2KO and Drd2loxP/loxP (F). Representative microphotographies of NOTCH1-3 and HES1 Immunohistochemistry performed in lacDrd2KO and Drd2loxP/loxP mice (G).
On the other hand, and opposite to the evidence we found in pituitary tumor cell lines, no differences were detected in the expression of pituitary Dll1 or Jagged1 Notch ligands between lacDrd2KO and control mice (Figure 6B). Instead, Hes1 target gene was significantly reduced in the pituitaries of lacDrd2KO in comparison with Drd2loxP/loxP mice while similar expression levels of the other Notch target genes measured, Hes5 and Hey2 were observed (Figure 6C).
Even though marked differences were found for gene and protein Notch expression levels, the cellular localization of the receptors was similar for both genotypes. NOTCH1-3 protein expression evidenced by immunohistochemistry was found in membranes and cytoplasms of both lacDrd2KO and Drd2loxP/loxP pituitary cells. Furthermore, no differences between genotypes were observed in the staining pattern of pituitary HES1, which was localized in the cytoplasm (Figure 6G).
DISCUSSION
The Notch system plays a fundamental role in normal development and cell-fate determination in a variety of multicellular organisms. It modulates specific cell cycle inhibitor expression promoting cell cycle exit and balancing proliferation and differentiation mainly acting through Hes1 [36]. In pituitary development it regulates cell precursor number, organ size, cell differentiation and fate [49].
Genes that are important during development or differentiation often contribute to tumor promotion and survival when they become deregulated. In this context, Notch signaling has been associated with several human cancers, including acute T cell lymphoblastic leukemia, cervical, breast and prostate cancers, and lung and hepatocellular carcinoma [50–52]. Interestingly, Notch activation can be growth-promoting or inhibitory, depending on the cellular context. For example, in non-small cell lung cancer, Notch function seems to be oncogenic, and γ-secretase inhibitors, which inhibit Notch receptor activation, show antitumor activity both in vitro and in vivo [53]. In human colon cancer Notch1-related signaling also positively regulates tumor growth by promoting proliferation and survival of cancer stem cells and colon cancer cells [54]. Conversely, in prostate cancer Notch can be considered a tumor suppressor particularly at the onset of tumorigenesis, when the Notch target gene Hey1 is activated. Hey1 as a co-factor of androgen receptor can inhibit androgen dependent targets, including those involved in prostate cancer progression [55]. Moreover, different Notch paralogs may have different actions in cancerous transformation; for example, it has been described that NOTCH1 and NOTCH3 have disparate roles from NOTCH2 in bladder cancer [56].
Notch signaling has also been implicated in the pathogenesis of human pituitary adenomas. Upregulation of NOTCH3 gene expression was described in non functioning pituitary adenomas [43, 46, 47], and in prolactinomas [44] compared to normal pituitaries. In accordance, increased expression of the Notch ligand Jagged1 was found in non functioning pituitary adenomas compared to normal human pituitary tissue [46]. Furthermore, NOTCH2 expression was found increased in the Hoechst high efflux population of stem cells named the side population (SP) in human pituitary adenomas [40], but reduced levels of NOTCH2 mRNA were described in pituitary adenomas compared to craniopharyngiomas [48]. These data posit Notch2 and Notch3 as active players in pituitary tumorigenesis. But no complete characterization of the Notch system has been reported in human pituitary adenomas.
In a more comprehensive manner we addressed the expression of several components of the system in human pituitary tumors. In accordance with Notch3 involvement in pituitary tumorigenesis, we found NOTCH3 immunopositive cells in every human adenoma sample tested with higher levels in corticotropinomas and somatotropinomas, and a remarkable disposition of positive cells in clusters in these two adenoma types. Higher NOTCH3 staining detected in corticotropinomas suggest a more active Notch pathway in this adenoma subtype in accordance with results obtained in ATt20 cells compared to prolactinoma cell lines. In concordance with Notch activation in this pituitary tumor type, an overexpression of the ligand Dll4 was found in corticotropinomas when compared to craniopharyngiomas [48].
Variable mRNA expression levels of NOTCH1-4, the Notch ligand JAGGED1 and the target gene HES1 were found in non functioning adenomas, corticotropinomas and somatotropinomas independently of tumor histotype. Interestingly, this is the first evidence of expression of not only NOTCH3 but also other components of this cell signaling in human corticotropinomas.
A salient feature was the positive correlation found for most Notch paralogs with HES1 expression when all adenomas were considered. Furthermore, and in accordance with other authors, a strong association between NOTCH3 and JAGGED1 was found in our cohort [46]. Both findings point to an activation of Notch signaling in some pituitary adenomas, in which the ligand JAGGED1 and the target HES1 would be paramount. Our results underscore the value of analyzing in each tumor sample a set of genes of the Notch system and their correlation, instead of performing single-gene analysis, in order to understand the participation of Notch in tumor generation. It remains to be elucidated if pituitary tumors with an active Notch system would benefit from a Notch targeted therapy. In that case, anti-Notch therapy could be an adjuvant to classic drug treatments like cabergoline or somatostatin analogs in low responder tumors with a demonstrated Notch activation.
On the other hand, we did not find any correlation between any component of the Notch system and the cellular proliferation index determined by Ki-67 levels. This feature could be associated to the many functions and processes regulated by Notch activation besides proliferation [8, 57].
Notch2 and Notch3 receptors are expressed during mouse pituitary embryogenesis and have been found to play key roles in cell specification during the gland development [35–37]. Furthermore, in adult mice, Vankelecom's group described high levels of transcripts of Notch1 and Hes1 in the SP of pituitary cells which display stem/progenitor characteristics [38] when compared to the expression in the main population. They also determined that Notch2-4 and Hes5 were predominantly accumulated in the SP of adult mouse pituitaries, and that Notch pathway activation in this compartment induced cell proliferation rate [39].
Our data in the mouse corticotropinoma cell line AtT20 reflects activation of the Notch pathway by the increased mRNA levels of the ligand Jagged1 and of the intracellular domains of NOTCH1-3 receptors when compared to normal mouse pituitary glands. In the light of the need of the free intracellular domain of Notch receptors to activate the pathway, the high NICD/membrane domain ratio found in the corticotropinoma cell line indicates greater cleavage of the receptors, a process which is fundamental for the effect of Notch on tumor growth or maintenance. Furthermore, higher expression of the Notch target gene Hes1, but lower of Hey2 and Hes5 in ATt20 cells compared to mouse pituitaries suggested that HES1 may be a better candidate in Notch signaling in pituitary corticotropinomas and that neither HEY2 nor HES5 would participate in Notch system actions in the tumoral corticotrope cell line.
There are also few data describing the Notch system in animal models of prolactinomas, even though several components of the Notch pathway such as NOTCH3, ASCL1 and HES1 are altered in human prolactinomas [44].
Our comparative study in prolactinoma cells, in vivo model and normal glands did not reveal a substantial activation status of the Notch system in GH3 and MMQ cells, as observed in AtT20 corticotropes, when compared to normal glands, even though a complete Notch signaling system was determined with the expression of ligands, receptors and target genes. Jagged1 levels were higher in GH3 somato-lactotrope cells than in MMQ and complete rat pituitaries; and Dll1 was also higher in GH3 cells than MMQ, GH3 tumors and normal pituitaries. It could be hypothesized that Jagged1 is the main ligand by which Notch acts in GH3 and ATt20 but not in MMQ cells, and that Dll1 participates mainly in GH3 cells and not in the pure prolactin or corticotrope cell line. In GH3 cells, for example, it was described that the non canonical Notch ligand Dlk1 is expressed in some clones, in which it represses GH expression and secretion but does not affect prolactin production [58]. In the light of exclusive prolactin production in MMQ cells in contraposition to GH and prolactin secretion in GH3 cells, it could be inferred that Jagged1 and/or Dll1 may be involved in specific hormone secretion or cell type behavior.
Additionally, we show that Notch target gene Hes1 and Hey2 levels were decreased in prolactinoma cell lines, in line with a previous finding in human prolactinomas [44], while on the contrary, Hey1 was overexpressed in GH3 cells. Interestingly, Hey2 was nearly absent in MMQ and GH3 cells, but present in GH3 xenografts and control pituitaries. According to our data and the fact that cell to cell contact is an important aspect of Notch signaling cascade, Hey2 expression may be, in some way, associated to the presence of extracellular matrix components, vasculature and/or cell to cell contacts which are found in the complete pituitary and in the GH3 xenografted tumor. This observation is also in agreement with the very low expression of this gene found in AtT20 cell line. Moreover, the extracellular matrix probably plays a role in the expression of different components of the Notch system, as can be inferred when comparing GH3 in vivo tumors generated by GH3 inoculation, and GH3 cells. GH3 tumors showed higher activation of NOTCH1 and lower of NOTCH2 receptor than isolated GH3 somatolactotropic cells. Differences in Dll1 ligand expression were also observed, pointing tumor vasculature and/or extracellular matrix components which are absent in cell lines, as important modulators of Notch signaling in somatoprolactinomas.
On the other hand, in prolactinomas harbored by lacDrd2KO female mice, not only Notch1 and Notch3 mRNA levels but also NOTCH2-3 membrane and NOTCH1 active domains were highly expressed in knockout mice compared to their control counterparts, the Drd2loxP/loxP mice. Even so similar subcellular location was found as specific NOTCH1-3 staining was found mainly in the cytoplasm and in some cell membranes both in lacDrd2KO and Drd2loxP/loxP mice, and neither pituitary Jagged1 nor Dll1 ligands showed any differences between genotypes. Similarly, Lu and co-workers found no differences when evaluating JAGGED1 mRNA levels comparing human prolactinomas and control pituitaries [46]. Instead, it has been described that other ligands, such as Dll1 and Dll4, were underexpressed in non functioning and prolactin secreting adenomas [43, 44], and overexpressed in corticotropinomas [48] respectively, pointing to a specific Notch system profile for different pituitary adenomas histotypes.
A significant reduction in Hes1 mRNA levels was found in the anterior pituitaries of the lacDrd2KO mice bearing prolactinomas compared to Drd2loxP/loxP mice. In concordance, decreased HES1 mRNA levels have been described both in prolactinomas and non functioning adenomas compared to normal human pituitaries [44].
Therefore, our results suggest that the generation of prolactinomas evoked by disruption of lactotrope dopamine D2R receptors is associated with high expression of some Notch components, particularly Notch receptors; these results should be highlighted in the search of new targets for dopamine agonist resistant prolactinomas.
Published data of Notch participation on pituitary adenoma generation and progression are scarce. Based on our present results of differential expression in pituitary adenomas, cell lines and normal pituitaries it becomes evident that Notch system involvement in pituitary tumorigenesis is dependent on each tumor type, and specially activated in corticotrope tumors. Further studies are needed to precisely unravel the most important components that participate in each adenoma histotype, and the processes affected by Notch pathways during pituitary tumor development and maintenance.
MATERIALS AND METHODS
Patient samples
Human pituitary adenoma samples were obtained from patients derived to pituitary neurosurgery at La Pequeña Familia Clinic in Junín and Santa Isabel Clinic, in Buenos Aires, Argentina. Adenomas were previously classified according to hormone production in somatotropinomas, prolactinomas, corticotropinomas and non functioning adenomas, when no hormone secretion was detected. Table 1 and Table 2 describe clinical features, tumor samples and their corresponding Ki67 values.
Samples were conserved in RNAlater (Ambion Inc) for mRNA determination or immediately fixed after surgery in 4 % neutral buffered formalin, dehydrated in graded ethanol and embedded in paraffin. Sections of 4 μm thickness were cut and immunohistochemistry for different antigens was performed.
The project and procedures were approved by the Ethical Committees of the La Pequeña Familia and Santa Isabel Clinics. The patient was informed of the studies to be undertaken and signed the respective consent. Patient privacy was always preserved.
Cell lines and culture
AtT20 mouse corticotrope and GH3 rat somato-prolactinoma cell lines were cultured in adhesion in DMEM/F12 medium, supplemented by 10% fetal bovine serum, 1% glutamine and 1% penicillin/streptomycin and maintained at 37°C and 5% CO2. Rat prolactinoma MMQ cells were cultured in suspension in DMEM/F12 medium with 10% horse serum, 5% fetal bovine serum, 1% glutamine and 1% penicillin/streptomycin and maintained at 37°C and 5% CO2.
Animals
Mice
Mice lacking expression of D2Rs in pituitary lactotropes were generated by crossing Drd2loxP/loxP mice [59] with transgenic mice expressing Cre recombinase driven by the mouse prolactin promoter (Tg(Prl-cre)1Mrub [60]) for two generations. Tissue specificity of Cre expression in (Tg(Prl-cre)1Mrub transgenic mice was analyzed by real time PCR and Cre mRNA levels were highly expressed in the pituitary lactotropes and very low or almost absent in the hypothalamus, liver, kidney, ovary and lung [61]. Functional Cre recombinase activity is present in most prolactin producing cells of the anterior pituitary in a highly selective manner as described [60]. LacDrd2KO and their Drd2loxP/loxP control littermates were congenic to C57BL/6J (n = 10).
Breeding pairs of female Drd2loxP/loxP and male Drd2loxP/loxP.Tg(Prl-Cre) mice were used to generate Drd2loxP/loxP (control) and Drd2loxP/loxP.Tg(Prl-Cre) (lacDrd2KO) littermates, which were included in each experiment. Female 10 month-old mice were used in the present experiments.
Normal pituitaries from adult female BalbC mice were used as controls in the determination of mRNA expression by qRT-PCR and protein expression by Western blot in comparison with AtT20 cells.
Xenotropic tumors were generated by sc injection of 500,000 GH3 cells in Nude/Nude mice. Tumors were allowed to develop for 21 days, thereafter animals were euthanized and tumors were excised and collected in RNA later for qRT-PCR or in lysis buffer for protein extraction and Western blot experiments
Rats
Female Sprague-Dawley rats were used. Animals were euthanized at two months of age. Pituitaries were used as controls in the determination of mRNA expression by qRT-PCR and protein expression by Western blot in comparison with rat tumor cell lines and GH3 in vivo tumors.
All animals (mice and rats) were housed in groups of 4 or 5 in a temperature-controlled room with light on at 0700 h and off at 1900 h, and had free access to tap water and laboratory chow. Experimental procedures were carried according to guidelines of the institutional animal care and use committee of the National University of the Northwest of Buenos Aires Province and Instituto de Biología y Medicina Experimental – CONICET.
Reagents
Unless otherwise specified, all chemicals were purchased from Sigma (St. Louis, MO).
RNA extraction and cDNA synthesis
Total RNA extraction from human pituitary tumors, mouse or rat pituitaries or from 6 × 105 tumor cells was recovered using TRI reagent (Molecular Research Center, Inc) as previously described [62]. The RNA concentration was determined on the basis of absorbance at 260 nm, its purity was evaluated by the ratio of absorbance at 260/280 nm (∼2.0 was considered appropriate), and its integrity was evaluated by agarose gel electrophoresis. RNA was kept frozen at −80°C until analyzed. One μg of RNA was reversed transcribed in 20 μl volume in the presence of 3 mM MgCl2, 50 mM Tris·HCl (pH 8.3), 75 mM KCl, 1 mM deoxy-NTPs, 0,01 mM DTT, 1 pM oligo(dT)15–18 primer (Biodynamics, Buenos Aires, Argentina), and 10 U of MMLV reverse transcriptase (Invitrogen, CA, USA). The reverse transcriptase was omitted in negative reactions.
Real time PCR
Quantitative PCR was performed as previously described in [63]. Sense and antisense oligonucleotide primers were designed by the use of PrimerBlast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Oligonucleotides were synthesized by Biodynamics SRL and their sequences and annealing temperatures are described in Supplementary Table 1.
Briefly, the reactions were performed by kinetic PCR using 7.5 ul FastStart SYBR Green Master Mix (containing FastStart Taq DNA Polymerase, Reaction Buffer, Nucleotides, and SYBR Green I, Roche Laboratories), 150 ng cDNA and 0.5 μM primers in a final volume of 15 μl. After denaturation at 95°C during 15 min, the cDNA products were amplified with 40 cycles 20 s at 95°C, 60 s at 55–65,5°C depending on the primer pair, and 40 s at 72°C. The accumulating DNA products were monitored by the LineGene9600 (Bioer, Binjiang, China), and data were stored continuously during the reaction. Results were validated on the basis of the quality of dissociation curves as described in [63], and target gene expression relative to Gapdh mRNA was calculated as previous published work [63]. When studying patient samples, qPCRs were performed sequentially, and in some samples not all Notch receptors, target or downstream genes could be measured due to limitations of cDNA availability.
Western blot
Pituitary samples and cells lysates were prepared in a motor microtissue mixer in 80 to 300 μL of lysis buffer (50 mM HEPES [pH 7.4], 140 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 100 mM sodium fluoride (NaF), and 10 μg/mL 1% Triton X-100) and 1-mM phenymethylsulfonyflouride (PMSF), and protease cocktail inhibitor (Roche Diagnostic, Mannheim, Germany) was added to the buffer just before use. The homogenate was then centrifuged at 12.000 rpm for 30 min at 4°C. An aliquot of the supernatant was taken to quantify proteins by the Qubit Quant-it protein assay kit (Invitrogen, Buenos Aires, Argentina).
Forty micrograms of proteins in 20 μl of homogenization buffer were mixed with 5 μl of 5x sample buffer (312 mM Tris.HCl, 10% SDS, 25% glycerol, 0.002% bromophenol blue and 1% Beta-mercaptoethanol, pH 6.8). Samples were heated 5 min at 95°C and separated by 10% sodium dodecyl sulphate-polyacrilamide gel electrophoresis (SDS–PAGE) and electrotransferred to nitrocellulose membranes (G&E, Little Chalfont, UK). After blocking with 3% nonfat dry milk solution in phosphate saline buffer - Tween (PBST) (10 mM sodium phosphate, 2 mM potassium phosphate [pH 7.4], 140 mM NaCl, 3 mM KCl, and 0.1% Tween 20) blots were incubated overnight at 4°C with primary antibodies. Rabbit polyclonal anti-Notch 1 [1:1000] and rabbit polyclonal anti-Notch 3 [1:1000], Santa Cruz Biotechnology Inc (Texas, USA) and rabbit polyclonal anti-Notch 2 [1:1000], Merck Millipore (Darmstadt, Germany) were used.
Membranes were washed with PBST and incubated with the corresponding horse raddish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology Inc), and protein bands were visualized on ImageQuant LAS 4000 mini (G&E). The monoclonal anti-β-actin (Santa Cruz Biotechnology Inc) antibody [1:1500] was used to validate equal amount of protein loaded and transferred.
For repeated immunoblotting, membranes were incubated in stripping buffer (62.5 mM Tris, 2% sodium dodecyl sulfate, and 100 mM mercaptoethanol, pH 6.7) along 40 min at 55°C and reprobed. Band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
NOTCH1-3 expression levels were evaluated by the semi-quantification of two bands, the active intracellular domain (NICD) of 80 KDa and the cleaved membrane domain of the receptor which is still at the cellular membrane (NTMIC) of 110 KDa.
Immunohistochemistry
Immunohistochemistry of paraffin embedded pituitaries from 10- month-old LacDrd2KO and Drd2loxP/loxP female mice or human pituitary tumor samples, was performed as previously described [64].
Briefly, antigen retrieval procedure was performed in citrate buffer (10 mM, pH = 6) and the microwave technique. Tissues were exposed to the primary antibody overnight at 4°C. Replacement of the primary antibody with phosphate buffer was employed as a negative control. Subsequently, slides were washed to eliminate antibody excess and incubated with the appropriate secondary antibody and then with streptavidin/biotin peroxidase complex. Diaminobenzidine was used as chromogen.
For mouse tissue the following antibodies were used: Rabbit polyclonal antibody against NOTCH1 and NOTCH2 (dilution 1:700, Merck Millipore), NOTCH3 (dilution 1:200, Santa Cruz Biotechnology Inc), HES1 (dilution 1:400, Merck Millipore). For human tissue the same rabbit polyclonal antibody against NOTCH3 (dilution 1:200, Santa Cruz Biotechnology Inc) was used. Biotin-conjugated secondary goat anti-rabbit IgG antibody (dilution 1:100; Santa Cruz Biotechnology Inc) was used.
Samples were counterstained with hematoxylin and mounted with permanent mounting medium. Morphometric analysis was performed using a Carl Zeiss transmitted light microscope at 400 and 1000 total magnification.
Statistical analyses
Normal data distribution and variance homogeneity was tested in all cases. Correlations between each NOTCH receptor and JAGGED1 or HES1 expression in human pituitaries adenomas were tested by Spearman non parametric test. Gene expression of Notch receptors, ligands and targets in AtT20 vs control pituitaries and lacDrd2KO vs Drd2loxP/loxP mice were analyzed by Student's t-test. Analysis of variance (ANOVA) followed by LSD Fisher test was used to analyze Notch receptor, ligand and target gene expression in rat tumoral models vs control pituitary.
Results are expressed as means ± SEM. p < 0.05 was considered significant.
SUPPLEMENTARY TABLE
Acknowledgments
Fundacion Williams (DBV).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica, Argentina: PICT 330-2013 to DBV, PICT 901-2013 to CC, Fundación Rene Barón to DBV, Consejo Nacional de Investigaciones Científicas y Técnicas and Universidad Nacional del Noroeste de la Provincia de Buenos Aires: PIO CONICET-UNNOBA 2015-2016 15720150100010CO and SIB UNNOBA 2015-3160 to CC.
REFERENCES
- 1.Ezzat S, Asa SL, Couldwell WT, Barr CE, Dodge WE, Vance ML, McCutcheon IE. The prevalence of pituitary adenomas: a systematic review. Cancer. 2004;101:613–619. doi: 10.1002/cncr.20412. [DOI] [PubMed] [Google Scholar]
- 2.Kopczak A, Renner U, Karl Stalla G. Advances in understanding pituitary tumors. F1000Prime Rep. 2014;6:5. doi: 10.12703/P6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mete O, Asa SL. Clinicopathological correlations in pituitary adenomas. Brain Pathol. 2012;22:443–453. doi: 10.1111/j.1750-3639.2012.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scangas GA, Laws ER., Jr Pituitary incidentalomas. Pituitary. 2014;17:486–491. doi: 10.1007/s11102-013-0517-x. [DOI] [PubMed] [Google Scholar]
- 5.Colao A, Pivonello R, Di Somma C, Savastano S, Grasso LF, Lombardi G. Medical therapy of pituitary adenomas: effects on tumor shrinkage. Rev Endocr Metab Disord. 2009;10:111–123. doi: 10.1007/s11154-008-9107-z. [DOI] [PubMed] [Google Scholar]
- 6.Di Sarno A, Landi ML, Cappabianca P, Di Salle F, Rossi FW, Pivonello R, Di Somma C, Faggiano A, Lombardi G, Colao A. Resistance to cabergoline as compared with bromocriptine in hyperprolactinemia: prevalence, clinical definition, and therapeutic strategy. J Clin Endocrinol Metab. 2001;86:5256–5261. doi: 10.1210/jcem.86.11.8054. [DOI] [PubMed] [Google Scholar]
- 7.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 10.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 11.Yavropoulou MP, Maladaki A, Yovos JG. The role of Notch and Hedgehog signaling pathways in pituitary development and pathogenesis of pituitary adenomas. Hormones (Athens) 2015;14:5–18. doi: 10.1007/BF03401377. [DOI] [PubMed] [Google Scholar]
- 12.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, Osborne BA. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113:1689–1698. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis HD, Leveridge M, Strack PR, Haldon CD, O’Neil J, Kim H, Madin A, Hannam JC, Look AT, Kohl N, Draetta G, Harrison T, Kerby JA, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2000;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol Cell Biol. 2000;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 18.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Semin Cell Dev Biol. 2012;23:473–480. doi: 10.1016/j.semcdb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, Lewis A, Watson S, Behrens A, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T, Difilippantonio MJ, Caplen NJ, Ried T. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer Res. 2013;73:2003–2013. doi: 10.1158/0008-5472.CAN-12-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Wang J, Shi Z, Franklin JL, Deane NG, Coffey RJ, Beauchamp RD, Zhang B. Deciphering genomic alterations in colorectal cancer through transcriptional subtype-based network analysis. PLoS One. 2013;8:e79282. doi: 10.1371/journal.pone.0079282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, Maru DM, Hawke DH, Rak J, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, Sticht C, Tomasi ML, Delogu S, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542.e12. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, Hashida H, Takabayashi A, Sasaki M, Robine S, Itoh K, Yoshioka K, Kakizaki F, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 31.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 32.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, Lonigro RJ, Quist M, Siddiqui J, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, Cardiff RD, Yoon K, Gaiano N, et al. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui H, Kong Y, Xu M, Zhang H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer Res. 2013;73:3451–3459. doi: 10.1158/0008-5472.CAN-12-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raetzman LT, Ross SA, Cook S, Dunwoodie SL, Camper SA, Thomas PQ. Developmental regulation of Notch signaling genes in the embryonic pituitary: Prop1 deficiency affects Notch2 expression. Dev Biol. 2004;265:329–340. doi: 10.1016/j.ydbio.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 36.Monahan P, Rybak S, Raetzman LT. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology. 2009;150:4386–4394. doi: 10.1210/en.2009-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raetzman LT, Wheeler BS, Ross SA, Thomas PQ, Camper SA. Persistent expression of Notch2 delays gonadotrope differentiation. Mol Endocrinol. 2006;20:2898–2908. doi: 10.1210/me.2005-0394. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Hersmus N, Van Duppen V, Caesens P, Denef C, Vankelecom H. The adult pituitary contains a cell population displaying stem/progenitor cell and early embryonic characteristics. Endocrinology. 2005;146:3985–3998. doi: 10.1210/en.2005-0185. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Crabbe A, Van Duppen V, Vankelecom H. The notch signaling system is present in the postnatal pituitary: marked expression and regulatory activity in the newly discovered side population. Mol Endocrinol. 2006;20:3293–3307. doi: 10.1210/me.2006-0293. [DOI] [PubMed] [Google Scholar]
- 40.Mertens F, Gremeaux L, Chen J, Fu Q, Willems C, Roose H, Govaere O, Roskams T, Cristina C, Becu-Villalobos D, Jorissen M, Poorten VV, Bex M, et al. Pituitary tumors contain a side population with tumor stem cell-associated characteristics. Endocr Relat Cancer. 2015;22:481–504. doi: 10.1530/ERC-14-0546. [DOI] [PubMed] [Google Scholar]
- 41.Tando Y, Fujiwara K, Yashiro T, Kikuchi M. Localization of Notch signaling molecules and their effect on cellular proliferation in adult rat pituitary. Cell Tissue Res. 2013;351:511–519. doi: 10.1007/s00441-012-1532-3. [DOI] [PubMed] [Google Scholar]
- 42.Batchuluun K, Azuma M, Yashiro T, Kikuchi M. Notch signaling-mediated cell-to-cell interaction is dependent on E-cadherin adhesion in adult rat anterior pituitary. Cell Tissue Res. 2017;368:125–33. doi: 10.1007/s00441-016-2540-5. [DOI] [PubMed] [Google Scholar]
- 43.Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65:10214–10222. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 44.Evans CO, Moreno CS, Zhan X, McCabe MT, Vertino PM, Desiderio DM, Oyesiku NM. Molecular pathogenesis of human prolactinomas identified by gene expression profiling, RT-qPCR, and proteomic analyses. Pituitary. 2008;11:231–245. doi: 10.1007/s11102-007-0082-2. [DOI] [PubMed] [Google Scholar]
- 45.Vankelecom H, Gremeaux L. Stem cells in the pituitary gland: A burgeoning field. Gen Comp Endocrinol. 2010;166:478–488. doi: 10.1016/j.ygcen.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Lu R, Gao H, Wang H, Cao L, Bai J, Zhang Y. Overexpression of the Notch3 receptor and its ligand Jagged1 in human clinically non-functioning pituitary adenomas. Oncol Lett. 2013;5:845–851. doi: 10.3892/ol.2013.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao Z, Miao Y, Lin Y, Lu X. Overexpression of the Notch3 receptor in non-functioning pituitary tumours. J Clin Neurosci. 2012;19:107–110. doi: 10.1016/j.jocn.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 48.Chang CV, Araujo RV, Cirqueira CS, Cani CM, Matushita H, Cescato VA, Fragoso MC, Bronstein MD, Zerbini MC, Mendonca BB, Carvalho LR. Differential Expression of Stem Cell Markers in Human Adamantinomatous Craniopharyngioma and Pituitary Adenoma. Neuroendocrinology. 2017;104:183–93. doi: 10.1159/000446072. [DOI] [PubMed] [Google Scholar]
- 49.Raetzman LT, Cai JX, Camper SA. Hes1 is required for pituitary growth and melanotrope specification. Dev Biol. 2007;304:455–466. doi: 10.1016/j.ydbio.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abravanel DL, Belka GK, Pan TC, Pant DK, Collins MA, Sterner CJ, Chodosh LA. Notch promotes recurrence of dormant tumor cells following HER2/neu-targeted therapy. J Clin Invest. 2015;125:2484–2496. doi: 10.1172/JCI74883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C, Qian J, Ma M, Zhang Y, Han B. Notch 3 protein, not its gene polymorphism, is associated with the chemotherapy response and prognosis of advanced NSCLC patients. Cell Physiol Biochem. 2014;34:743–752. doi: 10.1159/000363039. [DOI] [PubMed] [Google Scholar]
- 52.Lefort K, Ostano P, Mello-Grand M, Calpini V, Scatolini M, Farsetti A, Dotto GP, Chiorino G. Dual tumor suppressing and promoting function of Notch1 signaling in human prostate cancer. Oncotarget. 2016;7:48011–26. doi: 10.18632/oncotarget.10333. https://doi.org/10.18632/oncotarget.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116:5207–5218. doi: 10.1002/cncr.25449. [DOI] [PubMed] [Google Scholar]
- 55.Belandia B, Powell SM, Garcia-Pedrero JM, Walker MM, Bevan CL, Parker MG. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol Cell Biol. 2005;25:1425–1436. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi T, Gust KM, Wyatt AW, Goriki A, Jager W, Awrey S, Li N, Oo HZ, Altamirano-Dimas M, Buttyan R, Fazli L, Matsubara A, Black PC. Not all NOTCH Is Created Equal: The Oncogenic Role of NOTCH2 in Bladder Cancer and Its Implications for Targeted Therapy. Clin Cancer Res. 2016;22:2981–2992. doi: 10.1158/1078-0432.CCR-15-2360. [DOI] [PubMed] [Google Scholar]
- 57.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 58.Ansell PJ, Zhou Y, Schjeide BM, Kerner A, Zhao J, Zhang X, Klibanski A. Regulation of growth hormone expression by Delta-like protein 1 (Dlk1) Mol Cell Endocrinol. 2007;271:55–63. doi: 10.1016/j.mce.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bello EP, Mateo Y, Gelman DM, Noain D, Shin JH, Low MJ, Alvarez VA, Lovinger DM, Rubinstein M. Cocaine supersensitivity and enhanced motivation for reward in mice lacking dopamine D2 autoreceptors. Nat Neurosci. 2011;14:1033–1038. doi: 10.1038/nn.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noain D, Perez-Millan MI, Bello EP, Luque GM, Casas Cordero R, Gelman DM, Peper M, Tornadu IG, Low MJ, Becu-Villalobos D, Rubinstein M. Central dopamine D2 receptors regulate growth-hormone-dependent body growth and pheromone signaling to conspecific males. J Neurosci. 2013;33:5834–5842. doi: 10.1523/JNEUROSCI.5673-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez Millan MI, Luque GM, Ramirez MC, Noain D, Ornstein AM, Rubinstein M, Becu-Villalobos D. Selective disruption of dopamine D2 receptors in pituitary lactotropes increases body weight and adiposity in female mice. Endocrinology. 2014;155:829–839. doi: 10.1210/en.2013-1707. [DOI] [PubMed] [Google Scholar]
- 62.Cristina C, Diaz-Torga G, Baldi A, Gongora A, Rubinstein M, Low MJ, Becu-Villalobos D. Increased pituitary vascular endothelial growth factor-a in dopaminergic D2 receptor knockout female mice. Endocrinology. 2005;146:2952–2962. doi: 10.1210/en.2004-1445. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Tornadu I, Diaz-Torga G, Risso GS, Silveyra P, Cataldi N, Ramirez MC, Low MJ, Libertun C, Becu-Villalobos D. Hypothalamic orexin, OX1, alphaMSH, NPY and MCRs expression in dopaminergic D2R knockout mice. Neuropeptides. 2009;43:267–274. doi: 10.1016/j.npep.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Luque GM, Perez-Millan MI, Ornstein AM, Cristina C, Becu-Villalobos D. Inhibitory effects of antivascular endothelial growth factor strategies in experimental dopamine-resistant prolactinomas. J Pharmacol Exp Ther. 2011;337:766–774. doi: 10.1124/jpet.110.177790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


