ABSTRACT
CD226 is an activating receptor expressed on natural killer (NK) cells, CD8+ T cells, and other immune cells. Upon binding to its ligands expressed on target cells, CD226 activates intracellular signaling that triggers cytokine production and degranulation in NK cells. However, the role of CD226 in contact dynamics between NK and cancer cells has remained unclear. Our time-lapse images showed that individual wild-type CD226+ NK cells contacted B16F10 melanoma cells for 23.7 min, but Cd226−/− NK cells only for 12.8 min, although both NK cell subsets showed equal contact frequency over 4 h. On the surface of B16F10 cells, CD226+ cells stayed at the same site with oscillating movement (named stable contact), while Cd226−/− NK cells moved around at a velocity of 4 μm/min (named unstable contact). Consequently, Cd226−/− NK cells did not kill B16F10 cells in vitro and did not inhibit their metastasis into the lung in vivo. Taken together, our data demonstrate that CD226 enables prolonged stable interaction between NK and cancer cells, which is needed for efficient killing of cancer cells.
KEYWORDS: Contact duration, contact dynamics, contact stability, melanoma, NK cell-mediated cytotoxicity
Abbreviations
- APC
allophycocyanin
- calcein AM
calcein-acetoxymethyl
- DNAM-1
DNAX accessory molecule 1
- FITC
fluorescein isothiocyanate
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- ICAM-1
intercellular adhesion molecule 1
- IL-2
interleukin-2
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- KLRG1
killer cell lectin-like receptor G1
- LDH
lactate dehydrogenase
- LFA-1
lymphocyte function-associated antigen 1
- MHC
major histocompatibility complex
- MICA
MHC class I chain-related gene A
- MIP
macrophage inflammatory protein
- MULT-1
mouse ULBP-like transcript 1
- NK
natural killer
- NKG2A
NK group 2 member A
- NKG2D
NK group 2 member D
- PE
phycoerythrin
- PI
propidium iodide
- SEM
standard error of mean
- TIGIT
T-cell immunoglobulin and ITIM domain
- ULBP
UL16-binding protein
- WT
wild type
Introduction
Natural killer (NK) cells kill virus-infected and cancer cells while sparing autologous normal cells.1,2 NK cell cytotoxicity is regulated by the balance of activating and inhibitory signals received at the immunological synapse.3 The inhibitory signals are generated by the binding of MHC-I molecules on target cells to killer cell immunoglobulin-like receptors in humans, Ly49 in mice, and a CD94–NK group 2 member A (NKG2A) heterodimer in both species.3-5 NK cells sense signs of infection and malignant transformation through the activating receptors NKG2D, natural cytotoxic receptor, 2B4, and CD226 (also called DNAM-1).3,6 Activating receptors facilitate the conjugation of NK and cancer cells, and transduce signals that augment cytokine production by and degranulation of NK cells. The lack of inhibitory receptors reduces NK cell responsiveness by a process called NK cell education.7 Educated NK cells form stable conjugates with cancer cells via inside-out signaling to LFA-1 by activating receptors.7 Educated NK cells also display the coordinated regulation and colocalization of CD226 and LFA-1 at the immunological synapse, which are essential for tumor cell killing by educated NK cells.8
CD226 is a member of the immunoglobulin superfamily and is expressed on the majority of NK cells, T cells, and monocytes.9-11 CD155 (poliovirus receptor) and its family member CD112 (also called nectin-2) are ligands for human and mouse CD226.6,12,13 Although these ligands are broadly expressed on epithelial and endothelial cells in many tissues,14,15 they are overexpressed on colorectal, gastric, and ovarian cancer cells, neuroblastoma, leukemia, and myeloma.16-22 Upon ligation, Src kinase phosphorylates the cytoplasmic domain of CD226, which triggers the binding of the adaptor Grb2, activation of the Vav-1, phosphatidylinositol 3′ kinase, phospholipase C-γ1, and protein kinases Erk and Akt, as well as calcium influx.23 This signaling cascade increases actin polymerization and granule polarization, which is required for cytotoxic killing of target cells, and also augments IFN-γ production by NK cells.23 Anti-CD226 neutralizing antibodies inhibit NK cell cytotoxicity against many cancer cells expressing its ligand.9 CD226 deficiency in mice increases susceptibility to carcinogen-induced fibrosarcoma and papilloma.24 CD226 is also implicated in NK cell–mediated elimination of HIV-infected CD4+ T cells25 and plays a critical role in the expansion and maintenance of virus-specific memory NK cells in mouse cytomegalovirus–infected mice.26 Overall, these data suggest that CD226 promotes NK cell activation, at least in part, by activating intracellular signaling for survival, cytokine production, and cytotoxicity. CD226-deficient CD8+ T cells weakly conjugate with tumor cells and show a decreased recruitment of LFA-1 and lipid rafts to the immunological synapse, which correlates with reduced tumor cell killing in vitro, although these cells show normal production and degranulation of cytotoxic granules after T cell receptor stimulation.27 CD226 is also critical for the ability of CD8+ T cells to eliminate tumor cells during anti-TIGIT antibody therapy.28
In addition, CD226 is able to act as an adhesion molecule: CD226-transfected COS-7 cells bind Colo-205 cells,9 and a CD226-expressing NK cell line (YT-S) conjugates with CD155-expressing RMA-S cells.23 Yet, several questions remain. How many times does each CD226+ NK cell contact cancer cells? How long do such contacts last? Does CD226 affect contact stability? Do all CD226+ NK cells contact cancer cells? To address these issues, we examined the contact dynamics between CD226-deficient NK and cancer cells at a single-cell level using time-lapse imaging. Our data show that CD226-deficient NK cells fail to establish long-lasting stable contacts with cancer cells and have impaired antitumor activity in vitro and in vivo.
Results
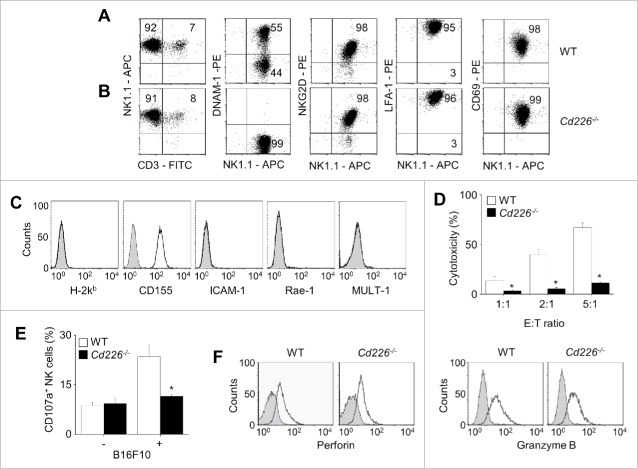
Cd226−/− NK cells show impaired cytotoxicity at the population level
Because NK cell cytotoxicity is largely regulated by interactions between activating receptors on NK cells and their respective ligands on cancer cells, we first examined the surface expression of activating receptors on NK cells. All effector NK cells were NK1.1+, LFA-1+, NKG2D+, and CD69+, but only half of them were CD226+ (Fig. 1A). Wild-type (WT) and Cd226−/− NK cells showed similar expression patterns except for CD226 (Fig. 1B). Target B16F10 cells expressed CD155 (CD226 ligand) but not ICAM-1 (LFA-1 ligand), Rae-1, H-2Kb, or MULT-1 (NKG2D ligands) (Fig. 1C), suggesting that the B16F10 cell line is a good tool to study the role of CD226 in the recognition of cancer cells by NK cells because it allows ruling out the interference from other receptors. In the in vitro LDH assay, WT NK cells destroyed B16F10 target cells better than did Cd226−/− NK cells (Fig. 1D). When co-cultured with B16F10 cells, WT NK cells showed higher CD107a+ expression (degranulation marker) than did Cd226−/− NK cells (Fig. 1E). However, WT and Cd226−/− NK cells had similar amounts of intracellular cytotoxic proteins, such as perforin and granzyme B (Fig. 1F).
Figure 1.
Cd226−/− NK cells show impaired cytotoxicity. (A) Phenotypes of wild-type (WT) NK cells (n = 3). (B) Phenotypes of Cd226−/− NK cells (n = 3). (C) Phenotypes of B16F10 cells (n = 3). (D) Cytotoxicity of NK cells against B16F10 cells was determined by the LDH assay (n = 3, *p < 0.01). (E) To analyze exocytosis, NK and B16F10 cells were co-cultured for 2 h in the presence of anti-CD107a-FITC antibody, and CD107+ expression level on NK cells was determined using flow cytometry (n = 3, *p < 0.01). (F) Intracellular perforin and granzyme B protein levels of wild-type and Cd226−/− NK cells (n = 3).
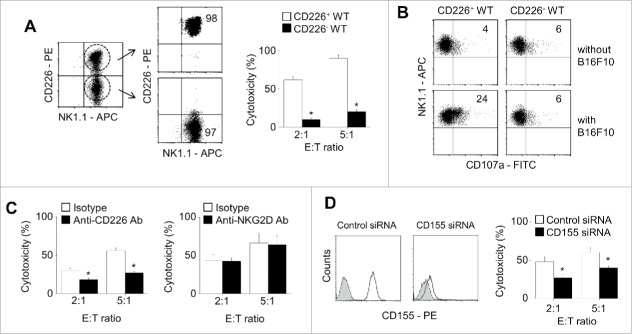
We also verified the role of CD226 in NK cell cytotoxicity by using purified CD226+ WT NK cells (Fig. 2A). These cells showed higher cytotoxicity against B16F10 cells (Fig. 2A) and better degranulation (Fig. 2B) than did WT CD226− NK cells. Neutralizing antibody against CD226 decreased WT NK cell cytotoxicity against B16F10 cells, but anti-NKG2D antibody did not (Fig. 2C). WT NK cells only weakly killed B16F10 cells transfected with CD155 siRNA compared with B16F10 cells transfected with negative-control siRNA (Fig. 2D). These data suggest that CD226 deficiency in NK cells impairs degranulation and cytotoxicity without affecting the levels of intracellular cytotoxic molecules.
Figure 2.
CD226-negative NK cells show impaired cytotoxicity. (A) CD226+ and CD226− NK cells were purified from total splenic NK cells using flow cytometry and their cytotoxicity against B16F10 cells was determined by the LDH assay (n = 3, *p < 0.01). (B) To analyze exocytosis, NK and B16F10 cells were co-cultured for 2 h in the presence of anti-CD107a-FITC antibody. CD107+ expression level on CD226+ and CD226− NK cells was determined using flow cytometry (n = 3). (C) NK cells were treated with isotype, anti-CD226, or anti-NKG2D neutralizing antibodies (10 μg/ml) for 2 h. Cytotoxicity of NK cells against B16F10 cells was determined by the LDH assay (n = 3, *p < 0.01). (D) B16F10 cells were treated with negative control or CD155 siRNAs. CD155 expression level was determined by using flow cytometry and cytotoxicity of NK cells against B16F10 cells was determined by the LDH assay (n = 3, *p < 0.01).
Cd226−/− NK cells show impaired cytotoxicity at the single-cell level
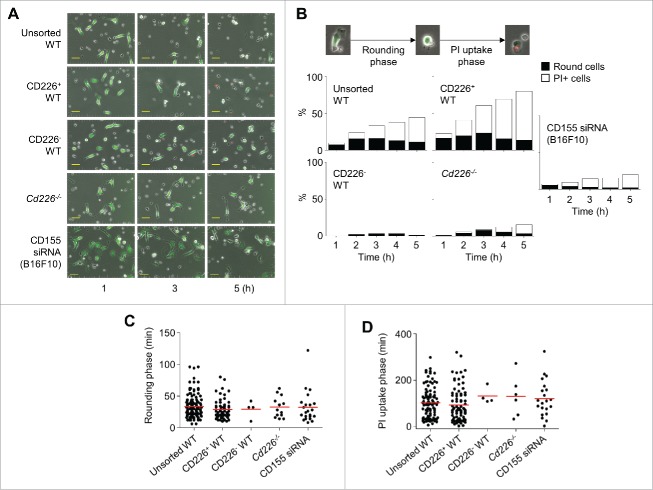
We also assessed the role of CD226 in NK cell cytotoxicity at the single-cell level by using time-lapse imaging. We mixed calcein AM–stained B16F10 cells and unsorted (Movie S1), CD226+ (Movie S2), or CD226− (Movie S3) WT NK cells or Cd226−/− NK cells (Movie S4). In addition, we mixed unsorted WT NK cells and calcein AM–stained B16F10 cells transfected with CD155 siRNA (Movie S5). Representative images collected at 2-h intervals are shown in Fig. 3A. B16F10 cells appeared sessile and extended and retracted pseudopods; these cells grew well and attached to dishes. Upon encountering NK cells, B16F10 cells detached and became rounded (called rounding phase), followed by the uptake of PtdIns (called PtdIns uptake phase). Unsorted and CD226+ WT NK cells efficiently killed B16F10 cells, but CD226− WT NK cells and Cd226−/− NK cells did not. In addition, unsorted WT NK cells weakly killed B16F10 cells transfected with CD155 siRNA (Fig. 3A and B). In the presence of unsorted or CD226+ WT NK cells, B16F10 cells became rounded within 29–33 min (Fig. 3C) and were stained with PtdIns within further 92–102 min (Fig. 3D). Only a few B16F10 cells died in the presence of CD226− WT NK cells or Cd226−/− NK cells although their dying kinetics was similar to those in the presence of unsorted or CD226+ WT NK cells. In addition, only a few B16F10 cells transfected with CD155 siRNA died in the presence of total WT NK cells.
Figure 3.
CD226-deficient NK cells show impaired cytotoxicity at the single cell level. (A) Unstained NK cells (unsorted WT, CD226+ WT, CD226− WT, or Cd226−/− NK cells) and calcein AM–stained B16F10 cells (transfected with CD155 siRNA or not) were imaged every 2 min from 1 h to 5 h. Representative photos are shown (magnification, 200 ×; scale bars, 50 μm, n = 10 movies of 3 independent experiments per group). (B) Representative images show dying of cancer cells (original magnification, 200 ×; electronically zoomed). Cancer cells underwent rounding and propidium (PtdIns) uptake phases. Ratios of rounding and PtdIns uptake cells were counted every 1 h. (C) Time needed for cells to become rounded morphology was calculated (n = 129, 59, 4, 13, and 22 from the left). (D) Time needed for rounded cells to uptake PtdIns was calculated (n = 101, 81, 4, 6, and 21 from the left).
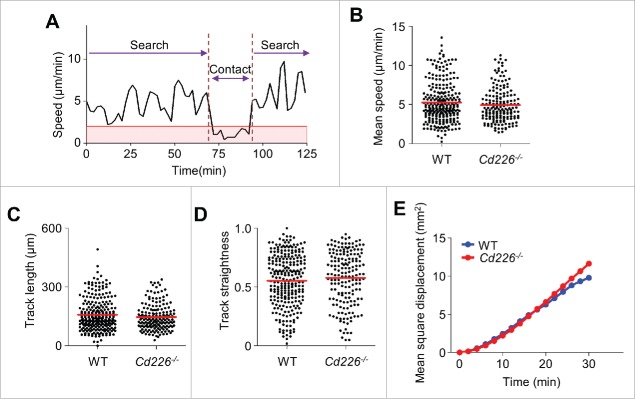
Next, we analyzed NK cell behavior. NK cell motility could be divided into 2 stages: search and contact (Fig. 4A). WT and Cd226−/− NK cells similarly migrated at approximately 5 μm/min during the search stage (Fig. 4B) and at <2 μm/min during the contact stage (data not shown). Both WT and Cd226−/− NK cells moved in all directions and showed similar track lengths (Fig. 4C), straightness scores (Fig. 4D), and mean square displacement (Fig. 4E), which indicates long-distance random migration of NK cells at the searching stage. These data suggest that CD226 does not affect NK cell migration.
Figure 4.
Migration modes of NK cells around cancer cells. (A) Representative profile of NK cell migration speed (n = 10 movies of 3 independent experiments per group). (B – E) Mean speed (B), track length (C), track straightness (D), and mean square displacement (E) of WT (n = 255) and Cd226−/− NK cells (n = 174).
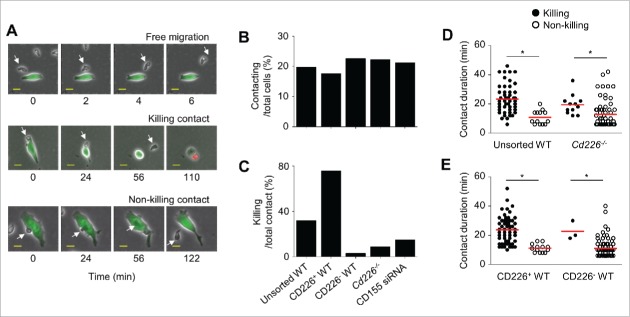
Cd226−/− NK cells show short unstable contacts with B16F10 cells
We next analyzed the contact modes of NK and B16F10 cells. We found 3 types of NK cells: (1) cells that moved freely without attraction to B16F10 cells; (2) cells that contacted and killed B16F10 cells; (3) cells that contacted B16F10 cells but did not kill them (Fig. 5A). Based on these differences, we analyzed contact dynamics of WT and Cd226−/− NK cells. WT and Cd226−/− NK cells contacted B16F10 cells at equal frequencies (1.3 times in 4 h, data not shown). The ratios of contacting cells per total number of cells were also similar: 19% for unsorted WT NK cells, 18% for purified CD226+ WT NK cells, 22% for purified CD226− WT NK cells, 22% for Cd226−/− NK cells with B16F10 cells, and 21% for unsorted WT NK cells with CD155 siRNA-transfected B16F10 cells (Fig. 5B). Nevertheless, only a few NK cells contacted cancer cells in all NK cell subsets; the outcomes of the contacts were markedly different depending on the presence of CD226: 80% of contacts with purified CD226+ NK cells but only 3% of contacts with purified CD226− NK cells led to the death of B16F10 cells (Fig. 5C). In addition, 5% of contacts with Cd226−/− NK cells killed B16F10 cells and 15% of contacts with unsorted WT NK cells killed B16F10 cells transfected with CD155 siRNA (Fig. 5C). Contact duration between cancer cells and NK cells differed: it was 23.3 min for most unsorted WT cells and 23.7 min for most CD226+ WT NK cells; 10.8 min for most CD226− WT cells and 12.8 min for most Cd226−/− NK cells (Fig. 5D and E). These data indicate that CD226 prolongs the duration of contact between NK and B16F10 cells but does not affect their contact frequency. These data indicate that CD226 prolongs the duration of contact between NK and B16F10 cells but does not affect their contact frequency.
Figure 5.
Classification of NK cells based on contact modes. (A) Unstained NK cells (unsorted WT, CD226+ WT, CD226− WT, or Cd226−/− NK cells) and calcein AM–stained B16F10 cells (transfected with CD155 siRNA or not) were imaged every 2 min from 1 h to 5 h. Representative images of the contact modes of CD226+ NK cells (arrows) showing free migration, killing contact, and non-killing contact (magnification, 200 ×; electronically zoomed; scale bars, 15 μm; n = 10 movies of 3 independent experiments per group). (B) Ratios of contacting NK cells to total NK cells (n = 1,413, 527, 521, 376, and 688 from the left). (C) Ratios of killing per total contacts (n = 279, 93, 118, 84, and 146 from the left). (D, E) Contact duration of killing and non-killing interactions between cancer cells and unsorted (n = 79), Cd226−/− (n = 191), CD226+ (n = 71), and CD226− (n = 145) NK cells. (n = 3, *p < 0.01).
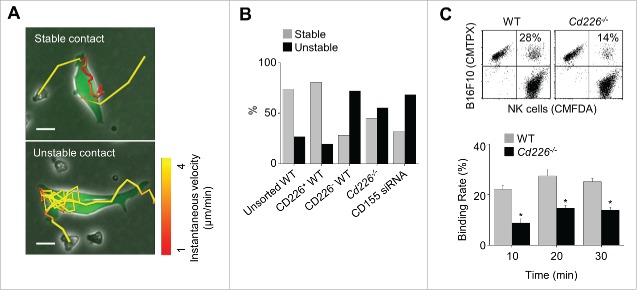
We also analyzed contact stability. Some NK cells showed oscillating movement on the surface of B16F10 cells at an average velocity of < 2 μm/min (called stable contact), while others were moving around on the surface of target cells at a speed of 4 μm/min (called unstable contact) (Fig. 6A and Movie S6). More unsorted or CD226+ WT NK cells showed stable contacts than did CD226− WT NK cells or Cd226−/− NK cells. In addition, unsorted WT NK cells showed unstable contacts with B16F10 cells transfected with CD155 siRNA (Fig. 6B). Our binding assay confirmed weaker binding of Cd226−/− NK cells than WT NK cells (Fig. 6C). Overall, these data suggest that CD226 enhances binding of NK cells to B16F10 cells and improves contact stability.
Figure 6.
Stability of contacts between NK and cancer cells. (A) Unstained NK cells (unsorted WT, CD226+ WT, CD226− WT, or Cd226−/− NK cells) and calcein AM–stained B16F10 cells (transfected with CD155 siRNA or not) were imaged every 2 min from 1 h to 5 h. Representative images of stable and unstable contacts (magnification, 200 ×; electronically zoomed; scale bar, 15 μm; n = 10 movies of 3 independent experiments per group). (B) Ratios of stable and unstable contacts between cancer cells and NK cells (n = 142, 84, 116, 111, and 146 from the left). (C) Binding rates of cancer cells and wild-type or Cd226−/− NK cells analyzed by flow cytometry (n = 3, mean ± SEM, *p < 0.01).
Cd226−/− NK cells only weakly inhibit tumor metastasis in vivo
Because of shorter and unstable contacts, we postulated that Cd226−/− NK cells might have low anti-metastatic activity in vivo. When injected intravenously into recipient mice, B16F10 cells formed 135 metastatic colonies on the surface of lung at 2 weeks after transfer (Fig. 7A and B). WT NK cells inhibited metastasis by 83%, whereas Cd226−/− NK cells inhibited it by 28%. (Fig. 7A and B). On histological assessment, WT NK cells efficiently inhibited the formation of tumor colonies in the deeper areas of the lung compared with the control group, but Cd226−/− NK cells weakly inhibit tumor metastasis to the lung (Fig. 7C). These data suggest that Cd226−/− NK cells have weak anti-metastatic activity in vivo, probably due to the impairment of their interaction with cancer cells.
Figure 7.
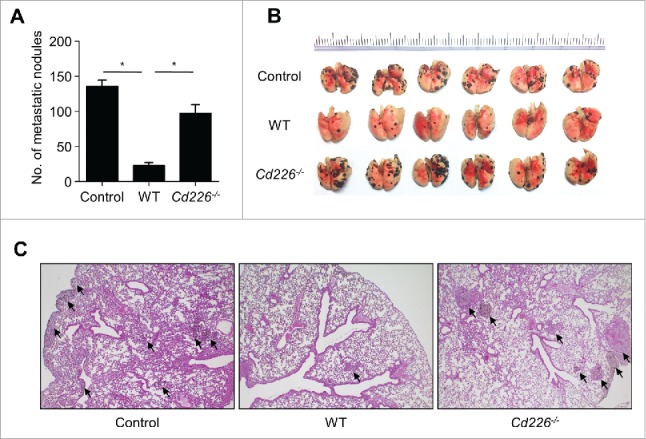
Cd226−/− NK cells weakly inhibit the lung metastasis of cancer cells in vivo. C57BL/6 mice (n = 6) were injected intravenously with B16F10 cells on day 0 and NK cells on day 2. Lungs were collected on day 14 and the metastatic nodules were counted. Mean ± SEM (A) and representative images are shown (B). Representative images of hematoxylin and eosin staining of lung sections (magnification, × 100) (C). *p < 0.01.
Discussion
Our study provides several insights into the cytotoxic mechanisms of NK cells. We found 3 CD226-independent events: (i) CD226 does not affect the speed of NK cell migration (∼5 μm/min); (ii) CD226 does not affect the frequency of NK cell contacts with cancer cells; and (iii) a small fraction (approximately 20%) of NK cells randomly contact cancer cells even if the former do not express CD226. In contrast, we found that (i) CD226 prolongs the duration of contacts between NK and cancer cells, (ii) it increases contact stability; (iii) it enhances the antitumor activity of NK cells in vitro and in vivo. Collectively, our data reveal how NK cells kill cancer cells in a CD226-dependent manner.
Our study extends the previous data on the dynamics of cancer cell killing by NK cells. NK cells kill target cells in a well-ordered manner: transport of the lytic granules to the microtubule-organizing center; polarization of the organizing center to the immunological synapse; and exocytosis of lytic granules into the immunological synapse.29-33 The released perforin and granzymes induces membrane protrusions in the target cells and their apoptotic and necrotic cell death.34 The dynamics of killing by NK cells has also been reported: NK cells can deliver the lytic hit within 10 min,29,35,36 individual NK cells sequentially kill up to 6 target cells over 16 h37; NK cells are able to kill more target cells if the cytotoxic granules are recycled38,39; eventual depletion of perforin in the granules is thought to contribute to the exhaustion of the NK cell killing capacity. In this study, we examined the dynamics of the NK cell contacts with cancer cells that precede killing of the latter. We found that if NK cells stably contact cancer cells for more than 23 min at a frequency of 2-3 over 4 h, they can efficiently kill cancer cells, and this ability depends on CD226 expressed on the NK cell surface.
Our data confirm the critical role of CD226 in cancer cell killing by NK cells. CD226 controls immunological synapse formation, cytotoxicity, cytokine secretion, and differentiation of NK cells.2 Our data show that CD226 also ensures the sufficient contact of NK and cancer cells required for efficient killing of cancer cells. In contrast, several studies suggest that CD226 does not directly affect NK cell functions. Anti-CD226 neutralizing antibodies do not alter cytokine production by NK cells activated by IL-12 and IL-18.2 However, it has not been tested whether these antibodies inhibit cytokine production by NK cells activated by CD226 ligands expressed on cancer cells. NK cells with mutated CD226 (Y319F and N321Q) conjugate with cancer cells normally, although these NK cells cannot kill cancer cells.23 However, since contact duration and stability were not examined in the latter study, it is difficult to conclude whether the CD226 mutations directly affected contact quality. Overall, the available information suggests that CD226 ensures sufficient contact of NK and cancer cells and its downstream signaling is obviously required for degranulation of perforin and granzymes and cancer cell killing.
It is becoming clear that considerable functional heterogeneity exists within the peripheral NK cells.40 Most NK cells (approximately 75%) are unable to kill cancer cells, whereas a minority of NK cells are responsible for most cancer cell death, and serial killers killing more than 3 cancer cells were observed in 12-h imaging experiments.41 However, the latter study did not characterize the phenotypes of killer and non-killer NK cells. Our data consistently suggest that a small fraction of NK cells (approximately 20%) are responsible for most cancer cell death. Furthermore, we suggest that CD226 might be one of the markers of killer NK cells. NK cells of various maturation stages exhibit differential responsiveness to cytokines and different levels of cytotoxicity and are found in various tissues, such as the spleen, lymph nodes, liver, and lung.42 CD226 is expressed in all NK cell progenitors, whereas it is downregulated as NK cells mature, generating CD226+ and CD226− NK cells in the periphery.2 Following stimulation with IL-12 and IL-18, CD226+ NK cells produce more IFN-γ, IL-6, CCL5, and GM-CSF than do CD226− NK cells.2 CD226+ NK cells also suppress tumor growth and metastasis in vivo more effectively than CD226− NK cells.2 Our data provide evidence that half of the splenic NK cells were CD226+ NK cells and that CD226+ have better antitumor activity than CD226− NK cells.
Further studies will be required to address the cytotoxic mechanisms of CD226-deficient NK cells. It will be interesting to study any compensatory signals in these cells. The CD155 and CD112 ligands expressed on cancer cells can bind not only the activating receptor CD226 but also the inhibitory receptors T cell immunoreceptor with Ig and ITIM domain (TIGIT) or CD96 on immune cells.43 Thus, the loss of CD226 would increase the availability of CD155 and CD112 for TIGIT, which might complicate the exact interpretation of the functional data for CD226− NK cells. Interestingly, we observed that CD226− NK cells expressed TIGIT and killer cell lectin-like receptor G1 (KLRG1) at much higher levels than did CD226+ NK cells (data not shown). In addition, a total of 561 and 305 genes were found to be over- and under-expressed, respectively, in CD226+ NK cells compared with CD226− NK cells.2 Thus, we cannot exclude that weak cytotoxicity and physical contacts of CD226− NK cells might be due to the enhanced expression of TIGIT, KLRG1, or other unknown factors. Our future study will help to reveal how NK cells gather and balance positive and negative information from cancer cells.
It will be interesting to examine why Cd226−/− NK cells have weak antitumor activity in vivo. Their failure to control lung metastasis could be due to impaired contacts with cancer cells, impaired homing to the tumor site, or decreased viability. It was previously reported that treatment of endothelial cells with anti-CD155 antibody or treatment of monocytes with anti-CD226 antibody prevented transendothelial migration of monocytes, although this was shown in an in vitro system.44 These data indirectly suggest that Cd226−/− NK cells might show impaired homing to the tumor site. The data from our preliminary immunofluorescence study show that more NKp46+ NK cells are present in the lung metastatic foci of mice injected with WT NK cells than in those of mice injected with PBS or Cd226−/− NK cells (data not shown). These data will help to reveal in greater detail why Cd226−/− NK cells show impaired control of tumor metastasis in vivo.
It will also be interesting to study whether CD226 is the main activating receptor in NK cells. NK cells simultaneously use several activating receptors, such as CD226 recognizing CD155 and CD112, LFA-1 binding to ICAM-1, and NKG2D binding to Rae-1 and MULT-1.45,46 Our unpublished observations suggest that CD226-deficient NK cells normally bind and kill C1498 and Yac-1 cells expressing ICAM-1, CD155, Rae-1, and MULT-1. Thus, we postulate that CD226-deficient NK cells can use other activating receptors, such as LFA-1 and NKG2D, to kill cancer cells. This study was designed with a very narrow scope: to examine the role of CD226 in contact dynamics of NK and cancer cells. To address this issue, we used CD226-deficient NK cells as effector cells and B16F10 as target cells; B16F10 selectively express CD155 but not ICAM-1, Rae-1, or MULT-1. Thus, it is difficult to conclude from our data whether CD226 is the main activating receptor in NK cells. Our next study will examine the possible hierarchy of several activating receptors in NK cell activation to reveal how NK cells gather positive information from cancer cells.
It will also be worth comparing the contacts between dendritic cells and CD226+ or CD226− NK cells. The proliferation and production of IFN-γ and TNF-α of CD226-deficient CD8+ T cells was reduced when stimulated with dendritic cells, although it was normal when stimulated with anti-CD3 and anti-CD28 antibodies.27 CD226+ NK cells would have access to dendritic cells given the role of CD226 in the functioning of the immunological synapse and generation of effector cytokines. CD226− NK cells may be positioned to alert other leukocytes through secretion of MIP-1 in a non-synapse-dependent but cytokine-dependent manner.2 Presumably, these secretion events occur after differentiation of CD226+ NK cells into CD226− NK cells. It will be important to assess the behavior of these CD226+ NK cells in the presence of dendritic cells in vitro and in vivo.
Our studies might have implications for the rational design of NK cell–based immunotherapy of cancer patients. Up-regulation of activating ligands on tumor cells is also a good strategy to improve antitumor activity of NK cells. Several studies reported that anticancer drugs such as inhibitors of proteasome, histone deacetylase, GSK3, and HSP-90 as well as genotoxic drugs increase surface expression of NKG2D and CD226 ligands on cancer cells.47-49 Treatment of multiple myeloma with nitric oxide donors increases the expression of CD155 on cancer cells, rendering these cells more susceptible to NK cell–mediated killing.48 Treatment of K-562, HCT-116, and Hep-G2 cells with cytochalasin D, nocodazole, or docetaxel increases cell surface expression of NKG2D and CD226 ligands such as MICA, ULBP, CD155, and CD122.42 CD226 is not expressed in a bimodal fashion on human NK cells, but its high to low/negative expression is observed.2 Thus, it is important to examine whether the same cytokine/cytotoxicity signatures can be found among human NK cells that express various levels or no CD226. If these functionally unique cells can be found, the prospect of translating these findings into clinical applications will increase. NK cell transfer is increasingly being considered in the treatment of cancer patients50 and may even have merit in protection from or resolution of some infections with viruses or other pathogens.
In summary, our time-lapse imaging demonstrates that NK cells use CD226 to increase their contacts with and killing of cancer cells. Optimizing the in vitro protocol for the enrichment of CD226+ NK cells may be a good approach to expand the clinical application of NK cells, if it is reasonably cost-effective. Our imaging data offer new opportunities to identify the parameters that regulate the efficacy of NK cell–based immunotherapy in cancer patients. A better understanding of how NK cells contact cancer cells can be ultimately used to enhance their antitumor activity in cancer patients.
Materials and methods
Mice and cells
Cd226−/− mice were provided by Dr. A. Shibuya (University of Tsukuba, Ibaraki, Japan) and C57BL/6 mice were purchased from Samtako (Gyeonggi, Korea). All animal studies were approved by the Chungbuk National University Animal Experimentation Ethics Committee and were performed in accordance with the approved guidelines. NK cells were isolated from mouse spleen cells by negative selection using an NK isolation kit (Miltenyi Biotec, Auburn, CA, USA). Purified NK cells were cultured in RPMI 1640 medium supplemented with 3,000 U/ml recombinant human IL-2 (Bayer HealthCare Pharmaceuticals, Emeryville, CA, USA), 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. Cell purity exceeded 90%. IL-2-activated NK cells were used from day 10 to 12. Activated NK cells were further sorted into CD226+ and CD226− cells using a FACSAriaII flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Purity of CD226+ cells exceeded 90%. B16F10 melanoma cells were purchased from American Type Culture Collection (Manassas, VA, USA).
Flow cytometry
Cells were stained for 15 min at 4°C with antibodies against mouse CD3, CD69, ICAM-1, H-2Kb, LFA-1, NK1.1, NKG2D (BD Biosciences), CD226, CD155 (BioLegend, San Diego, CA, USA), or Rae-1 (R&D Systems, Minneapolis, MN, USA). NK cells were also fixed using a CytoFix-CytoPerm Kit (BD Biosciences) according to the manufacturer's instructions and then were stained with anti-perforin–APC antibody and anti-granzyme B–FITC antibody (eBioscience, San Diego, CA, USA). To analyze exocytosis, NK cells (1 × 105) were mixed with B16F10 cells (1 × 105), centrifuged at 1,000 rpm for 1 min, and incubated for 2 h at 37°C in the presence of anti-CD107a–FITC antibody (BD Biosciences). The cells were then stained with anti-NK1.1–APC antibody. Cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences) and the data were processed using Cell Quest Pro software (BD Biosciences).
Lactate dehydrogenase (LDH) release assay
NK cells were incubated with B16F10 cells in 96-well plates at various effector-to-target cell ratios. After 4-h incubation, the plates were centrifuged and 100 μl of the supernatants was transferred to new 96-well plates. B16F10 cell death was determined using LDH release assay according to the manufacturer's instructions (Takara, Shiga, Japan). The percentage of specific lysis was calculated from LDH content as follows: (experimental release–target spontaneous release–effector spontaneous release) / (target maximum release–target spontaneous release) × 100%.51
RNA interference
A double-stranded small interfering RNA (siRNA) oligonucleotide targeting CD155 (GenBank accession number NM_027514), 5′-GAG CAU AAA GCA AGG UUG AdTdT-3′, 5′-CCA CUG CAC UUU UCU AGG UdTdT-3′, 5′-CUA GGC UAC AUC UUU CUU AdTdT-3′, was chemically synthesized (Bioneer, Daejon, Korea) and transfected into B16F10 cells (25 pmol per well of a 6-well plate) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's instructions. Cells were incubated with the siRNA–lipid complex in growth medium without antibiotics for 48 h. Control cells were transfected with a negative control siRNA oligonucleotide at a matching concentration.
Time-lapse imaging
B16F10 cells (5 × 104 cells/35 mm dish) were pre-cultured in serum-containing medium for 4 h at 37°C, washed twice with serum-containing medium, and stained with 1 μM calcein acetoxymethyl ester (Calcein AM, Thermo Fisher Scientific) in serum-free medium for 30 min at 37°C, and washed twice. NK cells (1 × 105 cells/35 mm dish) were then added to calcein AM–labeled B16F10 cells (target) in 35 mm dish. Propidium iodide (PtdIns, 2 μM) was added to medium. Time-lapse imaging was performed with a Biostation IM-Q microscope equipped with a 20 × magnification objective (numeric aperture 0.5) in an environmental chamber kept at 37°C and 5% CO2 (Nikon Inc., Melville, NY, USA). Dishes were preincubated for 1 h in the chamber and images were acquired every 2 min for 4 h.51 NK cells were manually tracked by using Imaris software version 7.2 (Bitplane, Zurich, Switzerland). Instantaneous velocity was calculated automatically by Imaris software. The number of contacts that lasted for >6 min was determined. PtdIns-stained and calcein AM–leaking B16F10 cells were considered dead cells.
Cell binding assay
NK cells (1 × 106 cells/ml) were stained with 0.5 μM 5-chloromethylfluorescein diacetate (CMFDA, Thermo Fisher Scientific)and B16F10 cells (1 × 106 cells/ml) with 5 μM 5-(and-6)-(((4-chloromethyl)benzoyl)amino) tetramethylrhodamine (CMTPX, Thermo Fisher Scientific) in serum-free medium for 15 min at 37°C. After staining, cells were washed twice in culture medium with 10% fetal bovine serum. NK cells (4 × 105) and B16F10 cells (1 × 105) were mixed in a 12 × 75-mm polystyrene tube (BD Biosciences) and centrifuged at 1,000 rpm for 1 min, and pellets were incubated at 37°C for 30 min. Cell mixtures were then gently suspended and analyzed by flow cytometry. The conjugation ratio was calculated as the portion of CMFDA/CMTPX double-positive events within the CMTPX-positive events.
In vivo metastasis model
B16F10 cells (4 × 105 cells/mouse) were injected intravenously into C57BL/6 mice on day 0. NK cells (4 × 106 cells/mouse) were injected intravenously on day 2. Lungs were collected on day 14 and the metastatic nodules were counted.52 For histologic analysis, lung tissues were fixed in 10% buffered formalin, embedded in paraffin and sectioned at 5 μm thickness. Sections were stained with hematoxylin and eosin.
Statistical analysis
Data represent the mean ± SEM of at least 3 independent experiments performed in triplicates (in vitro) or 6 mice (in vivo); p values were calculated using Student's t-test or one-way ANOVA in GraphPad Prism Software (San Diego, CA, USA). For NK or B16F10 cell counts, p values were calculated using Mann–Whitney test in GraphPad Prism Software.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr. A. Shibuya (University of Tsukuba, Ibaraki, Japan) for kindly providing Cd226−/− mice.
Funding
This research was supported by grants funded by the Korean government (NRF-MRC-2008–0062275 and KHIDI-HI15C0778).
References
- 1.Lakshmikanth T, Burke S, Ali TH, Kimpfler S, Ursini F, Ruggeri L, Capanni M, Umansky V, Paschen A, Sucker A, et al.. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest 2009; 119:1251-63; PMID:19349689; https://doi.org/ 10.1172/JCI36022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinet L, Ferrari De Andrade L, Guillerey C, Lee JS, Liu J, Souza-Fonseca-Guimaraes F, Hutchinson DS, Kolesnik TB, Nicholson SE, Huntington ND, et al.. DNAM-1 expression marks an alternative program of NK cell maturation. Cell Rep 2015; 11:85-97; PMID:25818301; https://doi.org/ 10.1016/j.celrep.2015.03.006 [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu Rev Immunol 2005; 23:225-74; PMID:15771571; https://doi.org/ 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- 4.Horst D, Verweij MC, Davison AJ, Ressing ME, Wiertz EJ. Viral evasion of T cell immunity: Ancient mechanisms offering new applications. Curr Opin Immunol 2011; 23:96-103; PMID:21146386; https://doi.org/ 10.1016/j.coi.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 5.Colucci F, Di Santo JP, Leibson PJ. Natural killer cell activation in mice and men: Different triggers for similar weapons? Nat Immunol 2002; 3:807-13; PMID:12205470; https://doi.org/ 10.1038/ni0902-807 [DOI] [PubMed] [Google Scholar]
- 6.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, et al.. Identification of PVR (CD155) and nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 2003; 198:557-67; PMID:12913096; https://doi.org/ 10.1084/jem.20030788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas LM, Peterson ME, Long EO. Cutting edge: NK cell licensing modulates adhesion to target cells. J Immunol 2013; 191:3981-5; PMID:24038086; https://doi.org/ 10.4049/jimmunol.1301159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enqvist M, Ask EH, Forslund E, Carlsten M, Abrahamsen G, Beziat V, Andersson S, Schaffer M, Spurkland A, Bryceson Y, et al.. Coordinated expression of DNAM-1 and LFA-1 in educated NK cells. J Immunol 2015; 194:4518-27; PMID:25825444; https://doi.org/ 10.4049/jimmunol.1401972 [DOI] [PubMed] [Google Scholar]
- 9.Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR, Lanier LL, et al.. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 1996; 4:573-81; PMID:8673704; https://doi.org/ 10.1016/S1074-7613(00)70060-4 [DOI] [PubMed] [Google Scholar]
- 10.Kojima H, Kanada H, Shimizu S, Kasama E, Shibuya K, Nakauchi H, Nagasawa T, Shibuya A. CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J Biol Chem 2003; 278:36748-53; PMID:12847109; https://doi.org/ 10.1074/jbc.M300702200 [DOI] [PubMed] [Google Scholar]
- 11.Vo AV, Takenaka E, Shibuya A, Shibuya K. Expression of DNAM-1 (CD226) on inflammatory monocytes. Mol Immunol 2016; 69:70-6; PMID:26675069; https://doi.org/ 10.1016/j.molimm.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 12.Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, Shibuya A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol 2004; 16:533-8; PMID:15039383; https://doi.org/ 10.1093/intimm/dxh059 [DOI] [PubMed] [Google Scholar]
- 13.Tahara-Hanaoka S, Miyamoto A, Hara A, Honda S, Shibuya K, Shibuya A. Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem Biophys Res Commun 2005; 329:996-1000; PMID:15752754; https://doi.org/ 10.1016/j.bbrc.2005.02.067 [DOI] [PubMed] [Google Scholar]
- 14.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood 1998; 92:4602-11; PMID:9845526. [PubMed] [Google Scholar]
- 15.Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: Implications for poliovirus infection. J Infect Dis 2002; 186:585-92; PMID:12195344; https://doi.org/ 10.1086/342682 [DOI] [PubMed] [Google Scholar]
- 16.Masson D, Jarry A, Baury B, Blanchardie P, Laboisse C, Lustenberger P, Denis MG. Overexpression of the CD155 gene in human colorectal carcinoma. Gut 2001; 49:236-40; PMID:11454801; https://doi.org/ 10.1136/gut.49.2.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahara-Hanaoka S, Shibuya K, Kai H, Miyamoto A, Morikawa Y, Ohkochi N, Honda S, Shibuya A. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood 2006; 107:1491-6; PMID:16249389; https://doi.org/ 10.1182/blood-2005-04-1684 [DOI] [PubMed] [Google Scholar]
- 18.Carlsten M, Bjorkstrom NK, Norell H, Bryceson Y, van Hall T, Baumann BC, Hanson M, Schedvins K, Kiessling R, Ljunggren HG, et al.. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res 2007; 67:1317-25; PMID:17283169; https://doi.org/ 10.1158/0008-5472.CAN-06-2264 [DOI] [PubMed] [Google Scholar]
- 19.Castriconi R, Dondero A, Corrias MV, Lanino E, Pende D, Moretta L, Bottino C, Moretta A. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: Critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res 2004; 64:9180-4; PMID:15604290; https://doi.org/ 10.1158/0008-5472.CAN-04-2682 [DOI] [PubMed] [Google Scholar]
- 20.Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, Falco M, Lanino E, Pierri I, Zambello R, et al.. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: Evidence for the involvement of the poliovirus receptor (CD155) and nectin-2 (CD112). Blood 2005; 105:2066-73; PMID:15536144; https://doi.org/ 10.1182/blood-2004-09-3548 [DOI] [PubMed] [Google Scholar]
- 21.El-Sherbiny YM, Meade JL, Holmes TD, McGonagle D, Mackie SL, Morgan AW, Cook G, Feyler S, Richards SJ, Davies FE, et al.. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res 2007; 67:8444-9; PMID:17875681; https://doi.org/ 10.1158/0008-5472.CAN-06-4230 [DOI] [PubMed] [Google Scholar]
- 22.Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, Sato T, Yoshikawa H, Hara H, Shibuya A, et al.. Increased soluble CD155 in the serum of cancer patients. PLoS One 2016; 11:e0152982; PMID:27049654; https://doi.org/ 10.1371/journal.pone.0152982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Wu N, Lu Y, Davidson D, Colonna M, Veillette A. DNAM-1 controls NK cell activation via an ITT-like motif. J Exp Med 2015; 212:2165-82; PMID:26552706; https://doi.org/ 10.1084/jem.20150792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, Yasui T, Kikutani H, Shibuya K, Shibuya A. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med 2008; 205:2959-64; PMID:19029379; https://doi.org/ 10.1084/jem.20081611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magri G, Muntasell A, Romo N, Saez-Borderias A, Pende D, Geraghty DE, Hengel H, Angulo A, Moretta A, Lopez-Botet M. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 2011; 117:848-56; PMID:21030563; https://doi.org/ 10.1182/blood-2010-08-301374 [DOI] [PubMed] [Google Scholar]
- 26.Nabekura T, Kanaya M, Shibuya A, Fu G, Gascoigne NR, Lanier LL. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 2014; 40:225-34; PMID:24440149; https://doi.org/ 10.1016/j.immuni.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsbottom KM, Hawkins ED, Shimoni R, McGrath M, Chan CJ, Russell SM, Smyth MJ, Oliaro J. Cutting edge: DNAX accessory molecule 1-deficient CD8+ T cells display immunological synapse defects that impair antitumor immunity. J Immunol 2014; 192:553-7; PMID:24337740; https://doi.org/ 10.4049/jimmunol.1302197 [DOI] [PubMed] [Google Scholar]
- 28.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, et al.. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014; 26:923-37; PMID:25465800; https://doi.org/ 10.1016/j.ccell.2014.10.018 [DOI] [PubMed] [Google Scholar]
- 29.Wulfing C, Purtic B, Klem J, Schatzle JD. Stepwise cytoskeletal polarization as a series of checkpoints in innate but not adaptive cytolytic killing. Proc Natl Acad Sci U S A 2003; 100:7767-72; PMID:12802007; https://doi.org/ 10.1073/pnas.1336920100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orange JS, Harris KE, Andzelm MM, Valter MM, Geha RS, Strominger JL. The mature activating natural killer cell immunologic synapse is formed in distinct stages. Proc Natl Acad Sci U S A 2003; 100:14151-6; PMID:14612578; https://doi.org/ 10.1073/pnas.1835830100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown AC, Oddos S, Dobbie IM, Alakoskela JM, Parton RM, Eissmann P, Neil MA, Dunsby C, French PM, Davis I, et al.. Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super-resolution microscopy. PLoS Biol 2011; 9:e1001152; PMID:21931537; https://doi.org/ 10.1371/journal.pbio.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths GM, Tsun A, Stinchcombe JC. The immunological synapse: A focal point for endocytosis and exocytosis. J Cell Biol 2010; 189:399-406; PMID:20439993; https://doi.org/ 10.1083/jcb.201002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, Podack ER, Zinkernagel RM, Hengartner H. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 1994; 369:31-7; PMID:8164737; https://doi.org/ 10.1038/369031a0 [DOI] [PubMed] [Google Scholar]
- 34.Trapani JA, Bird PI. A renaissance in understanding the multiple and diverse functions of granzymes? Immunity 2008; 29:665-7; PMID:19006688; https://doi.org/ 10.1016/j.immuni.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 35.Eriksson M, Leitz G, Fallman E, Axner O, Ryan JC, Nakamura MC, Sentman CL. Inhibitory receptors alter natural killer cell interactions with target cells yet allow simultaneous killing of susceptible targets. J Exp Med 1999; 190:1005-12; PMID:10510090; https://doi.org/ 10.1084/jem.190.7.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrod KR, Wei SH, Parker I, Cahalan MD. Natural killer cells actively patrol peripheral lymph nodes forming stable conjugates to eliminate MHC-mismatched targets. Proc Natl Acad Sci U S A 2007; 104:12081-6; PMID:17609379; https://doi.org/ 10.1073/pnas.0702867104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhat R, Watzl C. Serial killing of tumor cells by human natural killer cells–enhancement by therapeutic antibodies. PLoS One 2007; 2:e326; PMID:17389917; https://doi.org/ 10.1371/journal.pone.0000326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu D, Martina JA, Wu XS, Hammer JA 3rd, Long EO. Two modes of lytic granule fusion during degranulation by natural killer cells. Immunol Cell Biol 2011; 89:728-38; PMID:21483445; https://doi.org/ 10.1038/icb.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P, Zheng G, Yang Y, Zhang C, Xiong P, Xu Y, Fang M, Tan Z, Zheng F, Gong F. Granzyme B is recovered by natural killer cells via clathrin-dependent endocytosis. Cell Mol Life Sci 2010; 67:3197-208; PMID:20449762; https://doi.org/ 10.1007/s00018-010-0377-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanherberghen B, Olofsson PE, Forslund E, Sternberg-Simon M, Khorshidi MA, Pacouret S, Guldevall K, Enqvist M, Malmberg KJ, Mehr R, et al.. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood 2013; 121:1326-34; PMID:23287857; https://doi.org/ 10.1182/blood-2012-06-439851 [DOI] [PubMed] [Google Scholar]
- 41.Guldevall K, Brandt L, Forslund E, Olofsson K, Frisk TW, Olofsson PE, Gustafsson K, Manneberg O, Vanherberghen B, Brismar H, et al.. Microchip screening platform for single cell assessment of NK cell cytotoxicity. Front Immunol 2016; 7:119; PMID:27092139; https://doi.org/ 10.3389/fimmu.2016.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huntington ND, Martinet L, Smyth MJ. DNAM-1: Would the real natural killer cell please stand up! Oncotarget 2015; 6:28537-8; PMID:26471195; https://doi.org/ 10.18632/oncotarget.5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, Zhao W, Li H, Chen Y, Tian H, Li L, Zhang L, Gao C, Zheng J. Immunoreceptor TIGIT inhibits the cytotoxicity of human cytokine-induced killer cells by interacting with CD155. Cancer Immunol Immunother 2016; 65:305-14; PMID:26842126; https://doi.org/ 10.1007/s00262-016-1799-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan DP, Seidman MA, Muller WA. Poliovirus receptor (CD155) regulates a step in transendothelial migration between PECAM and CD99. Am J Pathol 2013; 182:1031-42; PMID:23333754; https://doi.org/ 10.1016/j.ajpath.2012.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene 2008; 27:5932-43; PMID:18836474; https://doi.org/ 10.1038/onc.2008.267 [DOI] [PubMed] [Google Scholar]
- 46.Gross CC, Brzostowski JA, Liu D, Long EO. Tethering of intercellular adhesion molecule on target cells is required for LFA-1-dependent NK cell adhesion and granule polarization. J Immunol 2010; 185:2918-26; PMID:20675589; https://doi.org/ 10.4049/jimmunol.1000761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fionda C, Soriani A, Zingoni A, Santoni A, Cippitelli M. NKG2D and DNAM-1 ligands: Molecular targets for NK cell-mediated immunotherapeutic intervention in multiple myeloma. Biomed Res Int 2015; 2015:178698; PMID:26161387; https://doi.org/ 10.1155/2015/178698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fionda C, Abruzzese MP, Zingoni A, Soriani A, Ricci B, Molfetta R, Paolini R, Santoni A, Cippitelli M. Nitric oxide donors increase PVR/CD155 DNAM-1 ligand expression in multiple myeloma cells: Role of DNA damage response activation. BMC Cancer 2015; 15:17; PMID:25609078; https://doi.org/ 10.1186/s12885-015-1023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acebes-Huerta A, Lorenzo-Herrero S, Folgueras AR, Huergo-Zapico L, Lopez-Larrea C, Lopez-Soto A, Gonzalez S. Drug-induced hyperploidy stimulates an antitumor NK cell response mediated by NKG2D and DNAM-1 receptors. Oncoimmunology 2016; 5:e1074378; PMID:27057443; https://doi.org/ 10.1080/2162402X.2015.1074378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berrien-Elliott MM, Romee R, Fehniger TA. Improving natural killer cell cancer immunotherapy. Curr Opin Organ Transplant 2015; 20:671-80; PMID:26414502; https://doi.org/ 10.1097/MOT.0000000000000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HK, Kim YG, Kim JS, Park EJ, Kim B, Park KH, Kang JS, Hong JT, Kim Y, Han SB. Cytokine-induced killer cells interact with tumor lysate-pulsed dendritic cells via CCR5 signaling. Cancer Lett 2016; 378:142-9; PMID:27216980; https://doi.org/ 10.1016/j.canlet.2016.05.020 [DOI] [PubMed] [Google Scholar]
- 52.Han SB, Lee CW, Kang MR, Yoon YD, Kang JS, Lee KH, Yoon WK, Lee K, Park SK, Kim HM. Pectic polysaccharide isolated from Angelica gigas Nakai inhibits melanoma cell metastasis and growth by directly preventing cell adhesion and activating host immune functions. Cancer Lett 2006; 243:264-73; PMID:16412568; https://doi.org/ 10.1016/j.canlet.2005.11.040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.