ABSTRACT
Neoantigens derived from tumor-specific somatic mutations are excellent targets for anti-tumor immune responses. In ovarian clear cell carcinoma (OCCC), checkpoint blockade yields durable responses in a subset of patients. To approach the question of why only some patients respond, we first investigated neoantigen loads and immune signatures using exome sequencing and expression array data for 74 OCCC patients treated conventionally. Neither the number of missense mutations nor total predicted neoantigens assessed in the tumor correlated with clinical outcomes. However, the number of neoantigens per missense mutation (“neoAg frequency”) did correlate with clinical outcomes. Cox multivariate regression analysis demonstrated that low neoAg frequencies correlated with increased progression-free survival (PFS) and was an independent predictive factor for PFS in OCCC (p = 0.032), especially at stage I-II (p = 0.0045). Immunity-associated genes including those related to effector memory CD8 T cells were dominantly expressed in tumors with low neoAg frequencies in stage I-II patients, suggesting CD8 T cell-mediated elimination of immunogenic sub-clones expressing neoantigens (immunoediting) had occurred. In contrast, we observed decreased HLA-A, -B, and -C expression (p = 0.036, p = 0.026, and p = 0.030, respectively) as well as increased ratios of CTLA-4, PD-1, Tim-3, and LAG3 to CD8A expression (p = 0.0064, p = 0.017, p = 0.033 and p = 0.0136, respectively) in stage I-II tumors with high neoAg frequencies. Constrained anti-tumor immunity may thus result in limited immunoediting, and poor prognosis. Our results show that neoAg frequency in OCCC is an independent prognostic factor for clinical outcome and may become a potential candidate biomarker for immunomodulatory agent-based treatments.
KEYWORDS: Immune signature, missense mutation, neoantigen, neoantigen frequency, ovarian clear cell carcinoma, prognostic factor
Introduction
Ovarian clear cell carcinoma (OCCC) represents the second most common histologic subtype of this tumor, comprising 5–25% of all ovarian carcinomas. While early-stage OCCC is known to have a survival outcome similar to high-grade serous ovarian cancer (HGSC), its prognosis is poorer in advanced-stage disease.1,2 Recent studies have shown that OCCC has a molecular phenotype distinct from other types of epithelial ovarian cancers such as HGSC.3 Available data suggest that current conventional treatment recommendations for maximal cytoreductive surgery followed by systemic chemotherapy with a taxane and platinum combination may be less effective in advanced OCCC.4 Therefore, development of new modalities such as immunotherapy are of importance in the management of women with OCCC. Recently, immune checkpoint inhibitors have shown promising results in a subset of OCCC patients in early-phase clinical trials.5,6 However, immunological biomarkers that predict OCCC treatment outcome have not yet been identified, although they have been well studied in the HGSC subtype.7
Tumors commonly exhibit multiple somatic mutations. Neoantigens derived from such tumor-specific mutations are good potential targets for anti-tumor immune responses because they are foreign to the immune system.8-11 In melanomas and non-small cell lung cancers (NSCLC), the mutational and neoantigen density has been reported to correlate with clinical benefit from immune checkpoint blockade therapy.12-14 In addition, colorectal cancer patients harboring high mutational burdens due to mismatch-repair deficiency experience a more favorable clinical course than patients with lower mutational loads.15 We also reported that the neoantigen load and the status of antigen presentation machinery determine the prognosis of clear cell renal cell carcinoma (ccRCC) patients on conventional therapy (i.e. not on immunomodulatory therapy).16 Recently, it has been demonstrated that predicted antigenic mutations are selectively depleted in some cancers, consistent with the notion that immunogenic sub-clones expressing neoantigens are eliminated by T cell-mediated immunoediting.17,18
In the present study, we investigated mutation burden, neoantigen load, the depletion of expected antigenic mutations in the tumor, and immune signatures in the tumor using data from exome sequencing and expression arrays in 74 OCCC patients. We further investigated the relationships of these factors with clinical outcomes to identify potential prognostic biomarkers in OCCC.
Results
The number of missense mutations does not correlate with clinical outcomes
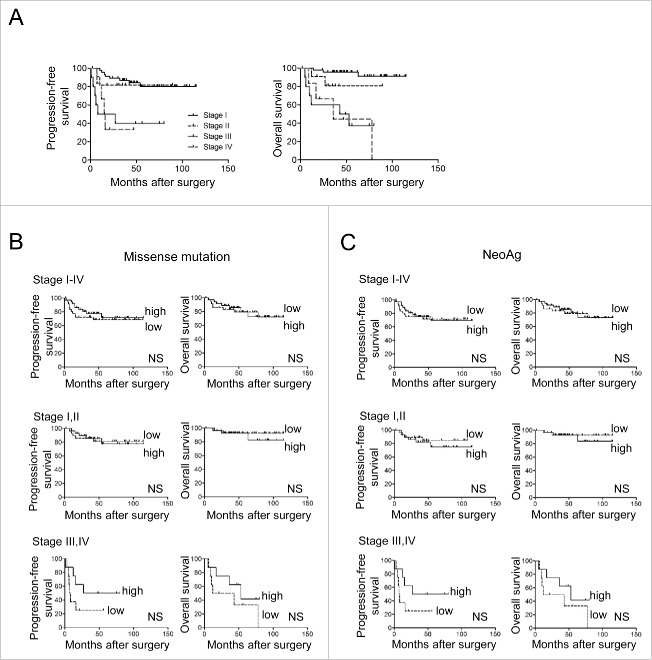
The mutation burden is reported to correlate with prognosis of certain cancers under checkpoint blockade immunotherapy.12-15 To examine whether it also correlated with the prognosis in patients with OCCC on conventional therapy, we investigated the number of missense mutations in this tumor type. The clinicopathological characteristics of the 74 OCCC patients in the study cohort are summarized in Table 1. Their prognoses including progression-free and overall survivals (PFS, OS) stratified according to FIGO stage are shown in Fig. 1A. The number of missense mutations in these patients ranged from zero to 494 (median value, 37) (Table 2 and Supplementary Table S1). We divided these stage I-IV patients into 2 groups with values above or below the median number of missense mutations and plotted Kaplan-Meier survival curves. There were no significant differences in either PFS or OS of patients whose tumors possessed numbers of missense mutations above or below the median value (Fig. 1B, upper). This remained the case when stratifying patients into subgroups of stage I and II (n = 58) and stage III and IV (n = 16) although there was a tendency for better survival of patients with more mutations in the latter group (Fig. 1B, middle and lower). These results document that the number of missense mutations fails to correlate significantly with survival in OCCC.
Table 1.
Summary of 74 ovarian clear cell carcinoma patients´ characteristics.
| N (%) | |
|---|---|
| Total patients | 74 (100) |
| Age | |
| >55 | 37 (50) |
| ≤ 55 | 37 (50) |
| FIGO stage | |
| I | 47 (63) |
| II | 11 (15) |
| III | 10 (14) |
| IV | 6 (8) |
| Ascites cytology | |
| positive | 24 (32) |
| negative | 50 (68) |
| Residual tumor | |
| Yes | 10 (14) |
| No | 64 (86) |
Figure 1.
Numbers of missense mutations and neoantigens are not correlated with prognosis in OCCC patients. A, Kaplan-Meier progression-free (left) and overall (right) survival curves in 74 OCCC patients stratified according to the FIGO stage. B, Kaplan-Meier progression-free (left) and overall (right) survival curves for OCCC patients stratified according to the number of missense mutations in stages I-IV (upper), I-II (middle), and III-IV (lower). C, Kaplan-Meier progression-free (left) and overall (right) survival curves for OCCC patients stratified according to the number of neoantigens in stages I-IV (upper), I-II (middle), and III-IV (lower). Low, high designates values below or above the median. Statistical analysis was by log-rank testing. NS, not significant.
Table 2.
The number of missense mutations and neoantigens, and neoAg freq in 74 OCCC.
| Median value | Range | Total no. | |
|---|---|---|---|
| Missense mutations | 37 | 0–494 | 3911 |
| Neoantigens | 21 | 0–297 | 2181 |
| NeoAg freq | 0.59 | 0–1 | — |
NeoAg freq, number of neoantigens per missense mutation.
Overall predicted number of neoantigens does not correlate with clinical outcomes
We next determined the neoantigen (neoAg) load in each patient based on MHC class I binding prediction scores according to NetMHCpan v2.8. Mutated peptides (9- and 10-mer) derived from missense mutations predicted to bind to each patient´s HLA molecules with high affinity (IC50<500 nM) were considered candidate neoepitopes (Supplementary Table S2). We defined a neoAg as an antigenic mutation producing any number of HLA-A, -B, and -C-restricted neoepitopes (Supplementary Table S3).
The estimated number of neoAg ranged from zero to 297 in these patients (median number, 21) (Table 2 and Supplementary Table S1). We stratified all patients (stage I-IV) according to their higher or lower than median numbers of predicted neoAg, designated neoAghi (with >21) or neoAglo (with ≤ 21). Here again, we found no significant survival difference between them either for PFS or OS (Fig. 1C, upper). Similar results were obtained in stage I and II (n = 58) and in stage III and IV (n = 16) patients analyzed separately as well, although there was again a tendency for better survival in the high group in the latter (Fig. 1C, middle and lower). These results indicate that the number of predicted neoAg was also not informative for OCCC clinical outcome.
A low frequency of neoantigens per mutation (neoAg freq) does correlate with better PFS as an independent prognostic factor
Recently, it has been shown that colorectal cancer and ccRCC exhibit preferential depletion of neoantigenic mutations within the totality of mutations in the tumor, providing circumstantial evidence of immunoediting in these types of cancer.17,18 To test whether the same may apply in patients with OCCC, we investigated the ratio of neoAg per missense mutation (designated the neoantigen frequency, neoAg freq) in each individual. One patient (case C-227) with no missense mutations was excluded from the analysis (Supplementary Table S1). As one might expect, the number of neoAg and missense mutations was significantly correlated (Fig. 2A, upper). However, the value of the neoAg freq (ranging from 0–1 with a median of 0.59, Table 2 and Supplementary Table S1) did not correlate with the number of missense mutations at all (Fig. 2A, lower).
Figure 2.
Neoantigen frequency (NeoAg freq) correlates with prognosis of OCCC patients. A, Correlations of log10 (missense mutation) and log10 (neoantigen) (upper) or neoAg freq (lower). B, Kaplan-Meier progression-free (left) and overall (right) survival curves in 73 OCCC patients stratified according to neoAg freq in stages I-IV (upper), I-II (middle), and III-IV (lower). Low, high designates values below or above the median. Statistical analysis was by log-rank testing. NS, not significant.
Next, we investigated the correlation of neoAg freq with prognosis. We stratified all 73 patients (stage I-IV) according to their possession of neoAg freq above or below the median, and plotted Kaplan-Meier survival curves. Patients with a lower than median neoAg freq (≤ 0.59) had longer PFS than those with a higher neoAg freq. This was statistically significant (p = 0.0112) (Fig. 2B, upper). Notably, especially stage I-II patients with a lower neoAg freq (≤ 0.58) had significantly longer PFS than those with higher neoAg freq (p = 0.0066) (Fig. 2B, middle). In stage III-IV patients, this became only a trend; thus, patients with lower than median neoAg freq (≤ 0.62 in this case) did have longer PFS, but this did not reach statistical significance (Fig. 2B, lower). However, this association with PFS did not translate into a benefit for OS.
We next compared these results with other clinical parameters contributing to OCCC prognosis. Univariate and multivariate survival analyses were performed using the Cox proportional hazards regression model. As shown in Table 3, age, FIGO stage, ascites cytology, residual tumor, and neoAg freq were all significantly associated with relapse according to univariate analysis of the entire cohort of patients. However, in multivariate analysis, only FIGO stage, residual tumor size, and neoAg freq remained significant for PFS. Strikingly, in patients with stage I and II disease, only neoAg freq was a strong predictor of PFS in both univariate and multivariate analyses. These results suggest that low neoAg freq is an independent favorable prognostic factor in patients with OCCC, at least in stage I and II patients for PFS.
Table 3.
Univariate and multivariate analyses of PFS for Stage I-IV and stage I-II OCCC.
| Univariate analysis |
Mulivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | P values | HR | 95% CI | P values |
| Stage I-IV | ||||||
| Age (> 55 vs ≤ 55) | 3.4 | 1.3–10.4 | 0.0109 | 2.2 | 0.8–7.0 | 0.1137 |
| FIGO stage (I,II vs III, IV) | 5.6 | 2.3–13.8 | 0.0003 | 3.9 | 1.5–9.8 | 0.0049 |
| Ascites cytology (negative vs positive) | 3.4 | 1.4–8.5 | 0.0069 | 2.5 | 1.0–6.5 | 0.0467 |
| Residual tumor (no vs yes ) | 7.5 | 2.9–18.8 | 0.0001 | |||
| NeoAg freq (low vs high) | 3.4 | 1.3–10.5 | 0.0103 | 2.9 | 1.1–8.9 | 0.0326 |
| Stage I-II | ||||||
| Age (> 55 vs ≤ 55) | 2.2 | 0.6–8.6 | 0.2189 | |||
| Ascites cytology (negative vs positive) | 2.2 | 0.6–7.8 | 0.2407 | 2 | 0.5–7.2 | 0.2906 |
| NeoAg freq (low vs high) | 10.1 | 1.9–186.7 | 0.004 | 9.9 | 1.9–182.9 | 0.0045 |
Robust immune responses in patients with low neoAg freq in stage I and II OCCC
To investigate whether a lower neoAg freq is potentially caused by anti-tumor immune responses resulting in immunoediting, we compared immune signatures between patients with lower and higher neoAg freq using microarray data. We interrogated the data set of stage I and II patients in whom neoAg freq had been shown to be a strong prognostic factor. We identified 942 probes that were differentially expressed between the 2 groups, based on a fold-expression change of > 1.5 and significance at P<0.05 (by Student t test) (Fig. 3A, left). A functional annotation analysis was performed with the DAVID Bioinformatics Resources 6.8 tool. Analysis of the 319 probes that were upregulated in patients with a higher neoAg freq yielded no significant DAVID gene clusters (DAVID Benjamini FDR). In contrast, analysis of the 623 probes that were upregulated in patients with a lower neoAg freq yielded 93 highly significant GO terms, mostly involving immune responses (Fig. 3A, right).
Figure 3.
Strong T cell immune signatures in stage I and II patients with low neoAg freq. Microarray analysis of 57 specimens (stage I and II) showing differential gene expression between patients with higher (high) or lower (low) than median neoantigen frequency. A, heatmap and cluster analysis based on differentially expressed genes detected by 941 Affymetrix probes satisfying the criteria of the Student t test for P <0.05 and fold-change >1.5. Data were analyzed with the Subio Platform and BasicPlug-in v1.19 (Subio Inc.). Red, high gene expression; green, low gene expression. The lists of top 5 and top 10 gene ontologies significantly differently expressed in the groups with higher (256 DAVID IDs) and lower (418 IDs) neoAg freq, respectively, are shown on the right. B, Gene set enrichment analysis (GSEA) using immune cell-related gene sets in patients with higher and lower neoantigen frequency. C, Correlation coefficients of expression of genes for cytotoxic molecules and immune cell markers. NES, normalized enrichment score; NS, not significant.
Using a second method to verify immune signatures, we performed gene set enrichment analysis (GSEA) for different immune cells. Gene expression matrix with 23,158 genes of 57 stage I-II OCCC patients was used for the analysis. We found that the gene sets for “effector memory CD8 T cells” (p = 0.031) and “T cells” (p = 0.046) were significantly enriched in patients with lower neoAg freq (Fig. 3B).
We also scrutinized the expression of genes for effector molecules such as perforin (PRF1), granzyme A (GZMA), and IFNγ, and their correlation with the expression of the representative cell markers CD8A, CD4, CD19, CD68, and KLRF1. As shown in Fig. 3C, GZMA (r = 0.7780), PRF1 (r = 0.4906), and IFNγ (r = 0.3932) were more strongly correlated with expression of CD8A than the other tested markers CD4, CD19, CD68, or KLRF1. This suggests that the cytolytic molecules detected may have originated from CD8 T cells. Together, these data support the interpretation that robust immune responses in the tumor had resulted in decreased neoAg freq by immunoediting, and that this is the reason for the better prognosis of these OCCC patients.
Factors affecting restricted CD8 T cell immune responses in patients with a high neoAg freq
In contrast to the patients with a low neoAg freq, immune responses seemed to be compromised in patients with a high neoAg freq. Therefore, we next investigated factors potentially restraining immune responses in these tumors. Because CD8 T cell recognition and activation is compromised by decreased expression of HLA class I heavy chains or β2-microglobulin by the tumor, we compared the expression of these molecules in patients with a higher or lower neoAg freq. No differences in β2-microglobulin expression were observed in these patients, but the expression of genes encoding HLA-A, -B and -C heavy chains was significantly lower in the group with a high neoAg freq (p = 0.036, p = 0.0264, and p = 0.0301, respectively, comparing lower to higher neoAg freq) (Fig. 4A). Therefore, low HLA expression may be one reason for reduced immune responses in patients with a high neoAg freq.
Figure 4.

Immunosuppression in patients with high neoAg freq. HLA class I gene expression in patients grouped as neoAg freq high vs. low (A). Expression of immune checkpoint molecule genes relative to the CD8A gene in patients with high vs. low neoAg freq (B).
Expression of immune checkpoint molecules on T cells can result in inhibition of anti-tumor immune responses. We found that ratios of CTLA-4:CD8A, PD-1:CD8A Tim-3:CD8A, and LAG-3:CD8A were significantly higher in patients with a high neoAg freq (p = 0.0064, p = 0.0172, p = 0.0333, and p = 0.0136, respectively) (Fig. 4B). Therefore, dominant gene expression of immune checkpoint molecules relative to CD8 T cells may be another reason for limited immune responses in patients with a high neoAg freq.
Taken together, these results suggest that several factors contributed to reduced immune responses in the tumor which result in the maintenance of the higher neoAg freq high and which lead to a poorer prognosis.
Discussion
In the present study, we performed whole-exome sequencing and expression array analysis of OCCC tumors from 74 patients. A major, perhaps unexpected, finding was that neither the number of missense mutations nor the number of potential neoantigens predicted to arise from these was associated with patient prognosis. Instead, we found that the neoantigen frequency defined as the number of neoantigens per individual missense mutation was an independent prognostic factor in patients with OCCC.
OCCC has been considered as a distinct entity among epithelial ovarian cancers (EOC) and constitutes a minority of cases of EOC in Western countries. However, its prevalence rises to 15–25% in Japan.2,19 Data on OCCC are limited even in The Cancer Genome Atlas (TCGA) database and therefore the mutation rate of OCCC has not been addressed in a large cohort. Our data demonstrate that the mutational burden and neoantigen load is not high in this disease and does not correlate with prognosis in OCCC patients receiving conventional therapy. Because a subset of OCCC patients was reported to respond to immune checkpoint blockade,5,6 it will be interesting to determine whether mutation/neoantigen burden, combined with analysis of HLA expression, shows any association with prognosis in OCCC patients who do receive checkpoint immunotherapies.
T cell-mediated immunoediting leading to elimination of immunogenic tumor cell clones presenting neoantigens has been reported.20,21 Moreover, genetic evidence of immunoediting showing lower numbers of predicted antigenic mutations relative to total mutations has been reported in certain human cancers. Particularly, depletion of antigenic determinants has been observed in colorectal cancer with microsatellite instability (MSI), known to be sensitive to immune checkpoint blockade.18 In our study, we found that OCCC patients with a lower neoAg freq at baseline examination have a better prognosis than those with a higher frequency. The former is accompanied by a strong T cell immune signature including memory CD8 T cells, especially in stage I-II tumors. This suggests that immune responses previously provoked in the tumor had eliminated the neoantigen-positive cells, resulting in a lower neoAg freq, and that this influences the prognosis of OCCC.
In contrast, we found reduced immune responses in patients with a high neoAg freq. It is clear that many factors may limit or suppress immune responses in the tumor, but because CD8 T cells recognize complexes of antigenic peptide with HLA class I molecules and exert cytotoxic activity, down regulation of HLA class I heavy chains or β2-microglobulin is a major mechanism to facilitate immune escape.22 Therefore, HLA downregulation in patients with high neoAg freq is an obvious factor restricting immune responses. Consistent with this, we also found high immune checkpoint gene expression relative to CD8A in these patients. It is possible that insufficient activation of T cells resulting from HLA downregulation induces relatively strong expression of immune checkpoint genes, because the number of candidate neoantigens in patients with higher or lower neoAg freq did not differ (Supplementary Table S1). From these findings, it should be a good strategy to negate those factors restricting immune responses in patients with high neoAg freq to enhance anti-tumor immunity against existing neoantigens. This might include IFN-based therapy to upregulate HLA class I23 as well as multiple immune checkpoint blockades. Or, chimeric antigen receptor (CAR) T-cell therapy targeting surface antigens (e.g. mesothelin) independent of HLA restriction would be another choice.24
The present study has some limitations. First, because we obtained tumor tissues only at the time of surgery, any changes thereafter in the number of neoantigens or their frequency, and immune signatures, cannot be determined. Evolution of the neoantigen landscape might well be expected, and has indeed been observed over the course of TIL therapy or immune checkpoint blockade.25,26 Evaluations based on sequential sampling are likely to result in better prognostic biomarkers. Second, we were unable to detect mutated gene expression in this study, because microarray data cannot discriminate wild-type from mutant alleles. Mouse studies have not only demonstrated the elimination of sub-clones expressing neoantigens (natural selection),21 but also suppression of neoantigen expression by epigenetic gene silencing.27 Whether this occurs in humans is not known. Third, we only considered neoantigens derived from missense mutations, but conceivably, there are other sources such as frame-shift mutations that have been reported as targets of immune responses.18
Currently, mutation burden and neoantigen load are proposed as prognostic biomarkers in some cancers with high mutation rates, such as melanoma, non-small cell lung cancer and MMR-deficient tumors under immune checkpoint blockade therapies. We previously showed that a combined score of neoantigen load and HLA expression was a predictor of overall survival in ccRCC patients with medium or low mutation burdens under conventional therapy. Here we have demonstrated that neoAg freq in stage I and II OCCC is an independent prognostic factor for clinical outcome under standard therapy. Therefore, it should now be investigated whether neoAg freq is informative as a possible biomarker in other types of cancer, for certain tumor stages under standard therapy or immunomodulatory-based treatment.
Materials and methods
Sample description and preparation
OCCC tumors and peripheral blood from the same patients were collected by the University of Tokyo Hospital and Saitama Medical University International Medical Center following the approval of the institutional review board and patients´ written informed consent (ID: G3531 and 13–098, respectively). Genomic DNA and total RNA were extracted from frozen tumor samples after cryostat sectioning, using DNA and AllPrep DNA/RNA Mini Kits (Qiagen, Hilden, Germany). Genomic DNA was isolated from matched peripheral blood samples using QIAamp DNA Mini Kits (Qiagen).
Whole-exome sequencing, read mapping and detection of somatic mutations
Paired tumor and blood genomic DNA libraries were constructed according to the protocol provided with the KAPA Hyper Prep Kit (Kapa Biosystems). Whole-exome capture was performed with the SureSelect Human All Exon kit v4 and v5 (Agilent Technologies) following the manufacturer's protocols. We sequenced exome capture libraries on the HiSeq 2000 platform according to the manufacturer's instructions, and 2 × 100-bp paired-end reads were generated. Image analysis and base calling were performed using the Illumina pipeline with default settings.28 Exome reads were independently mapped to the human genome (GRCh37/hg19) using Burrows-Wheeler Aligner (BWA) and Novoalign software. Reads with a minimal editing distance to the reference genome were taken to represent optimal alignments. Bam files were then locally realigned with SRMA. Normal-tumor pair bam files were processed using an in-house genotyper karkinos (https://sourceforge.net/projects/karkinos/). OxoG artifacts were removed by D-ToxoG program.29 RefSeq gene annotation was performed by ANNOVAR.30
Microarray analysis
OCCC tissues were analyzed on HG-U133 Plus 2.0 arrays (Affymetrix) containing 54,675 probe sets for human genes. Microarray analysis was performed as described previously.31,32 RNA targets were prepared according to the manufacturer's protocol using GeneChip 3′ IVT Express Kits (Affymetrix). Total RNA (100 ng) was converted into double-stranded cDNA templates for transcription. In vitro transcription synthesized amplified RNA (aRNA) and incorporated a biotin-conjugated nucleotide. Ten micrograms of fragmented cRNA were hybridized (45°C, 16 hours). Hybridization was controlled using GeneChip Eukaryotic Hybridization Control Kits (Affymetrix). Washing and staining was performed in a Fluidics Station 450 (Affymetrix) using the protocol EukGE-WS2v5. Scanning was performed in an Affymetrix GeneChip Scanner 3000 7G. The initial gene expression analysis data files (CEL files) were generated using Affymetrix GeneChip Operating Software. CEL files were processed using the standard MAS5 normalization technique.33 The signals were normalized to align at the 75th percentile, and then converted to log2 ratio against the mean in each probe set. There were 54,675 probes initially. We removed unreliable probes flagged as ‘Absent’ in 90% of samples, and then filtered out log2 ratio values not varying (between -2 and 2) among all samples. We applied hierarchical clustering (standard correlation, UPGMA) on the data set. All data analysis was performed using the Subio Platform and Basic Plug-in v1.19 (Subio Inc., Amami, Japan).
Gene Set Enrichment Analysis (GSEA) was performed to determine whether immune cells were enriched in the tumor microenvironment.34 GSEA was performed with GSEA v2.2.1 software with default parameters. The immune metagene list consisted of 31 immune cell-related gene sets provided by Angelova et al. was used and the association with each immune cell gene set was represented by a normalized enrichment score (NES).35 An immune cell type was considered significantly enriched when familywise error rate (FWER, p-value) <0.05.
HLA typing and MHC class I epitope binding prediction from whole exome sequencing and expression array data
HLA-typing was performed in the 74 patients from whom whole exome sequencing data were available. HLA types were assigned from exome sequencing data of normal tissues or peripheral blood mononuclear cells using HLA typing software (Omixon target HLA). Mutated peptides derived from missense mutations from exome sequencing data of tumor were used for MHC class I binding prediction as previously reported.16,36 Briefly, long peptides containing the predicted mutation or of wild-type were assessed using the Immune Epitope Database and Analysis Resource (http://www.iedb.org/) offline; 9- and 10-mer peptides were selected, each predicted to bind to a specific HLA allele for each patient. Predicted binding scores were ranked by MHC class I epitope binding algorithms (NetMHCpan v2.8). Mutated peptides which had an IC50 value below 500 nM were regarded as candidate neoepitopes.
Statistical analysis
Overall survival (OS) times were calculated as the number of days from surgery to death, or the last time the patient was known to be alive. Progression-free survival (PFS) times were calculated as the time elapsed between surgery and tumor progression or death from any cause. They were plotted as Kaplan-Meier survival curves. The log rank test was used to examine the significance of differences in survival between groups. The impact of various factors on OS and PFS was calculated by the univariate and multi-variate Cox proportional hazards regression model using JMP11.2.0 (SAS institute, Cary, NC). Comparison of results was by an unpaired, 2-tailed Student t test using GraphPad Prism 5 (GraphPad Software, Inc.). A value of p < 0.05 was considered significant. Univariate and multivariate survival analyses were performed using the Cox proportional hazards regression model.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The work was supported by a Grant-in-Aid for P-DIRECT and P-CREATE by Japan Agency for Medical Research and Development (Kosei Hasegawa) under grand numbers 11114014 and 16cm0106502h0001; a Grant-in-Aid for Scientific Research of the Ministry of Education, Culture, Sports, Science and Technology of Japan (Hirokazu Matsushita, Kosei Hasegawa, Kazuhiro Kakimi) under grant numbers 16K07162, 16K11152, and 16H04708.
References
- 1.del Carmen MG, Birrer M, Schorge JO. Clear cell carcinoma of the ovary: a review of the literature. Gynecol Oncol 2012; 126:481-90; PMID:22525820; https://doi.org/ 10.1016/j.ygyno.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 2.Sugiyama T, Kamura T, Kigawa J, Terakawa N, Kikuchi Y, Kita T, Suzuki M, Sato I, Taguchi K. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer 2000; 88:2584-9; PMID:10861437; https://doi.org/ 10.1002/1097-0142(20000601)88:11%3c2584::AID-CNCR22%3e3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- 3.Mabuchi S, Sugiyama T, Kimura T. Clear cell carcinoma of the ovary: molecular insights and future therapeutic perspectives. J Gynecol Oncol 2016; 27:e31; PMID:27029752; https://doi.org/ 10.3802/jgo.2016.27.e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anglesio MS, Carey MS, Kobel M, Mackay H, Huntsman DG, Vancouver Ovarian Clear Cell Symposium S . Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol 2011; 121:407-15; PMID:21276610; https://doi.org/ 10.1016/j.ygyno.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 5.Hamanishi J, Mandai M, Ikeda T, Minami M, Kawaguchi A, Murayama T, Kanai M, Mori Y, Matsumoto S, Chikuma S, et al.. Safety and antitumor activity of Anti-PD-1 antibody, Nivolumab, in patients with Platinum-Resistant Ovarian cancer. J Clin Oncol 2015; 33:4015-22; PMID:26351349; https://doi.org/ 10.1200/JCO.2015.62.3397 [DOI] [PubMed] [Google Scholar]
- 6.Disis ML, Patel MR, Pant S, Infante JR, Lockhart AC, Kelly K, Beck JT, Gordon MS, Weiss GJ, Ejadi S, et al.. Avelumab (MSB0010718C), an anti-PD-L1antibody, in patients with previously treated, recurrent or refractory ovarian cancer: A phase Ib, open-label expansion trial. J Clin Oncol 2015; 33:(suppl; abstr 5509); PMID:25918278; https://doi.org/ 10.1200/jco.2015.33.15_suppl.550925918278 [DOI] [Google Scholar]
- 7.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol 2012; 124:192-8; PMID:22040834; https://doi.org/ 10.1016/j.ygyno.2011.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava PK. Neoepitopes of Cancers: Looking Back, Looking Ahead. Cancer Immunol Res 2015; 3:969-77; PMID:26342008; https://doi.org/ 10.1158/2326-6066.CIR-15-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348:69-74; PMID:25838375; https://doi.org/ 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 10.Schumacher TN, Hacohen N. Neoantigens encoded in the cancer genome. Curr Opin Immunol 2016; 41:98-103; PMID:27518850; https://doi.org/ 10.1016/j.coi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 11.Ward JP, Gubin MM, Schreiber RD. The role of neoantigens in naturally occurring and therapeutically induced immune responses to cancer. Adv Immunol 2016; 130:25-74; PMID:26922999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al.. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371:2189-99; PMID:25409260; https://doi.org/ 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Geukes Foppen MH, Goldinger SM, et al.. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015; 350:207-11; PMID:26359337; https://doi.org/ 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al.. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348:124-8; PMID:25765070; https://doi.org/ 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al.. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372:2509-20; PMID:26028255; https://doi.org/ 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita H, Sato Y, Karasaki T, Nakagawa T, Kume H, Ogawa S, Homma Y, Kakimi K. Neoantigen load, antigen presentation machinery, and immune signatures determine prognosis in clear cell renal cell carcinoma. Cancer Immunol Res 2016; 4:463-71; PMID:26980598; https://doi.org/ 10.1158/2326-6066.CIR-15-0225 [DOI] [PubMed] [Google Scholar]
- 17.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015; 160:48-61; PMID:25594174; https://doi.org/ 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, et al.. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016; 44:698-711; PMID:26982367; https://doi.org/ 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 19.Itamochi H, Kigawa J, Terakawa N. Mechanisms of chemoresistance and poor prognosis in ovarian clear cell carcinoma. Cancer Sci 2008; 99:653-8; PMID:18377417; https://doi.org/ 10.1111/j.1349-7006.2008.00747.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331:1565-70; PMID:21436444; https://doi.org/ 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 21.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al.. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012; 482:400-4; PMID:22318521; https://doi.org/ 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol 2002; 3:999-1005; PMID:12407407; https://doi.org/ 10.1038/ni1102-999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 2006; 6:836-48; PMID:17063185; https://doi.org/ 10.1038/nri1961 [DOI] [PubMed] [Google Scholar]
- 24.Fesnak AD, June CH, Levine BL. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Cancer 2016; 16:566-81; PMID:27550819; https://doi.org/ 10.1038/nrc.2016.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verdegaal EM, de Miranda NF, Visser M, Harryvan T, van Buuren MM, Andersen RS, Hadrup SR, van der Minne CE, Schotte R, Spits H, et al.. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 2016; 536:91-5; PMID:27350335; https://doi.org/ 10.1038/nature18945 [DOI] [PubMed] [Google Scholar]
- 26.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, et al.. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 2017; 7:264-276; PMID:28031159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 2012; 482:405-9; PMID:22318517; https://doi.org/ 10.1038/nature10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, et al.. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014; 46:583-7; PMID:24816255; https://doi.org/ 10.1038/ng.2984 [DOI] [PubMed] [Google Scholar]
- 29.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, Fostel JL, Friedrich DC, Perrin D, Dionne D, et al.. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 2013; 41:e67; PMID:23303777; https://doi.org/ 10.1093/nar/gks1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38:e164; PMID:20601685; https://doi.org/ 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara T, Hiramatsu M, Isagawa T, Ninomiya H, Inamura K, Ishikawa S, Ushijima M, Matsuura M, Jones MH, Shimane M, et al.. ASCL1-coexpression profiling but not single gene expression profiling defines lung adenocarcinomas of neuroendocrine nature with poor prognosis. Lung Cancer 2012; 75:119-25; PMID:21737174; https://doi.org/ 10.1016/j.lungcan.2011.05.028 [DOI] [PubMed] [Google Scholar]
- 32.Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S, Yamamoto S, Asada K, Sone K, Kurikawa R, Makii C, et al.. Integrated copy number and expression analysis identifies profiles of whole-arm chromosomal alterations and subgroups with favorable outcome in ovarian clear cell carcinomas. PLoS One 2015; 10:e0128066; PMID:26043110; https://doi.org/ 10.1371/journal.pone.0128066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim WK, Wang K, Lefebvre C, Califano A. Comparative analysis of microarray normalization procedures: effects on reverse engineering gene networks. Bioinformatics 2007; 23:i282-8; PMID:17646307; https://doi.org/ 10.1093/bioinformatics/btm201 [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545-50; https://doi.org/ 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, et al.. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015; 16:64; https://doi.org/ 10.1186/s13059-015-0620-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karasaki T, Nagayama K, Kawashima M, Hiyama N, Murayama T, Kuwano H, Nitadori J, Anraku M, Sato M, Miyai M, et al.. Identification of Individual Cancer-Specific Somatic Mutations for Neoantigen-Based Immunotherapy of Lung Cancer. J Thorac Oncol 2016; 11:324-33; https://doi.org/ 10.1016/j.jtho.2015.11.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.