Abstract
Background
While limited data suggest that the fecal microbiota in healthy people are stable over time, the intra-individual variability of the fecal microbiota in constipated patients is unknown.
Methods
This study evaluated the intra-individual reproducibility of fecal microbiota analysed with 16S rRNA gene sequencing in two stool samples collected without and after a laxative, respectively, in 25 healthy people and 25 constipated women. Participants completed a food record for 3 days before the stool collection. Colonic transit was measured with scintigraphy.
Key Results
The constipated patients were older (48±15 versus 39±10 years, P=.02) than healthy participants but had a similar BMI. The total daily caloric intake was less (P=.005) in constipated (1265±350 kcal) than healthy participants (1597±402 kcal). Fourteen patients but only 2 controls (P<.005), had delayed colonic transit. For most measures of alpha (e.g., Observed OTU number, Shannon index) and beta diversity (e.g., Bray-Curtis dissimilarity, UniFrac, phyla level abundance), the ICCs between 2 stool samples were high, indicating moderate or strong agreement, and similar in healthy people and constipated patients. The ICC for the weighted UniFrac distance, which is weighted by abundance, was lower than its unweighted counterpart, indicating that the unweighted measure is more robust and reproducible.
Conclusions & Inferences
The intra-individual reproducibility of fecal microbiota in constipated patients is high and comparable to healthy participants. For most purposes, evaluating the fecal microbiota in a single stool sample should generally suffice in adequately powered studies of healthy and constipated patients.
Keywords: constipation, breath methane, irritable bowel syndrome, lactulose, methane, microbiome, microbiota, reproducibility, transit
Abbreviated abstract
The intra-individual reproducibility of fecal microbiota in constipated patients is high and comparable to healthy participants. For most purposes, evaluating the fecal microbiota in a single stool sample should generally suffice in adequately powered studies of healthy and constipated patients.
Alterations in the gut microbiome have been associated with or implicated in the pathophysiology of several gastrointestinal and liver diseases such as antibiotic-associated diarrhea, chronic constipation, irritable bowel syndrome (IBS), inflammatory bowel disease, obesity, colorectal cancer, and liver inflammation.1–5 While it has been suggested that each individual has a unique gut microbiota profile,6 the evidence suggesting that the fecal microbiota profile is stable within individuals over time is limited and mixed. For example, in 9 healthy participants studied twice over 3 months, the inter-individual variability exceeded the intra-individual variability.7 In another study, fecal samples evaluated with the low-error amplicon sequencing (LEA-Seq) technique were stable in 37 healthy adults sampled 2 to 13 times up to 296 weeks apart.8
By contrast, the fecal microbiota profile varied considerably over 6 and 15 months in 2 healthy people.9 Among 43 healthy individuals, single nucleotide polymorphism variations, but not the bacterial species in 88 fecal metagenomes, were stable over 37–378 days.10 Longitudinal sampling in 85 healthy adults at weekly intervals over 3 months disclosed considerable temporal variability.11 Similar to the composition of the individual microbiome, this too is personalized. Data from the Human Microbiome Project indicate that only approximately 30% of samples, as defined by taxa abundance-based metagenomic ‘codes’ and a measure of beta diversity, were stable for up to 450 days.6 Conceivably, temporal variations in the microbiota profile may be related not only to the disease state but also to alterations in diet, drugs, colonic transit, body mass index (BMI), and antibiotic use.3, 12–15 Hence, longitudinal assessments may be useful to more rigorously characterize fecal microbiota profiles in disease states.16 In IBS, particularly in diarrhea-predominant IBS, the fecal microbiota evaluated with older methods, not 16S rRNA techniques, were unstable over time.17–19 Therefore, it is necessary to evaluate the intra-individual stability of the fecal microbiota with 16S rRNA techniques in IBS and chronic constipation. Indeed, because there is an association between stool consistency and the composition of microbiota,20 it is conceivable that alterations in the fecal microbiota at least partly explain the temporal fluctuation in symptoms, most frequently from constipation- or diarrhea-predominant IBS to mixed type IBS or vice versa.21
In view of this, the objectives of this study were to (i) compare the intra-individual variability in human fecal microbiota over 2 samples taken one week apart, in healthy people and constipated patients, and separately, in people with normal or slow colonic transit and (ii) assess the reproducibility of the association between fecal microbiota and diet and physiological parameters (e.g., colonic transit and breath methane production). Our hypotheses were that (i) assessments of fecal microbiota in healthy people, patients with constipation, normal, and slow colonic transit are highly reproducible within individuals; (ii) the intra-individual variability of fecal microbiota in patients with constipation, and separately, with normal or slow colonic transit is not significantly different to that in healthy people; and (iii) the associations between the fecal microbiota and physiologic parameters (e.g., breath methane production and colonic transit) are also reproducible.
MATERIALS AND METHODS
Study Design and Participants
From February 2013 through April 2014, 25 female patients with chronic constipation and 25 healthy women, all non-smokers and aged between 18–80 years, consented to participate in this study that had been approved by the Institutional Review Board at Mayo Clinic. Patients had Rome III symptom criteria for a lower functional gastrointestinal disorder with significant constipation 22. Twenty of 25 patients reported having symptoms for at least 4 years before the study. Twenty-four of the 25 patients lived within 350 miles of Mayo Clinic, Rochester, MN; 21 of these lived within 150 miles. Neither the healthy participants nor the constipated patients had clinical evidence of significant systemic (e.g., cardiovascular) disease that could potentially interfere with the objectives of the study and/or pose safety concerns; prior gastric, intestinal, or colonic resection; inflammatory bowel disease; gastrointestinal cancer; antibiotic or probiotic use within 3 months prior to or during the study.
Assessment of Dietary Intake
Before starting study procedures, a registered dietitian advised participants to maintain a stable diet for 1 week before and throughout the study; and to follow a low fiber diet and avoid dairy products, high fructose corn syrup, fruits, fruit juices, honey, "sugar-free" candies, gums or products containing sorbitol, and live or active cultured-containing foods for 24 hours prior to the lactulose breath test. Participants were instructed to complete a food record for 3 days before the stool collection. Nutrition analysis of the food records was performed using the ESHA Food Processor software (Version 10.14, ESHA Research, Salem, OR). All study participants were instructed to not use pre or probiotics during the study. Only 1 patient was on a probiotic of doubtful efficacy that was discontinued 1 week before the study.
Scintigraphic Assessment of Colonic Transit
Colonic transit was measured with a standard, validated scintigraphic method 23, 24, using a methacrylate-coated, delayed-release capsule containing indium-111 (111In) adsorbed on activated charcoal particles. Colonic transit was summarized as the colonic geometric center (GC), which is the weighted average of counts in the different colonic regions at 24 (GC24) and 48 (GC48) hours. A higher GC reflects a faster colonic transit.
Stool Collection
Standardized instructions and stool kits were provided to patients for collecting stool samples, which were frozen and stored in a −80°C freezer. The first stool sample was collected without a laxative in all participants, with the exception of one patient who required an enema and another patient who required a laxative before providing the first sample. The second stool sample was collected within 7 days of the first sample; magneisum hydroxide (60 mL) and bisacodyl (5 mg) was administered to all participants the night before sample collection.
Sequencing and Analytical Methods
DNA was extracted from stool with a commercial kit (MoBio DNA extraction kit, Carlsbad, CA) following standard Human Microbiome Project guidelines.25 After extraction, total DNA was quantified using a Qubit assay kit (Life Technologies Corporation, NY, USA); in all cases, this was >100 ng/μL, with an average yield of 3799 (range 134 – 40,800) ng/μL for the first stool samples, and 2543 (range 87–11,600) ng/μL for the second. 16S-based sequencing was performed with an Illumina MiSeq sequencer (Illumina, San Diego, CA). Phylotype profiles of the microbiota from healthy and constipated populations were generated using deep rDNA hypervariable tag sequencing of the hypervariable V3–V5 region of the small subunit (SSU) rRNA gene, which has been validated for use with human microbiota and is the preferred technique by the Human Microbiome Project. With the longer reads from the MiSeq (300×300 paired end reads), sequencing included the V3–V5 regions, thereby optimizing the phylogenetic analysis 26; the 300 base pair reads ensured optimal phylogenetic identification. Barcoding of samples prior to sequencing yielded an average of 49,186 reads for the first stool sample (range 9438 – 161,117) and 48,237 for the second stool sample (range 17,465 – 164,321), ensuring detection of both dominant (core microbiota) and poorly-represented taxa (variable microbiota). Paired end reads were stitched, aligned, and classified using a custom pipeline (TORNADO v2.0) 27. Briefly, low base quality reads were either trimmed or discarded,28 and these reads were not classified as a bacteria kingdom29 or matched to the bacteria 16S rRNA secondary structure.30 To evaluate the microbial diversity and abundance, UPARSE was used for Operational Taxonomical Units (OTU) clustering,31 and FastTree was used for phylogeny.32 The 16S data were clustered into OTUs at 97% sequence similarity, and the taxonomy was assigned using the Ribosomal Database Project classifier and Greengenes database (v13.5)
Statistical analysis
The temporal stability of the stool microbiome over the two visits was quantified with an intraclass correlation coefficient (ICC) for established parameters of microbiota, i.e., (i) measures of alpha diversity (number of observed OTUs and Shannon index); (ii) measures of beta diversity (top principal coordinates analysis [PCoA] component for unweighted UniFrac, generalized UniFrac,33 weighted UniFrac, and Bray-Curtis distance); and (iii) Taxa relative abundance at the phylum and genus level (square-root transformed). The ICC is defined as
where represents the biological variability, i.e., individual-to-individual variability, and represents the temporal variability. The estimates of and could be obtained by fitting a linear mixed effects model
where yij is the measurment (e.g. alpha-diversity) of i th subject (i = 1 … n) and j th visit (j = 1, 2), bi is subject-level random effects and εij is the random error, which in our case represents the subject-specific temporal variation. Specifically, the ICC values were estimated using the R package ‘ICC’ along with the 95% confidence interval (CI) for each subgroup of samples, and were truncated at 0 in case of negative values. The strength of agreement was based on ICC values and defined as follows: ICC values of 0–0.2 indicate poor agreement, 0.3–0.4 indicates fair agreement, 0.5–0.6 indicates moderate agreement, 0.7–0.8 indicates strong agreement, and >0.8 indicates excellent agreement. While these parameters for agreement are widely used, they have not been specifically defined for evaluating microbiota. The assessment of statistical significance for the difference in ICC was based on the standard errors of the estimates using normal approximation. Boxplots of Spearman correlations between the OTU abundance of the two samples were also used to assess the OTU-level reproducibility. Further, differences in genus-level abundance between the first and second sample was assessed with a Wilcoxon signed rank test with false discovery rate control based on the Benjamini-Hochberg procedure to correct for multiple testing. The Wilcoxon signed rank test is a non-parametric test that does not assume a parametric distribution of the abundance data and thus is more robust than parametric tests.
To investigate the reproducibility of association between the fecal microbiota and variable clinical and demographic variables, PERMANOVA on the distance matrices was conducted using the first and second sample, respectively. The permutational multivariate analysis of variance (PERMANOVA) approach (‘adonis’ function in the R package ‘vegan’) was used to identify associations between microbial community profiles and variables of interest (e.g., constipation status, colonic transit). For all variables, statistical significance of the PERMANOVA results was assessed using 1,000 random microbial community/variable permutations.
To assess the impact of different ICC values, we estimated the sample size required to detect a weak, medium, and strong difference (i.e., effect size) in microbiota measurements with 80% power at different ICC values based on two sample t-test. All statistical analyses were performed in R-3.0.2 (R Development Core Teams, Vienna, Austria).
RESULTS
Patient Characteristics
The demographic and clinical characteristics of the study participants have been detailed previously.3 Salient findings are as follows (Table 1). Of the 25 patients, 13 had symptoms of functional constipation, 6 had IBS-C, and 6 had mixed IBS, predominantly constipation, by Rome III criteria. The constipated patients were older (48±15 versus 39±10 years, P=.02) than healthy participants but had a similar BMI. The total daily caloric intake was less (P=.005) in constipated (1265±350 kcal) than healthy participants (1597±402 kcal). Compared to patients, healthy participants also consumed more carbohydrate (P=.054), protein (P=.002), fat (P=.03), and fiber (P=.01) when expressed as an absolute amount, but not as a proportion of the total calorie intake. Gastric emptying at 2 hours, but not at 4 hours, was lower (P<.01) in constipated patients than in healthy participants. There was no difference in small intestinal transit between the two groups. Fourteen patients (9 with functional constipation, 3 with IBS-C, and 2 with mixed IBS) but only 2 controls (P<.005), had delayed colonic transit. Six healthy participants but no patients had rapid colonic transit. Colonic transit (GC24) was directly correlated with total calorie intake (r=0.31, P<.05) and total fiber intake (r=0.36, P<.05), and inversely correlated with age (r= −0.32, P<.05).
Table 1.
Summary of Patient Characteristics *
| Variable | Healthy (N=25) | Constipated (N=25) | P value |
|---|---|---|---|
| Age, y | 39±10 | 48±15 | .02 |
| BMI, kg/m2 | 26±4 | 25±4 | .13 |
| Total caloric intake, kcal | 1597±402 | 1265±350 | .005 |
| Carbohydrate, g | 188±54 | 155±62 | .054 |
| Protein, g | 75±19 | 60±26 | .002 |
| Fat, g | 60±26 | 46±15 | .03 |
| Fiber, g | 17±13 | 12±4 | .01 |
| Carbohydrate (% of total calories) | 47±9 | 49±12 | .24 |
| Protein (% of total calories) | 20±6 | 20±7 | .88 |
| Fat (% of total calories) | 33±6 | 32±6 | .85 |
| Breath methane (AUC, ppm* min) | 1488±2895 | 4100±6656 | .20 |
| Colonic transit, GC24 | 2.6±1.1 | 1.6±0.8 | .0006 |
| Colonic transit, GC48 | 3.9±0.9 | 2.8±1.0 | .001 |
Abbreviations: AUC, area under the curve; BMI, body mass index; GC24, geometric center of colonic transit at 24 hours; GC48, geometric center of colonic transit at 48 hours
All data presented as mean±SD
Reproducibility of Overall Composition of Fecal Microbiota
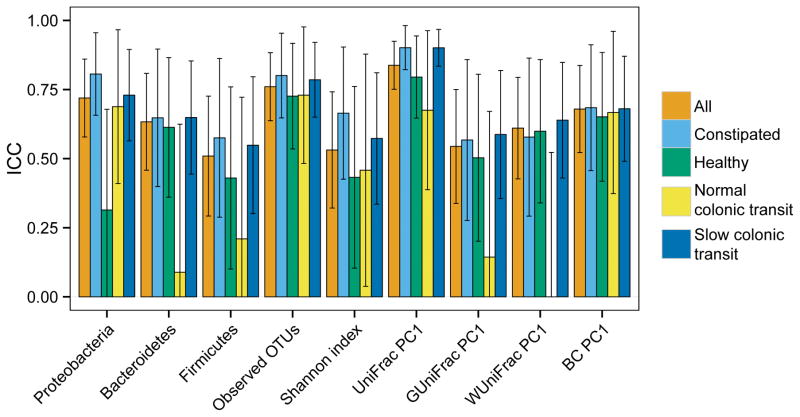
With some exceptions, the ICCs for most measures of alpha and beta diversity were high and similar in healthy participants and constipated patients (Table 2, Figure 1). The alpha diversity measures the species richness and evenness of microbiota within a sample. In all cohorts (i.e., healthy and constipated participants, normal and slow colonic transit), the ICC for number of observed OTUs, which is an unweighted measure that reflects richness, was numerically greater than the Shannon index, which also measures the relative abundance of different species. To be noted, the ICC for the Shannon index was numerically lower in healthy people (ICC 0.43, 95% CI 0.10–0.76) than in constipated patients (ICC 0.67, 95% CI 0.43–0.90).
Table 2.
Intraclass Correlation Coefficient (ICC) Between the Two Stool Samples for Different Measures of Microbiota Composition * †
| Subgroup | All-comers (N=50) | Healthy (N=25) | Constipated (N=25) | Normal colonic transit (N=19) | Slow colonic transit (N=31) |
|---|---|---|---|---|---|
| Alpha diversity | |||||
| Observed OTUs | 0.76 (0.64–0.88) | 0.73 (0.54–0.92) | 0.80 (0.65–0.95) | 0.73 (0.48–0.98) | 0.79 (0.65–0.92) |
| Shannon index | 0.53 (0.32–0.74) | 0.43 (0.10–0.76) | 0.67 (0.43–0.90) | 0.46 (0.04–0.88) | 0.57 (0.34–0.81) |
| Beta diversity | |||||
| BC dissimilarity | 0.68 (0.52–0.84) | 0.65 (0.42–0.88) | 0.68 (0.46–0.91) | 0.67 (0.37–0.96) | 0.68 (0.49–0.87) |
| Unweighted UniFrac | 0.84 (0.75–0.93) | 0.80 (0.65–0.94) | 0.90 (0.82–0.98) | 0.68 (0.39–0.96) | 0.90 (0.84–0.97) |
| Weighted UniFrac | 0.61(0.43–0.79) | 0.60 (0.34–0.86) | 0.58 (0.29–0.86) | 0.0 (0.0–0.52)‡ | 0.64 (0.43–0.85) |
| Generalized UniFrac | 0.54 (0.34–0.75) | 0.50 (0.20–0.81) | 0.57 (0.28–0.86) | 0.14 (0.0–0.67) | 0.59 (0.36–0.82) |
| Phylum level abundance | |||||
| Proteobacteria | 0.72 (0.58–0.86) | 0.31 (0.0–0.68) | 0.81 (0.66–0.96) | 0.69 (0.41–0.97) | 0.73 (0.56–0.90) |
| Bacteroidetes | 0.63 (0.46–0.81) | 0.61 (0.36–0.87) | 0.65(0.40–0.90) | 0.09 (0.0–0.62)‡ | 0.65 (0.44–0.85) |
| Firmicutes | 0.51 (0.29–0.73) | 0.43 (0.10–0.76) | 0.58 (0.29–0.86) | 0.21 (0.0–0.72)‡ | 0.55 (0.30–0.80) |
All values are intraclass correlation coefficients (ICC) (confidence intervals [CI])
Values in bold represent ICCs that are significantly different from each other (P<.05)
Lower bound of these CIs were truncated at zero. ICC=0 indicates no correlation between the two visits.
Figure 1.
Comparison of intraclass correlation coefficients (ICC) between two stool samples for different measures of fecal microbiota composition. The bars represent the absolute value of the ICC, i.e., the degree of similarity between the two stool samples for the corresponding measure microbiota composition. Most measures of alpha and beta diversity were similar across the 2 stool samples, and this was comparable in healthy participants and constipated patients.
In contrast to alpha diversity, the beta diversity reflects the diversity between samples. The ICC for the Bray-Curtis dissimilarity, which measures the species overlap between two populations, and for the unweighted UniFrac distance, which measures the shared OTU membership on a phylogenetic tree, was high in all cohorts (Table 2). By comparison, the ICC for the weighted UniFrac distance, which is a more quantitative assessment and weights the OTUs by their abundance, was generally lower than its unweighted counterpart, indicating that unweighted measures was more robust and reproducible. The ICC for weighted UniFrac was noticeably lower (P=.03) in participants with normal (ICC 0.0, 95% CI 0.0 – 0.52) as opposed to slow colonic transit. Similarly, the ICC for the Generalized UniFrac distance, which attenuates the weights on the dominant OTUs, was also lower (P=.1) in participants with normal compared to slow colonic transit.
Reproducibility of Taxonomic Composition of Fecal Microbiota
The ICCs for taxonomic abundances were moderate to high. At the phylum level, the ICC for Proteobacteria was lower (P=.01) in healthy participants than in constipated patients. By contrast, the ICCs for Bacteroidetes and Firmicutes were not significantly different between constipated patients and healthy participants, and between participants with normal and slow colonic transit. The taxa which accounted for these lower ICCs at the genus level were also identified (Table 3).
Table 3.
Comparison of Intraclass Correlation Coefficient (ICC) Between the Two Stool Samples for Abundance of Taxa at the Genus Level a
| Genus | All-comers | Healthy | Constipated | Normal colonic transit | Slow colonic transit |
|---|---|---|---|---|---|
| Bacteroidetes;Bacteroides | 0.73(0.59–0.87) | 0.69(0.46–0.91) | 0.75(0.58–0.93) | 0.06(0–0.60) | 0.78(0.64–0.92) |
| Bacteroidetes;Butyricimonas | 0.76(0.63–0.88) | 0.66(0.42–0.90) | 0.92(0.87–0.98) | 0.66(0.36–0.960) | 0.85(0.74–0.95) |
| Bacteroidetes;Odoribacter | 0.79(0.68–0.90) | 0.72(0.52–0.93) | 0.84(0.71–0.96) | 0.71(0.45–0.97) | 0.82(0.71–0.94) |
| Bacteroidetes;Parabacteroides | 0.79(0.68–0.90) | 0.75(0.56–0.94) | 0.80(0.65–0.94) | 0.90(0.80–1) | 0.72(0.55–0.89) |
| Bacteroidetes;unclassified | 0.77(0.65–0.89) | 0.57(0.28–0.86) | 0.89(0.81–0.97) | 0.50(0.10–0.90) | 0.84(0.74–0.95) |
| Firmicutes;Anaerofilum | 0.47(0.24–0.70) | 0.73(0.52–0.93) | 0.42(0.08–0.75) | 0.68(0.39–0.96) | 0.44(0.16–0.73) |
| Firmicutes;Anaerofustis | 0.61(0.43–0.79) | 0.74(0.54–0.93) | 0.32(0–0.68) | 0.59(0.24–0.93) | 0.62(0.40–0.84) |
| Firmicutes;Anaerostipes | 0.80(0.70–0.91) | 0.78(0.61–0.95) | 0.79(0.64–0.94) | 0.29(0–0.78) | 0.84(0.73–0.94) |
| Firmicutes;Anaerotruncus | 0.50(0.28–0.72) | 0.48(0.14–0.81) | 0.41(0.07–0.74) | 0.38(0–0.84) | 0.52(0.26–0.78) |
| Firmicutes;Blautia | 0.51(0.20–0.73) | 0.48(0.15–0.81) | 0.44(0.11–0.76) | 0.21(0–0.72) | 0.51(0.24–0.77) |
| Firmicutes;Butyrivibrio | 0.56(0.35–0.76) | 0.74(0.54–0.93) | 0.47(0.15–0.79) | 0.81(0.63–0.99) | 0.46(0.18–0.74) |
| Firmicutes;Christensenella | 0.93(0.89–0.97) | 0.94(0.89–0.99) | 0.92(0.86–0.98) | 0.87(0.75–1.0) | 0.96(0.93–0.99) |
| Firmicutes;Clostridium | 0.80(0.70–0.91) | 0.89(0.81–0.98) | 0.66(0.43–0.89) | 0.88(0.76–1.0) | 0.72(0.55–0.89) |
| Firmicutes;Coprobacillus | 0.57(0.38–0.77) | 0.73(0.53–0.93) | 0.33(0–0.69) | 0.39(0–0.84) | 0.62(0.40–0.84) |
| Firmicutes;Coprococcus | 0.50(0.28–0.72) | 0.50(0.18–0.82) | 0.35(0–0.70) | 0.27(0–0.77) | 0.51(0.25–0.77) |
| Firmicutes;Dialister | 0.69(0.54–0.84) | 0.86(0.75–0.97) | 0.59(0.33–0.85) | 0.92(0.83–1.0) | 0.64(0.42–0.85) |
| Firmicutes;Dorea | 0.44(0.20–0.68) | 0.44(0.09–0.79) | 0.44(0.12–0.77) | 0.35(0–0.82) | 0.44(0.15–0.73) |
| Firmicutes;Eubacterium | 0.68(0.53–0.84) | 0.62(0.36–0.89) | 0.77(0.61–0.94) | 0.59(0.24–0.94) | 0.70(0.52–0.88) |
| Firmicutes;Faecalibacterium | 0.57(0.37–0.77) | 0.60(0.33–0.88) | 0.47(0.15–0.78) | 0.39(0–0.84) | 0.61(0.39–0.83) |
| Firmicutes;Holdemania | 0.51(0.29–0.73) | 0.66(0.42–0.90) | 0.20(0–0.59) | 0.61(0.28–0.94) | 0.44(0.16–0.73) |
| Firmicutes;Lachnospira | 0.43(0.19–0.67) | 0.40(0.04–0.76) | 0.43(0.11–0.76) | 0.29(0–0.78) | 0.53(0.27–0.78) |
| Firmicutes;Lactococcus | 0.18(0–0.47) | 0.05(0–0.49) | 0.20(0–0.58) | 0.19(0–0.70) | 0.15(0–0.50) |
| Firmicutes;Moryella | 0.71(0.56–0.85) | 0.79(0.63–0.95) | 0.61(0.36–0.86) | 0.69(0.41–0.97) | 0.72(0.55–0.89) |
| Firmicutes;Oscillospira | 0.44(0.21–0.68) | 0.48(0.15–0.81) | 0.44(0.11–0.76) | 0.24(0–0.74) | 0.52(0.26–0.78) |
| Firmicutes;Phascolarctobacterium | 0.54(0.33–0.75) | 0.43(0.08–0.78) | 0.70(0.49–0.91) | 0.71(0.45–0.97) | 0.50(0.23–0.76) |
| Firmicutes;Roseburia | 0.66(0.49–0.82) | 0.64(0.39–0.89) | 0.61(0.35–0.86) | 0.66(0.37–0.96) | 0.64(0.43–0.85) |
| Firmicutes;Ruminococcus | 0.57(0.37–0.77) | 0.55(0.25–0.85) | 0.59(0.33–0.85) | 0.52(0.13–0.91) | 0.58(0.34–0.81) |
| Firmicutes;SMB53 | 0.70(0.56–0.85) | 0.67(0.43–0.91) | 0.73(0.54–0.92) | 0.50(0.10–0.90) | 0.73(0.57–0.90) |
| Firmicutes;Shuttleworthia | 0.69(0.54–0.84) | 0.44(0.10–0.79) | 0.82(0.68–0.95) | 0.58(0.22–0.93) | 0.71(0.54–0.89) |
| Firmicutes;Streptococcus | 0.36(0.10–0.62) | 0.39(0.03–0.76) | 0.29(0–0.66) | 0.10(0–0.62) | 0.39(0.09–0.69) |
| Firmicutes;Turicibacter | 0.44(0.20–0.68) | 0.60(0.33–0.88) | 0.26(0–0.63) | 0.74(0.49–0.98) | 0.24(0–0.58) |
| Firmicutes;Veillonella | 0.58(0.38–0.77) | 0.38(0.01–0.75) | 0.59(0.33–0.85) | 0.45(0.03–0.87) | 0.58(0.35–0.82) |
| Firmicutes;unclassified | 0.54(0.33–0.75) | 0.61(0.34–0.88) | 0.50(0.20–0.80) | 0.41(0–0.85) | 0.60(0.38–0.83) |
| Proteobacteria;Erwinia | 0.74(0.60–0.87) | 0.81(0.66–0.96) | 0.42(0.08–0.75) | 0.71(0.45–0.97) | 0.74(0.59–0.90) |
| Proteobacteria;Sutterella | 0.58(0.39–0.77) | 0.59(0.31–0.87) | 0.52(0.23–0.82) | 0.60(0.26–0.94) | 0.58(0.34–0.81) |
All values are intraclass correlation coefficients (ICC) (confidence intervals [CI]); values in bold represent ICCs that are significantly different from each other (P<.05)
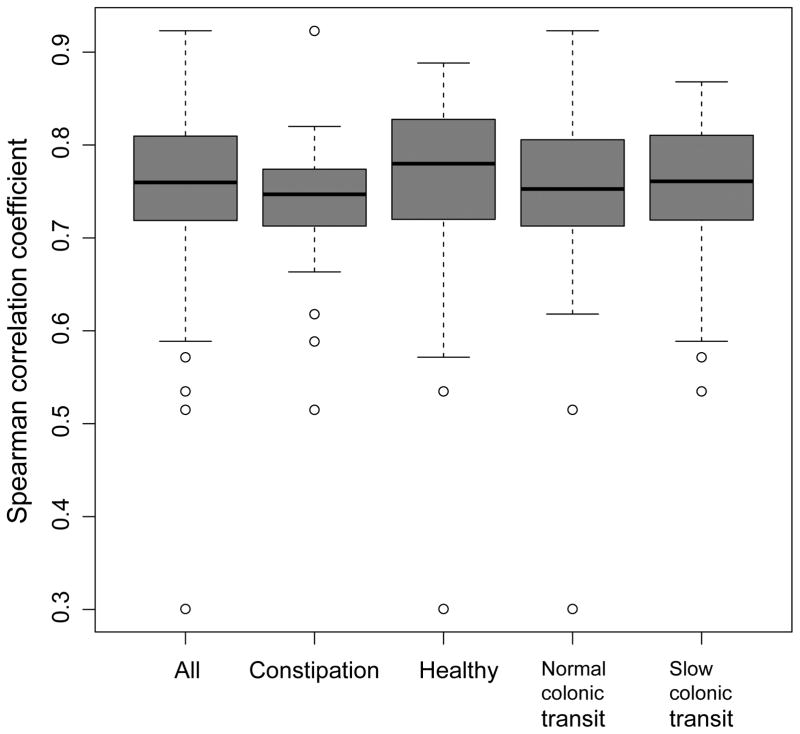
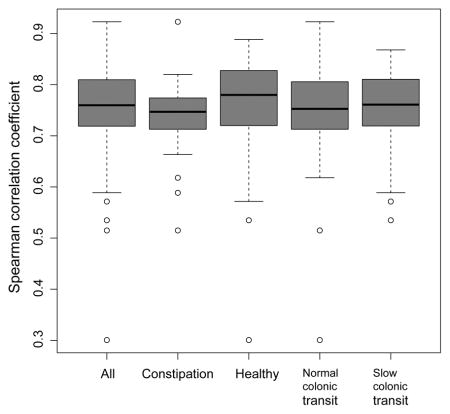
The OTU-level abundance profile in the first and second samples was significantly correlated in the entire cohort as well as in subgroups (i.e., healthy participants, constipated patients, and participants with normal or slow colonic transit) (Figure 2).
Figure 2.
Spearman correlations between observed operational taxonomic unit (OTU) abundances in the second stool sample compared to the first stool sample in healthy controls, constipated patients, normal colonic transit, and slow colonic transit.
We next tested for the effect of laxative treatment on the microbiota composition. Based on PERMANOVA, we did observe a significant effect (P=0.05, unweighted UniFrac), indicating that the temporal variance could be partially explained by the laxative treatment. Paired Wilcoxon tests suggested that at the genus level, Oscillospira was more abundant in the first sample (before laxative treatment), while Phascolarctobacterium, Veillonella, and Erwinia were more abundant in the second sample (after laxative treatment). These differences were significant before (P<.05) but not after adjusting for multiple testing using FDR control.
Reproducibility of Associations between Fecal Microbiota and Other Variables
We next studied the reproducibility of the associations we observed in our previous study using the first sample.3 We focused on the univariate association tests based on PERMANOVA. With a few exceptions, univariate associations between the fecal microbiota and other parameters were comparable in the first and second stool samples (Table 4). The fecal microbiota profile was associated with total calorie intake in the first but not in the second stool sample.
Table 4.
Associations Between Microbiota and Other Variables of Interest
| Variable | Stool-1 | Stool-2 | ||
|---|---|---|---|---|
| Unweighted UniFrac * | Weighted UniFrac * | Unweighted UniFrac * | Weighted UniFrac * | |
| Univariate analyses | ||||
| Age, y | .03 | .02 | .02 | .09 |
| BMI | .62 | .15 | .71 | .08 |
| Calories | .04 | .14 | .15 | .16 |
| Carbohydrate | .26 | .09 | .20 | .30 |
| Fat | .31 | .07 | .07 | .48 |
| Constipation status | .049 | .09 | .01 | .03 |
| Colonic transit | .22 | .008 | .05 | .04 |
| Breath methane | <.001 | .02 | <.001 | .12 |
| Multiple variable analyses | ||||
| Constipation status (adjusted for age, BMI, diet, and colonic transit) | .50 | .39 | .38 | .66 |
| Colonic transit (adjusted for age, BMI, diet, and constipation status) | .92 | .08 | .72 | .57 |
| Breath methane (age, BMI, constipation status, colonic transit, diet) | <.001 | .03 | .03 | .30 |
P values
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); UniFrac, a distance metric for comparing two microbial communities.
Sample Size Requirements
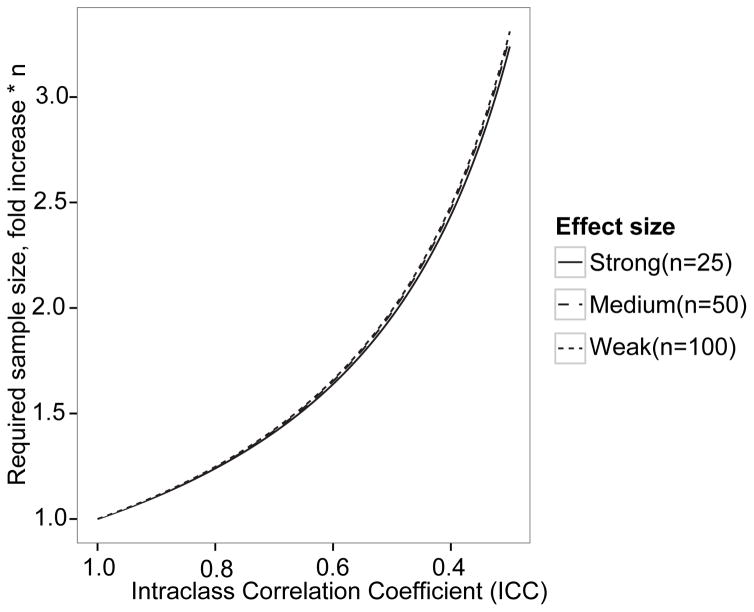
The sample size required to identify differences between fecal microbiota measurements in two groups with a two-sample t-test was first estimated assuming no measurement error (ICC=1.0). At 80% power and an alpha level of 0.05, 25, 50 and 100 individuals per group are required to detect hypothetically defined large, intermediate, and small differences (i.e., effect sizes) between groups. When the ICC for the reproducibility of microbiota parameters is incorporated into the sample size assessment, a greater sample size is required, as the ICC declines (Figure 3) to a comparable extent for all 3 hypothetical effect sizes. For example, to identify a strong difference, 25 samples per group are required when the ICC is 1.0. As the ICC (i.e., reproducibility) declines, the required sample size increases, for example to 63 samples per group for an ICC of 0.4.
Figure 3.
Effect of ICC on sample size requirements. As reproducibility decreases, i.e. ICC is lower, a larger sample size is required to detect all 3 effect sizes with 80% power at an α level of 0.05. For example, when the ICC is 0.6, a sample size of 1.5*25= x, 1.5*50= x, and 1.5*100= x is sufficient to detect strong, medium, and weak effects, respectively.
DISCUSSION
In this study, we used ICC to quantify the temporal stability of the stool microbiota based on several microbiota measurements. Besides the ICC measure, other measures that define the temporal variation are also possible. For example, Jalanka-Tuovinen et al. used the coefficient of variation (CoV) to study the temporal stability of the core gut microbial taxa in healthy subjects.34 The reason we chose the ICC measure over other measures is that ICC is directly related to statistical power, a key issue in the study design. Similar approaches have also been used to study the reproducibility and stability of various fecal sampling methods.35–39
Overall, the agreement between two fecal samples for all measures of alpha and beta diversity in healthy people and patients with chronic constipation was moderate or strong. The ICC for number of observed OTUs, which reflect the richness of microbiota, indicated strong agreement. For the Shannon index, which reflects the richness and evenness of fecal microbiota, the agreement between two stool samples was moderate. Similarly, for most measures of beta diversity, the ICCs suggested moderate or strong agreement. Extending previous studies,7 these observations suggest that for most diversity indices of microbiota composition, a single sample is cost-effective, not only in healthy people but also in constipated patients. However, if the sample size is the major constraint, statistical power could be improved moderately by using several technical replicates. The study also suggested that the composition of fecal microbiota was affected by a laxative.
For some measures of β diversity, i.e., the unweighted UniFrac which reflects the presence or absence rather than their abundance in OTUs, and the Bray-Curtis metric, which attaches an equal weight to all OTUs, the agreement was moderate or strong in all cohorts, including patients with normal colonic transit. Agreement for the weighted UniFrac and the generalized Unifrac distance was also moderate or strong in healthy people, constipated patients, and patients with slow colonic transit. However, the agreement for the weighted UniFrac and generalized Unifrac distances in patients with normal colonic transit was poor, possibly because of stochastic fluctuations related to a small sample size; only 19 participants had normal colonic transit. An alternative or additional explanation is that the weighted UniFrac and generalized Unifrac distances are influenced, respectively, predominantly and partly, by the most abundant lineages. Confirming this, the reproducibility of the phyla Bacteroidetes and Firmicutes was also lower in patients with normal colonic transit. These observations are consistent with the observation that bowel cleansing with a purgative prior to colonoscopy reduced the abundance of taxa belonging to Bacteroidetes and Firmicutes but not the overall diversity of microbiota.40 At the genus level, variations in Bacteroides, Lactococcus, and Streptococcus explained the low ICC in participants with normal colonic transit. Conceivably, this poor reproducibility may be partly explained by the effects of laxatives on colonic transit and stool consistency, which, to speculate, may be more pronounced in people with normal compared to slow colonic transit. The fecal microbiota, stool consistency, and colonic transit are known to be univariately associated with each other. Alterations in colonic transit affect stool consistency.41, 42 Stool consistency is associated with several measures of microbiota composition, i.e., species richness, enterotypes and taxonomic composition.43 Indeed, in a study of 3948 people, stool consistency had the largest effect size on the variation of fecal microbiota.44 Polyethylene glycol significantly increased the relative abundance of Bacteroides in ex-germ free mice colonized with human fecal microbiota.45
The univariate associations between the fecal microbiota and other variables were mostly comparable for the first and second stool samples. This provides further evidence for the intra-individual reproducibility of fecal microbiota and also supports the association between fecal microbiota and colonic transit as well as with breath methane production.3 However, minor differences were observed, which can again be explained by stochastic fluctuation related to a small sample size. However, alternative explanations are also possible. For example, in contrast to the first stool sample, total calorie intake was not significantly associated with the unweighted Unifrac distance, perhaps because the caloric intake was recorded for 3 days before the first stool sample and not thereafter. Acute changes in caloric intake are known to affect the stool microbiota.14, 46
These data will be useful for selecting the sample size in future studies.16, 47 The sample size depends on the effect size of the proposed intervention of disease state and the intra-subject variability.47 For a conservative ICC of approximately 0.5, which is lower than the observed ICC for most parameters in this study, the ideal sample size required to detect a given effect size will be approximately twice of that required for a perfect ICC (ICC=1.0). However, constrained by the absence of a standardized approach to report effect sizes or unknown effect sizes, pilot studies may often be required to estimate effect size.16
These measurements were obtained using state-of-the-art techniques from a carefully characterized cohort of healthy participants and constipated patients. However, the two stool samples were collected at 7-day intervals. Additional studies are necessary to ascertain if the fecal microbiota is stable over longer time periods, particularly since bowel symptoms may change over time, most frequently from constipation- or diarrhea-predominant IBS to mixed type or vice versa.21 Similar to a previous study,48 dietary caloric intake was lower in constipated than healthy participants. However, the reported average daily caloric intake in constipated participants was 1265 Kcal. We cannot exclude the possibility that this 3-day dietary assessment period was not representative of their regular caloric intake.
In conclusion, measurements of fecal microbial diversity are mostly reproducible in healthy people and constipated patients. A few measures of microbial composition were not reproducible in people with normal colonic transit.
KEY POINTS.
The intra-individual reproducibility of fecal microbiota in constipation is incompletely understood.
The agreement between two fecal samples for all measures of alpha and beta diversity in healthy people and patients with chronic constipation was moderate or strong.
For most purposes, evaluating the fecal microbiota in a single stool sample is generally sufficient in adequately powered studies.
Acknowledgments
Guarantor of the article: Adil E. Bharucha
Financial Support: Dr. Bharucha was supported in part by grant R01 DK078924 from the National Institutes of Health, US Department of Health and Human Services. This project was supported by the Center for Individualized Medicine at Mayo Clinic, Mayo Clinic-University of Illinois Strategic Alliance for Technology-Based Healthcare, and in part by grant number UL1 TR000135 from the National Center for Advancing Translational Sciences.
Competing Interests: The authors have no competing interests.
Specific Author contributions:
Study concept and design: Bharucha and Gaskins
Acquisition of data: Bharucha, O’Connor
Analysis and interpretation of data: Bharucha, Chen, Chia, Gaskins, O’Connor, Parthasarathy, Wolf.
Drafting of the manuscript: Bharucha, Chen, and Parthasarathy,
Critical revision of the manuscript for important intellectual content: All authors.
Statistical expertise: Chen
Obtained funding and study supervision: Bharucha and Gaskins.
References
- 1.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, Quraishi MN, Kinross J, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohman L, Tornblom H, Simren M. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 2015;12:36–49. doi: 10.1038/nrgastro.2014.200. [DOI] [PubMed] [Google Scholar]
- 3.Parthasarathy G, Chen J, Chen X, Chia N, O'Connor HM, Wolf PG, Gaskins HR, Bharucha AE. Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation. Gastroenterology. 2016;150:367–79. e1. doi: 10.1053/j.gastro.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sartor RB. Gut microbiota: Optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol. 2015 doi: 10.1038/nrgastro.2015.46. [DOI] [PubMed] [Google Scholar]
- 5.Simren M, Barbara G, Flint HJ, Spiegel BMR, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–76. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A. 2015;112:E2930–8. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vazquez-Baeza Y, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 13.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology - Human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 14.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. Conducting a microbiome study. Cell. 2014;158:250–62. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durban A, Abellan JJ, Jimenez-Hernandez N, Artacho A, Garrigues V, Ortiz V, Ponce J, Latorre A, et al. Instability of the faecal microbiota in diarrhoea-predominant irritable bowel syndrome. FEMS Microbiol Ecol. 2013;86:581–9. doi: 10.1111/1574-6941.12184. [DOI] [PubMed] [Google Scholar]
- 18.Matto J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–22. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Maukonen J, Satokari R, Matto J, Soderlund H, Mattila-Sandholm T, Saarela M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–33. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert JA, Alverdy J. Stool consistency as a major confounding factor affecting microbiota composition: an ignored variable? Gut. 2016;65:1–2. doi: 10.1136/gutjnl-2015-310043. [DOI] [PubMed] [Google Scholar]
- 21.Garrigues V, Mearin F, Badia X, Balboa A, Benavent J, Caballero A, Dominguez E, Diaz-Rubio M, et al. Change over time of bowel habit in irritable bowel syndrome: a prospective, observational, 1-year follow-up study (RITMO study) Aliment Pharmacol Ther. 2007;25:323–32. doi: 10.1111/j.1365-2036.2006.03197.x. [DOI] [PubMed] [Google Scholar]
- 22.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–91. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Camilleri M, Iturrino J, Bharucha AE, Burton D, Shin A, Jeong ID, Zinsmeister AR. Performance characteristics of scintigraphic measurement of gastric emptying of solids in healthy participants. Neurogastroenterol Motil. 2012;24:1076–e562. doi: 10.1111/j.1365-2982.2012.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deiteren A, Camilleri M, Bharucha AE, Burton D, McKinzie S, Rao AS, Zinsmeister AR. Performance characteristics of scintigraphic colon transit measurement in health and irritable bowel syndrome and relationship to bowel functions. Neurogastroenterol Motil. 2010;22:415–23. e95. doi: 10.1111/j.1365-2982.2009.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol. 2011;13:3000–9. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 27.Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, White B, Nelson H, Kocher JP, et al. IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries. PloS One. 2014;9:e114804. doi: 10.1371/journal.pone.0114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1. 0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28:2106–13. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalanka-Tuovinen J, Salonen A, Nikkila J, Immonen O, Kekkonen R, Lahti L, Palva A, de Vos WM. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE [Electronic Resource] 2011;6:e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flores R, Shi J, Yu G, Ma B, Ravel J, Goedert JJ, Sinha R. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33. doi: 10.1186/s40168-015-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loftfield E, Vogtmann E, Sampson JN, Moore SC, Nelson H, Knight R, Chia N, Sinha R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol Biomarkers Prev. 2016;25:1483–90. doi: 10.1158/1055-9965.EPI-16-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, Flores R, Sampson J, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer Epidemiol Biomarkers Prev. 2016;25:407–16. doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogtmann E, Chen J, Amir A, Shi J, Abnet CC, Nelson H, Knight R, Chia N, et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. Am J Epidemiol. 2017;185:115–23. doi: 10.1093/aje/kww177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogtmann E, Chen J, Kibriya MG, Chen Y, Islam T, Eunes M, Ahmed A, Naher J, et al. Comparison of Fecal Collection Methods for Microbiota Studies in Bangladesh. Appl Environ Microbiol. 2017;83:15. doi: 10.1128/AEM.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jalanka J, Salonen A, Salojarvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562–8. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 41.Degen LP, Phillips SF. How well does stool form reflect colonic transit? Gut. 1996;39:109–13. doi: 10.1136/gut.39.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saad RJ, Rao SS, Koch KL, Kuo B, Parkman HP, McCallum RW, Sitrin MD, Wilding GE, et al. Do stool form and frequency correlate with whole-gut and colonic transit? Results from a multicenter study in constipated individuals and healthy controls. Am J Gastroenterol. 2010;105:403–11. doi: 10.1038/ajg.2009.612. [DOI] [PubMed] [Google Scholar]
- 43.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 45.Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–77. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly BJ, Gross R, Bittinger K, Sherrill-Mix S, Lewis JD, Collman RG, Bushman FD, Li H. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31:2461–8. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandler RS, Jordan MC, Shelton BJ. Demographic and dietary determinants of constipation in the US population. Am J Public Health. 1990;80:185–9. doi: 10.2105/ajph.80.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]