Abstract
We compared three implementations of single‐shot arterial spin labeled (ASL) perfusion magnetic resonance imaging: two‐dimensional (2D) pulsed ASL (PASL), 2D pseudocontinuous ASL (PCASL), and background‐suppressed (BS) 3D PCASL obtained in a cohort of patients with mild cognitive impairment (MCI) and elderly controls. Study subjects also underwent 18F‐fluorodeoxyglucose positron emission tomography (18F‐FDG PET). While BS 3D PCASL showed the lowest (P < 0.001) gray matter–white matter cerebral blood flow (CBF) contrast ratio, it provided the highest (P < 0.001) temporal signal‐to‐noise ratio. Mean relative CBF estimated using the PCASL methods in posterior cingulate cortex (PCC), precuneus, and hippocampus showed hypoperfusion in the MCI cohort compared to the controls consistent with hypometabolism measured by 18F‐FDG PET. BS 3D PCASL demonstrated the highest discrimination between controls and patients with effect size comparable to that seen with 18F‐FDG PET. 2D PASL did not demonstrate group differentiation with relative CBF in any ROI, whereas 2D PCASL demonstrated significant differences only in PCC and hippocampus. Mean global CBF values did not differ across methods and were highly correlated; however, the correlations were significantly higher (P < 0.001) when either the same labeling (PCASL) or the same acquisition strategy (2D) was used as compared to when both the labeling and readout methods differed. In addition, there were differences in regional distribution of CBF between the three modalities, which can be attributed to differences in sequence parameters. These results demonstrate the superiority of ASL with PCASL and BS 3D readout as a biomarker for regional brain function changes in MCI. Hum Brain Mapp 38:5260–5273, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: pulsed ASL, pseudocontinuous ASL, background suppression, 3D spiral acquisition, positron emission tomography, mild cognitive impairment
INTRODUCTION
Arterial spin labeled (ASL) perfusion magnetic resonance imaging (MRI) uses magnetically labeled arterial blood water as an endogenous tracer to measure regional cerebral blood flow (CBF) [Alsop et al., 2015; Detre et al., 1992; Williams et al., 1992], which is tightly coupled to regional brain metabolism [Raichle, 1998]. A number of different strategies exist for magnetic labeling of arterial blood and for measuring the effects of labeling on brain signals, and combinations of these techniques can give rise to various acquisition protocols. Among the labeling approaches, pseudocontinuous ASL (PCASL) [Dai et al., 2008] is currently the recommended strategy [Alsop et al., 2015] because of its superior efficiency and compatibility with modern MR hardware. On the other hand, pulsed ASL (PASL) [Wong et al., 1998] has been available for almost 2 decades and is more widely employed due to its ease of implementation. ASL MRI has frequently been carried out using a two‐dimensional (2D) single‐shot echo planar imaging (EPI) readout because of its speed and sensitivity, though the effects of ASL can be measured with any method. Three‐dimensional (3D) readout schemes such as 3D GRASE [Gunther et al., 2005] and 3D spiral [Dai et al., 2008] can be optimally combined with background suppression (BS) techniques of static brain water [Fernandez‐Seara et al., 2008; Garcia et al., 2005; Maleki et al., 2012; Ye et al., 2000] to increase ASL temporal signal‐to‐noise ratio [Vidorreta et al., 2013; Ye et al., 2000] by reducing physiological noise [Wu et al., 2009].
ASL MRI has demonstrated sensitivity to regional alterations in brain function in patients with Alzheimer's disease (AD) [Alsop et al., 2008, 2000; Chao et al., 2009; Chen et al., 2011b; Dai et al., 2009; Du et al., 2006; Wang et al., 2013; Xu et al., 2007]. Significant hypoperfusion has been demonstrated in posterior cingulate cortex (PCC), precuneus, inferior parietal, and lateral prefrontal cortices and the effects are largely independent of underlying gray matter atrophy [Hu et al., 2010; Johnson et al., 2005; Mattsson et al., 2014; Wolk and Detre, 2012]. Hypoperfusion has also been reported in patients with amnestic mild cognitive impairment (MCI) [Johnson et al., 2005; Wang et al., 2013; Xekardaki et al., 2015], which is a heterogeneous clinical category consisting of individuals with prodromal AD, and a significant minority of subjects who have cognitive deficits from other etiologies and will not demonstrate pathological evidence of AD in the future. However, hypoperfusion in this population is less pronounced than in AD, and a few studies have also reported increased perfusion in the medial temporal lobes of patients with MCI and early AD [Alsop et al., 2008; Dai et al., 2009]. Areas of hypoperfusion largely overlap with regions of hypometabolism reported for 18F‐fluorodeoxyglucose positron emission tomography (18F‐FDG PET) [Chen et al., 2011b; Verfaillie et al., 2015], leading to the notion that ASL MRI might provide a less invasive and less costly substitute for 18F‐FDG PET as a measure of regional brain function in AD.
Previous studies have compared various ASL implementations. PCASL was shown to have higher SNR than PASL and higher tagging efficiency than continuous ASL (CASL) [Wu et al., 2007]. It was also reported to have superior test–retest reliability than CASL and PASL [Chen et al., 2011a]. Vidorreta et al. [2013] compared 2D PCASL with 3D PCASL methods (GRASE and spiral acquisitions, with and without BS), demonstrating that BS 3D methods provide higher temporal and spatial SNR in healthy subjects. More recently, PCASL with 3D GRASE demonstrated better reproducibility than 2D EPI acquisition for scans acquired 4 weeks apart, though 2D EPI demonstrated better, although not statistically significant spatial correlation with 15O H2O PET CBF [Kilroy et al., 2014].
Comparisons have also been made between ASL with 18F‐FDG PET. Perfusion measured by 2D PCASL was found to correlate well with glucose metabolism measured by PET in a healthy cohort, albeit with significant regional variability [Cha et al., 2013]. In patients with AD, good agreement in hypoperfusion and hypometabolism measured by 2D PCASL and 18F‐FDG PET, respectively, was demonstrated [Chen et al., 2011b; Musiek et al., 2012]. High agreement was also reported between 18F‐FDG PET hypometabolism and BS PCASL with 3D spiral acquisition hypoperfusion patterns in patients with frontotemporal dementia and Alzheimer's disease compared to controls [Verfaillie et al., 2015].
In this study, we compared ASL MRI data acquired from patients with MCI and elderly control subjects using single‐shot PASL acquired with 2D EPI using parameters matched to the ASL data in the Alzheimer's Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu), PCASL with 2D EPI readout using parameters previously demonstrated to be sensitive to regional CBF changes in Alzheimer's disease [Chen et al., 2011b; Musiek et al., 2012; Xie et al., 2016; Zhang et al., 2012], and PCASL with BS 3D spiral imaging, from here on referred to as 2D PASL, 2D PCASL, and BS 3D PCASL, respectively. The goal of this study was to assess the effects of different labeling and acquisition strategies on individual subject data and the sensitivity of the methods in detecting group differences between MCI patients and controls. The methods were also compared to 18F‐FDG PET, which represents a more established method for distinguishing between control and patient groups based on regional neural activity.
MATERIALS AND METHODS
Cohort
A sample of 24 amnestic MCI patients and 22 controls were recruited from the Penn Memory Center specifically for this study. As part of their evaluation, each participant underwent an extensive evaluation, including medical history and physical examination, neurological history and examination, and psychometric assessment. All patients had at least the following neuropsychological measures: Mini‐Mental State Examination (MMSE) [Folstein et al., 1975] (reported in Table 1); Digit Span subtest of the Wechsler Adult Intelligence Scale III [Wechsler, 1987]; Category fluency (animals) [Spreen and Strauss, 1998]; Consortium to Establish a Registry for Alzheimer's Disease (CERAD) Word List Memory (WLM) test [Morris et al., 1989]; Trail Making Test (TMT) A and B [Reitan, 1958]; and a 30‐item version of the Boston Naming Test (BNT) [Kaplan et al., 1983]. Additionally, relevant blood work and brain imaging studies were evaluated. Almost all patients had structural imaging available, usually MRI, that was used as part of the clinical decision‐making, largely to rule out other causes of MCI. Clinical diagnosis was determined by review of the above data at a consensus conference attended by neurologists, psychiatrists, and neuropsychologists. The scans obtained as part of the study presented here were not used for making the clinical diagnosis.
Table 1.
Demographic and clinical characteristics of the study sample (values reported as mean ± standard deviation)
| Control | MCI | |
|---|---|---|
| Number | 22 | 24 |
| Age, years | 70.9 ± 7.0 | 74.0 ± 7.4 |
| Gender (F/M) | 17/5 | 7/17 |
| Education, years | 15.2 ± 3.1 | 16.9 ± 2.7 |
| MMSEa | 29.3 ± 1.3 | 27.4 ± 1.8 |
Statistically significant difference, P < 0.001.
The diagnosis of amnestic MCI was made following the criteria outlined by Petersen and others [Petersen, 2004; Petersen et al., 2009; Winblad et al., 2004]. Patients had to have a memory problem, generally intact cognitive functioning and activities of daily living, objective evidence of memory impairment on cognitive testing, and not qualify for a diagnosis of dementia. There was no strict cutoff for the degree of memory impairment as clinical judgment accounting for the premorbid status of the patient and performance on other cognitive tests weighs into decisions of objective impairment [Petersen, 2004]. However, in general, these patients performed around 1.5 standard deviations (SDs) below age‐adjusted means on verbal and/or nonverbal memory tests. Controls were defined by an absence of significant cognitive complaints, normal performance on age‐adjusted cognitive measures, and consensus conference designation as “normal.” Inclusion criteria included age between 50 and 85, >7 years of education, and English speaking at an early age. Participants were excluded if they had a history of clinical stroke, significant traumatic brain injury, alcohol or drug abuse/dependence, prior electroconvulsive therapy, and any significant disease or medical/psychiatric condition that was felt to impact neuropsychological performance.
The human subjects' research in this study was performed in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and the standards established by the Institutional Review Board and the National Institutes of Health. All subjects provided informed consent for this study.
MRI and PET Data Acquisition
18F‐FDG PET and MRI scans of 19 patients and 6 controls were obtained on the same day. One patient and one control had their MRI scan obtained 6 days after and 13 days before their PET scans, respectively. ASL MRI from the remaining 4 patients and 15 controls were obtained with mean ± SD intervals of 430.5 ± 36.7 days and 411 ± 41.3 days, respectively, after their PET scan.
MR Imaging and Processing
MR images were acquired on a 3 T Siemens Trio MRI scanner (Erlangen, Germany) equipped with a product 32‐channel head coil. We did not control for modulators of CBF such as coffee, smoking, or time of the day. This could be a potential confound, although is unlikely to impact comparisons across ASL methods as they were performed in the same scanning session. High‐resolution sagittal T1 images were collected using 3D magnetization‐prepared rapid gradient echo (MPRAGE) with the following parameters: repetition time (TR) = 1.9 s, echo time (TE) = 2.89 ms, inversion time = 900 ms, flip angle = 9°, bandwidth = 170 Hz/px, voxel size = 1 × 1 × 1 mm3, field‐of‐view (FOV) = 256 × 256 mm2, 176 slices. ASL data were collected with the three selected methods after instructing subjects to remain still to the best of their ability. Each ASL scan had an approximate duration of 6 min. Details of the ASL sequences are as follows.
2D PASL
PASL data were acquired following the ADNI protocol (http://adni.loni.usc.edu/methods/documents/mri-protocols/) using a FAIREST scheme [Lai et al., 2001] combined with QUIPSSII [Luh et al., 1999] with TI/TI1 = 1.9 s/700 ms for accurate bolus definition. A 2D EPI readout was employed with TR/TE = 3.4 s/12 ms, FOV = 256 × 256 mm2, matrix size = 64 × 64, in plane resolution = 4 × 4 mm2, bandwidth = 2368 Hz/px, phase partial Fourier = 6/8, EPI factor = 64. Twenty four slices of 4 mm‐thickness and a distance factor of 25% were acquired. Fifty‐two label–control pairs were acquired. Unlike in ADNI, a separate M0 image was not acquired, and hence the mean of the control images (images acquired with control labeling) was used as M0 image for CBF quantification.
2D PCASL
The PCASL labeling was implemented using a labeling time = 1.5 s, postlabeling delay (PLD) = 1.5s, and labeling plane offset = 9cm. Two‐dimensional EPI readout parameters for 2D PCASL were TR/TE = 4 s/18 ms, FOV = 220 × 220 mm2, matrix size = 64 × 64, in‐plane resolution = 3.4 × 3.4 mm2, bandwidth = 3004 Hz/px, EPI factor = 64. Eighteen slices with 6 mm thickness and a 20% distance factor were acquired. Forty‐five label–control pairs were acquired. Similar to 2D PASL data, the mean of the control images was used as M0 image.
BS 3D PCASL
The PCASL labeling was implemented using a labeling time = 1.5 s, PLD = 1.5 s, and labeling plane offset = 8 cm. 3D‐PCASL data were obtained with TR/TE = 4.5 s/11 ms, FOV = 220 × 220 mm2, matrix size = 64 × 64, voxel size = 3.4 × 3.4 × 5 mm3. Twenty‐six nominal partitions were acquired with a centric encoding scheme and slice Partial Fourier = 6/8, using a stack of 2‐interleaf spirals with maximum slew rate = 120 T/m/s and maximum gradient amplitude = 36 mT/m. The background suppression scheme consisted of a presaturation module at the beginning of each TR, followed by a slice‐selective C‐FOCI pulse [Ordidge et al., 1996] and two additional nonselective hyperbolic‐secant pulses played during the PLD time, with timings optimized to suppress the static tissue magnetization signal to 10% of its equilibrium value [Vidorreta et al., 2013]. Forty label–control pairs were acquired. A separate M0 image was obtained without any magnetization preparation for CBF quantification.
MR Image Processing
Image processing was performed using Statistical Parametric Mapping (SPM8, Wellcome Department of Imaging Neuroscience, London, UK), ASL toolbox [Wang et al., 2008] and custom MATLAB (The Mathworks Inc., Natick, MA) scripts. First, the raw time series volumes for each method were realigned to correct for head motion using the method proposed by Wang [2012]. Briefly, the method consists of estimating a 6‐parameter rigid‐body transformation for each volume to align it to the first volume of the series. Thereafter, the spurious motion component caused by systematic label/control switching is regressed out from the estimated parameters before applying the transformation to the volumes. Next, the mean control images were automatically coregistered to the high‐resolution T1 images using SPM8 with normalized mutual information objective function, and the registrations were manually checked and corrected using a boundary‐based approach [Greve and Fischl, 2009] in the case of errors. Misregistrations were mostly noted in the case of 3D PCASL (∼43%) and were much less common in 2D PCASL (∼20%). The PASL mean control images showed poor tissue contrast and unidentifiable boundaries, making manual modification challenging. Instead, results of the automated coregistration algorithm were used. The resulting transformations were then applied to the corresponding label and control images. In parallel, the structural images were segmented into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) tissue probability maps (TPMs) using the “New Segment” tool in SPM8. The GM and WM TPMs were subsequently resliced to the native space, smoothed by an isotropic Gaussian kernel with FWHM = 5 mm and then thresholded to 0.75 to construct binary masks to be used to extract mean CBF in GM and WM. In addition, to create a mask to extract whole‐brain CBF, the sum of smoothed gray matter and white matter TPMs was thresholded to 0.75. Finally, the Diffeomorphic Anatomical Registration using Exponentiated Lie algebra (DARTEL) method [Ashburner, 2007] in SPM8 was implemented to create a group‐specific template combining each subject's segmented gray and white matter TPMs. The created template was subsequently registered to the MNI152 space using a linear affine transformation. The two transformations were later used to map the CBF maps and gray matter TPM to the MNI space.
CBF quantification for each control–label pair was obtained using
and
for PASL and PCASL, respectively. In these equations, ΔM is the control–label difference, λ is the brain/blood partition coefficient, ω is the post labeling delay, T 1blood is the T 1 of blood, α is the labeling (tagging) efficiency, M 0 is the equilibrium magnetization of the brain, and τ is the labeling duration. Assumed parameter values followed the recommendations in Alsop et al. [2015]. In particular, λ = λmean = 0.9 ml/g (mean λ for gray and white matter), T 1blood = 1650 ms. For PASL, ω = 1.9 s, τ = 700 ms and α = 0.98 [Alsop et al., 2015], and M 0 was made equal to the mean of the control images. For the PCASL techniques, ω = τ = 1.5 s. The labeling efficiencies for 2D PCASL and BS 3D PCASL were assumed to be 0.85 [Alsop et al., 2015] and 0.72 [Vidorreta et al., 2013], respectively. Similar to PASL, M 0 was made equal to the mean of the control images for 2D PCASL. In the case of BS 3D PCASL, the separately acquired M 0 was coregistered to the mean control image. In each case, the M0 images were smoothed using a 5 mm isotropic kernel to avoid misregistration errors [Alsop et al., 2015] before being used in the CBF calculation. The resulting CBF time series data was then averaged over time to obtain the mean CBF map. A binary mask comprising of gray matter, white matter, and ventricular CSF was created and applied to the mean CBF map to set out‐of‐brain voxels to zero. The CBF maps and the GM TPMs were mapped to the MNI space using the DARTEL template and affine transformation. Images in MNI space were smoothed with a FWHM = 8mm isotropic Gaussian kernel to better account for errors due to voxel mismatching with normalization across subjects. The MNI‐normalized smoothed GM TPM was thresholded to 0.4 to create a gray matter mask and region of interest (ROI) analyses for each subject were performed only within these masks to restrict ROI values to voxels composed of predominantly gray matter.
To account for incomplete T1 recovery [Alsop et al., 2015] for the 2D acquisition strategies and T2*/T2 decay [Cavusoglu et al., 2009] in the case of 2D/3D acquisitions, respectively, extracted ROI CBFs were further multiplied by
for 2D acquisitions and
for 3D acquisitions. The goal of this is to correct for variable TR and TE across ASL sessions. Tissue specific parameters were chosen based on whether the ROIs were in GM or WM. We used 0.98/0.82 [Herscovitch and Raichle, 1985], T 1,GM/T 1,WM = 1331/832 ms, T 2,GM/T 2,WM/T 2,blood = 80/110/186 ms, T 2*,GM/ ,WM/ ,blood = 44.2/44.7/43.6 ms [Cavusoglu et al., 2009; Chen and Pike, 2009; Wansapura et al., 1999]. For global CBF measurements, average of the parameters for GM and WM were used.
PET Imaging and Processing
18F‐FDG PET/CT was acquired on a Philips Gemini TF PET/CT scanner (Amsterdam, Netherlands). Participants were instructed not to eat for at least 4 h prior to their PET scan. Blood glucose levels were tested and individuals with blood glucose level >180 mg/dl were excluded from the study. Participants then received an intravenous injection of 5.0 ± 0.5 mCi of 18F‐FDG as they rested quietly in a dimly lit room with their eyes open but with minimal sensory stimulation. Approximately 30 min after the injection, a 30‐min 3D emission scan was obtained (256 mm FOV, 128 × 128 matrix, 2 × 2 × 2 mm3 voxel size). Line‐of‐response row‐action maximum likelihood algorithm reconstruction using sharp setting was performed followed by CT attenuation correction.
PET images were coregistered to the T1 images using SPM8 with normalized mutual information objective function similar to the ASL data and the quality of the coregistration was manually verified. Thereafter, the images were normalized to the MNI space using the DARTEL template following a procedure similar to the mean CBF maps described above. This involves smoothing by an isotropic Gaussian kernel with FWHM = 8 mm and use of subject‐specific gray matter masks to restrict the analysis within the gray matter. Finally, SUVR maps were generated using whole cerebellum as reference tissue. Regional uptake was measured from SUVR maps using the same methods applied to the CBF maps.
Details of Statistical Analysis
Comparison of temporal signal‐to‐noise ratio (tSNR)
For each subject, the mean CBF within the whole‐brain mask was computed for each CBF volume of the time series. The tSNR was computed as the ratio of the mean and standard deviation of these values. The tSNR measures for each acquisition method and for each group were compared statistically using a two‐way mixed ANOVA with the within‐subject factor of “ASL modality” and the between‐subject factor of “diagnosis.” Post‐hoc comparisons were carried out using Bonferroni correction. The statistical analysis was performed using SPSS Statistics, version 23 (IBM Corp). To assess the effect of subject motion on tSNR, average translation and rotation was estimated from the parameters of the rigid‐body realignment of the image time series for each subject and Spearman's correlation coefficient of the quantities with tSNR were computed and reported. Motion in controls and patients was compared using a mixed ANOVA model with “ASL methods” and “type of motion” (translation or rotation) as repeated measures and “diagnosis” as between‐group variable.
Comparison of presence of outliers
While tSNR assesses the overall variability in the CBF time series, it does not indicate the number of outliers in the series that are markedly different from the remaining volumes and therefore likely dominated by artifacts. Hence, in addition to computing tSNR, we counted the number of outliers present in the CBF time series and compared the results across methods. A volume was considered an outlier if its mean CBF was 1.5 interquartiles below the first quartile or above the third quartile of the time series [Tukey, 1977]. The goal of this analysis was twofold: First, we wanted to assess if non‐BS techniques are more susceptible to have outlier label/control pairs compared to BS techniques. The second goal was to assess the rate of outliers in the BS 3D implementation, as that provides an indication of potential challenges in multishot 3D acquisitions, where there is less flexibility of discarding contaminated volumes. The number of outliers for each acquisition method was compared using Friedman test. Post hoc pairwise comparisons were conducted using Wilcoxon signed rank test.
Comparison of gray matter to white matter contrast ratio
The mean CBF within gray matter and white matter was extracted from the mean CBF maps in native space and their ratio was computed. Similar to tSNR, the contrasts for each method were also compared with a two‐way mixed ANOVA.
Comparison of global CBF obtained using each ASL method
As all ASL methods are designed to quantify the same physiological parameter (CBF), we compared the global CBF values obtained using each method. Mean CBF values for each method and diagnostic condition were reported and compared using a two‐way mixed ANOVA similar to tSNR. Bland–Altman plots were also shown for pairwise comparisons of the global CBFs obtained by the three methods. Additionally, pairwise correlations were computed and reported. Statistical tests for determining the significance of the difference in correlation coefficients were performed using the method described by Steiger [1980] and evaluated using the software developed by Lee and Preacher [2013]. The method converts each correlation coefficient into a z‐score using Fisher's r‐to‐z transformation, computes the asymptotic covariance of the estimates, and subsequently uses the quantities in an asymptotic z‐test. We also compared the regional distribution of CBF between the three modalities based on the control subjects, as prior studies—e.g., Lovblad et al. [2015]—showed that differences in labeling can lead to regional variability and difference in signal intensity.
Comparison of sensitivity to differentiate MCI patients from controls between the three ASL methods and PET
Standardized uptake value ratio (SUVR) for PET images were computed by dividing each voxel's SUV by the mean SUV within the cerebellum. In the case of ASL, cerebellum is often not in the area of coverage and, hence, the mean CBF within the cortical GM was used for computing the relative CBF. The mean relative CBF and mean SUVR within PCC, precuneus, and hippocampus were extracted for ASL and PET for each subject. These ROIs have most consistently been shown to be sensitive to AD [Wolk and Detre, 2012]. Statistical significances of group differences were evaluated using nonparametric Mann–Whitney U‐tests. Effect sizes were calculated by converting the Mann–Whitney U to a z‐score and then dividing by the square root of the total number of subjects.
RESULTS
Demographic characteristics of the study cohort are summarized in Table 1. There were no significant differences in age or education between MCI and control subjects. Figure 1 shows the mean CBF maps of a representative control and MCI patient obtained using the three ASL methods. For comparison, the SUVR maps of the same subjects are also shown.
Figure 1.
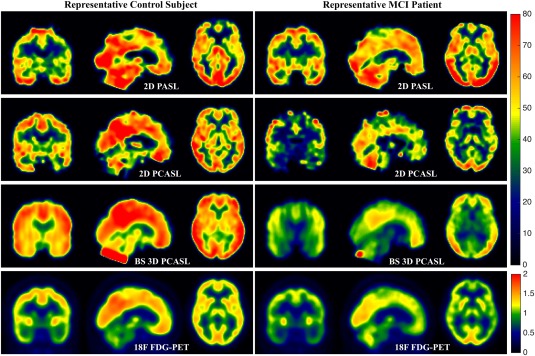
Triaxial view of mean CBF maps using 2D PASL (Row 1), 2D PCASL (Row 2), and BS 3D PCASL (Row 3) of a representative control subject on the left and MCI patient on the right. The bottom row shows the SUVR maps of the same subjects. [Color figure can be viewed at http://wileyonlinelibrary.com]
Comparison of Temporal Signal‐to‐Noise Ratio (tSNR)
TSNR values for the three ASL methods and for the two cohorts are reported in Table 2. BS 3D PCASL had an almost 2.5‐fold increase in tSNR as compared to the 2D methods. A two‐way mixed ANOVA demonstrated that there was a statistically significant main effect of method (F(1.43,62.79) = 152.4, P < 0.001, using Greenhouse–Geisser correction). Post‐hoc tests using Bonferroni correction for multiple comparisons showed that BS 3D PCASL had significantly higher (P < 0.001) tSNR compared to the two 2D methods, and 2D PCASL tSNR was also significantly higher (P = 0.022) compared to 2D PASL. There was also a significant main effect of diagnosis on tSNR (F(1,44) = 4.328, P = 0.043) with higher tSNR in control subjects than MCI for all the three methods. There was no significant interaction between method and diagnosis. Mean translations in 2D PASL, 2D PCASL, and BS 3D PCASL were all significantly anticorrelated with corresponding tSNRs, with Spearman's correlation coefficients of −0.42 (P = 0.004), −0.53 (P < 0.001), and −0.36 (P = 0.013), respectively. However, the mean rotation was only significantly correlated with tSNR in the case of 2D PCASL with Spearman's correlation coefficient of −0.40 (P = 0.006). Translations and rotations in patients were higher than in controls, but the differences did not reach statistical significance (P = 0.137).
Table 2.
TSNR, #Outlier CBF volumes/subject, GM–WM contrast ratio and global CBF for the control and MCI cohort
| 2D PASL | 2D PCASL | BS 3D PCASL | ||
|---|---|---|---|---|
| TSNR (mean ± SD) | Control | 2.28 ± 0.96 | 2.83 ± 1.56 | 6.44 ± 2.12 |
| MCI | 1.74 ± 0.70 | 2.05 ± 1.23 | 5.54 ± 2.14 | |
| Total | 2.00 ± 0.87 | 2.42 ± 1.44 | 5.97 ± 2.15 | |
| #Outliers/subject (mean ± SD (range)) | Control | 1.05 ± 1.29 (0–4) | 0.68 ± 0.72(0–2) | 0.14 ± 0.47(0–2) |
| MCI | 1.83 ± 1.66 (0–6) | 1.29 ± 1.12(0–4) | 0.67 ± 1.01(0–4) | |
| Total | 1.46 ± 1.53 (0–6) | 1.00 ± 0.99 (0–4) | 0.41 ± 0.83 (0–4) | |
| GM–WM contrast ratio (mean ± SD) | Control | 2.42 ± 0.72 | 2.29 ± 0.48 | 1.39 ± 0.13 |
| MCI | 2.41 ± 0.57 | 2.31 ± 0.46 | 1.32 ± 0.13 | |
| Total | 2.41 ± 0.64 | 2.30 ± 0.46 | 1.35 ± 0.14 | |
| Global CBF (mean ± SD) | Control | 36.51 ± 9.25 | 38.41 ± 8.77 | 39.69 ± 11.07 |
| MCI | 34.83 ± 7.62 | 33.28 ± 7.69 | 35.68 ± 10.32 | |
| Total | 35.63 ± 8.39 | 35.73 ± 8.53 | 37.60 ± 10.76 |
TSNR, temporal signal‐to‐noise ratio; GM, grey matter; WM, white matter; CBF, cerebral blood flow; MCI, mild cognitive impairment.
Comparison of Presence of Outliers
The mean ± SD and the range of detected outlier CBF volumes are reported in Table 2 for each method and diagnostic condition. There was a statistically significant difference in number of outliers, = 19.141, P < 0.001. Post‐hoc analyses with Wilcoxon signed rank test and Bonferroni correction demonstrated that the 2D PASL and 2D PCASL had significantly higher number of outliers compared to BS 3D PCASL (P = 0.004 and P = 0.029, respectively). Both the PCASL methods exhibited a similar range (0–4) of outliers, whereas the range was higher (0–6) in 2D PASL.
Comparison of Gray Matter to White Matter CBF Contrast Ratio
The GM–WM CBF ratios are shown in Table 2. The GM–WM CBF ratio was lowest in BS 3D PCASL almost certainly due to markedly increased blurring, which is evident in Figure 1. The blurring is the result of the T2 magnetization decay in time resulting in broadening of the point spread function in kz space. This blurring also reduces apparent CBF in basal ganglia and thalamus. A two‐way mixed ANOVA demonstrated a significant main effect of method (F(1.45,60.21) = 75.8, P < 0.001, with Greenhouse–Geisser correction). Post‐hoc tests showed that GM–WM contrast in 2D PCASL and 2D PASL was significantly higher than BS 3D PCASL (P < 0.001). There was no effect of diagnosis or any interaction between method and diagnosis.
Comparison of Global CBF Obtained Using Each ASL Method
The global CBF values from the three methods are listed in Table 2. There was no statistically significant difference in CBF between methods (P = 0.124), diagnosis (P = 0.194), or interaction between methods and diagnosis (P = 0.292). Variability in CBF derived from BS 3D PCASL was the highest across the cohorts. Figure 2 shows the Bland–Altman plots for pairwise comparisons of the global CBFs obtained with the three methods. The differences in CBF values obtained using 2D PASL and BS 3D PCASL had higher variance compared to the differences obtained using either the same labeling or same acquisition strategies. Mean CBF values for whole brain obtained using the three methods were also highly correlated. The correlation between the two PCASL methods (2D and 3D) was highest with Pearson's correlation coefficients of 0.79, P ≪ 0.0001. The correlation between the two 2D methods (PASL and PCASL) was also high, with a coefficient of 0.75, P ≪ 0.0001. The correlation between BS 3D PCASL and 2D PASL was considerably lower (0.49, P < 0.001). When the correlations were compared, both BS 3D PCASL vs 2D PCASL and 2D PCASL versus 2D PASL correlations were significantly higher compared to BS 3D PCASL versus 2D PASL (P < 0.001). Figure 3 shows the differences in relative CBFs in controls between the three modalities. 2D PASL had higher relative CBF in the cerebellum, occipital cortex and basal ganglia, and lower perfusion in the frontal lobe compared to the other two modalities. On the other hand, the distribution of relative CBF with BS 3D PCASL was opposite. The distribution of 2D PCASL was somewhere in between the two modalities.
Figure 2.
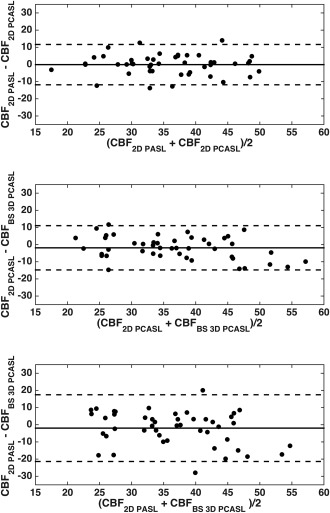
Bland–Altman plots comparing the global CBF measurements obtained using (top) 2D PASL and 2D PCASL, (middle) 2D PCASL and BS 3D PCASL, and (bottom) 2D PASL and BS 3D PCASL. The three lines in each case show the mean difference and the 95% confidence intervals.
Figure 3.
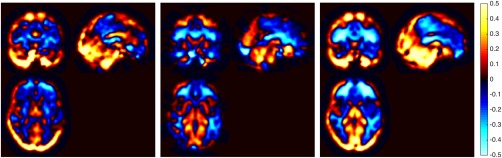
Difference in relative CBF in control subjects between (left) 2D PASL and 2D PCASL, (middle) 2D PCASL and BS 3D PCASL, and (right) 2D PASL and BS 3D PCASL. [Color figure can be viewed at http://wileyonlinelibrary.com]
Comparison of Sensitivity to Differentiate MCI Patients and Controls Between the Three ASL Methods and PET
Figure 4 shows the average CBF maps across the cohort separately for controls and MCI patients and their differences. The bottom row of the same figure shows the average SUVR in controls and MCI patients and their difference. Glucose metabolism in the MCI cohort was lower in the posterior cingulate cortex, precuneus, lateral parietal and the lateral prefrontal cortices. The hypometabolism in PCC was consistent with the hypoperfusion in both the PCASL methods, whereas that in the precuneus was consistent only with hypoperfusion pattern in 3D PCASL. The hypometabolism in the frontal lobe in the MCI cohort was not observed with the PCASL methods, but was reflected by PASL (although not statistically significantly).
Figure 4.
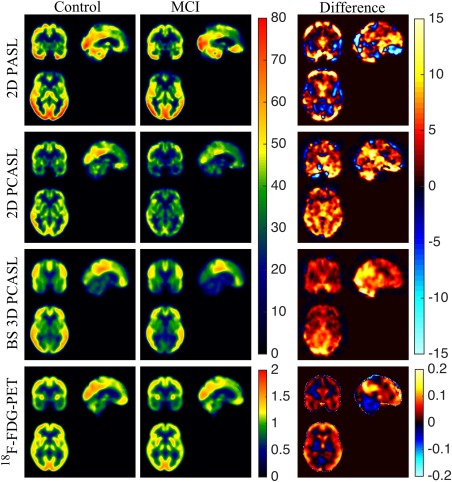
Triaxial view of (left to right) average CBF maps of the controls, patients, and their differences for 2D PASL (Row 1), 2D PCASL (Row 2), and BS 3D PCASL (Row 3). The bottom row shows the average SUVR maps of the controls, patients, and their differences (left to right). [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5 shows the box plots of the mean relative CBF values for each ASL methods and mean SUVR for PET, and Table 3 summarizes the effect sizes in each of the three a‐priori ROIs: PCC, precuneus, and hippocampus. 18F‐FDG PET displayed significant differences in metabolism between control and MCI groups for all three regions. Similarly, both PCASL methods displayed relative hypoperfusion in the MCI cohort compared to controls. Group differences in BS 3D PCASL were statistically significant in all three ROIs, while only the PCC and hippocampus were significant with 2D PCASL. No regions displayed a significant difference with 2D PASL. In fact, the precuneus was the only region in which MCI was associated with lower perfusion than the controls in absolute terms. As evident from the box plots, the variability of the relative mean CBF values in BS 3D PCASL was much lower compared to the 2D methods, and was comparable to PET.
Figure 5.
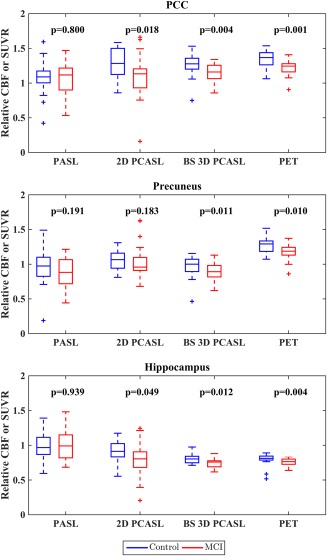
Box plots of mean relative CBFs and mean SUVR in different ROIs shown in blue for controls and red for MCI patients. Within each box, the three horizontal lines represent the first quartile, the median, and the third quartile (bottom to top). The two horizontal lines outside each box represent the minimum and maximum values ignoring outliers. Outliers, determined as 1.5 times the interquartile range above (or below) the third (or first) quartile, are shown separately with “+” symbols. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 3.
Table showing the effect sizes for Mann–Whitney U test for differentiating controls and MCI patients in different ROIs
| Effect size | |
|---|---|
| PCC | |
| 2D PASL | 0.04 |
| 2D PCASL | 0.35 |
| BS 3D PCASL | 0.42 |
| 18F–FDG PET | 0.47 |
| Precuneus | |
| 2D PASL | 0.19 |
| 2D PCASL | 0.20 |
| BS 3D PCASL | 0.37 |
| 18F–FDG PET | 0.38 |
| Hippocampus | |
| 2D PASL | 0.01 |
| 2D PCASL | 0.29 |
| BS 3D PCASL | 0.37 |
| 18F–FDG PET | 0.42 |
DISCUSSION
Background‐suppressed 3D PCASL provided superior sensitivity for detecting group differences in regional CBF between MCI patients and elderly controls than the other ASL methods tested, with effect sizes comparable to those seen with 18F‐FDG PET imaging. PCASL methods demonstrated higher temporal signal‐to‐noise ratios than PASL, likely due to improved labeling in the case of 2D PCASL, along with BS in the case of 3D PCASL. The sequence performance results obtained in this clinical population are generally consistent with those previously reported in young controls [Vidorreta et al., 2013], though, as expected, the tSNR values were lower, which is likely attributable to lower CBF and higher physiological noise in patients and elderly controls considered for this study. Across control and patient cohorts, there was good agreement between global CBF values obtained from each method. However, agreement on an individual subject level for different ASL schemes were found to be higher when using either the same labeling strategy (PCASL) or the same imaging strategy (2D EPI), and was significantly reduced when both the labeling and the data acquisition strategy differed. Thus, while data acquired with different ASL implementations are in general agreement, as they all measure the same physiological parameter (i.e., CBF), differences in labeling and imaging strategies cumulatively impact the degree of agreement. Multisite and multiplatform studies using ASL as a biomarker for neurodegeneration would clearly benefit from closely matched or identical ASL implementations, as has previously been suggested for normative data [Mutsaerts et al., 2015]. In particular, absolute CBF from significantly different ASL protocols should be compared, combined, and interpreted with caution.
BS 3D PCASL showed higher variability in CBF across subjects. A possible explanation is that the labeling efficiency at the labeling offset of 8 cm used in 3D may be more variable than the offset of 9 cm used in the 2D protocol. More recent data suggested that a lower labeling location provides more consistent results [Vidorreta et al., 2017]. Note that this higher variability does not affect the group discrimination analysis, which was performed using relative CBF.
Observed hypoperfusion in PCC, precuneus, and hippocampus in the MCI cohort using the PCASL methods was consistent with the levels of hypometabolism as measured by PET, although this difference did not reach significance with 2D PCASL in the precuneus. Overall, PCC seemed to be the most sensitive ROI for differentiating MCI patients and elderly controls with PCASL. A similar observation was made previously [Xekardaki et al., 2015], where PCC predicted cognitive deterioration in controls destined to develop MCI. In the case of PASL, ROI data failed to show group differences within the sample, likely reflecting the relatively poor sensitivity of this ASL variant, potentially compounded by coregistration errors due to poor tissue contrast that precluded manual correction. On the other hand, BS 3D PCASL demonstrated the largest effect sizes among the ASL variants (Table 3). The dispersion of the relative CBF values in BS 3D PCASL from Figure 5 appeared lower compared to 2D PCASL and 2D PASL methods and comparable to PET. Effect sizes were mostly medium (0.3–0.5), which is likely due to the low severity of disease in the MCI cohort studied. Indeed, effect sizes of the BS 3D PCASL were very similar to the “gold standard” 18F‐FDG PET measurements.
TSNR in the control cohort was significantly higher than in the patient group, which we attribute to a combined effect of higher signal (CBF) in controls and higher physiological noise in patients, likely due to motion. Although motion differences across groups did not reach significance, the translations and rotation values were indeed higher in the MCI patients and the observed correlations between tSNR and motion parameters further suggests its impact on tSNR. Higher tSNR in controls versus patients has also been reported previously in stroke patients [Fernandez‐Seara et al., 2008]. Outlier volumes were also detected in the CBF time series of each acquisition protocol, with greater numbers in the patient group, again likely due to increased physiological noise, and despite greatly increased tSNR in BS 3D PCASL data, outlier volumes were still present. While many 3D ASL methods use multishot acquisitions to improve image quality and resolution, an advantage of single‐shot acquisitions is the increased ease and efficiency of identifying and excluding outlier volumes in postprocessing of ASL time series.
Manual coregistration was needed to correct errors in the automatic coregistration produced by SPM, particularly for 3D PCASL data. SPM8 relies on intensity‐based methods for intermodal image registration using a normalized mutual information criterion. To provide optimal registration, the algorithm requires different tissues to have a distinct mean intensity. In EPI readouts, the between‐tissue contrast is primarily driven by the amount of T2* contrast accumulated at the echo time (TE). In ASL, the typically employed TE in EPI is shorter than in conventional blood oxygenation level‐dependent (BOLD) EPI acquisitions, resulting in lower contrast between tissues, which can occasionally cause errors in the registration output as observed in our data. In instances where tissue contrast was insufficient for successful automatic coregistration, registration was manually corrected using a boundary‐based approach [Greve and Fischl, 2009]. For 3D PCASL, the through‐plane blurring presents additional challenges for the coregistration algorithm and hence required more manual correction. On the other hand, in PASL the effect of the FAIR inversion pulse together with the shorter TE (compared to 2D PCASL) resulted in poorly identified tissue boundaries precluding manual registration, so automatic coregistration results from SPM were used without additional correction. As a result, the PASL data may have had greater coregistration errors than the other methods, which could ultimately have influenced ROI results and the discrimination between controls and MCI patients.
We conducted our analyses using relative CBF to facilitate comparison with PET, for which standard analysis uses normalization of 18F‐FDG uptake to cerebellum to account for differences in dose and overall cerebral delivery of 18F‐FDG between subjects. In addition, mean CBF in gray matter or whole brain is highly variable even in healthy populations [Dolui et al., 2016], and use of relative CBF eliminates this variability. Moreover, absolute CBF change within a specific ROI reflects both global and local CBF effects. While use of relative CBF obviously precludes detection of absolute CBF changes, which have also been observed with ASL in AD and MCI [Asllani et al., 2008], it enables comparing regional changes in CBF in disease‐specific patterns [Chen et al., 2011b]. In particular, reduction of relative CBF within specific ROI demonstrates lower metabolic function in that region that cannot be explained by global reduction in CBF. While cortical normalization of PET may have been more equivalent to the CBF measurements, this reference yielded a lower effect size for group differences within a priori ROIs compared to cerebellar normalization, as has been reported previously [Dukart et al., 2010]. As 18F‐FDG was being used in this context as a “gold standard,” we chose the more optimized cerebellar normalization.
The nominal PLD in this study was shorter than the recently recommended PLD of 1.8–2 s in an ASL white paper [Alsop et al., 2015]. It should be noted that the recommended PLD was for BS ASL acquisitions, which have higher signal compared to their non‐BS counterparts. In the case of non‐BS 2D acquisitions, the choice of a shorter PLD represents a tradeoff between two factors: (i) vascular transit artifacts and/or underestimation of CBF if the PLD is too short compared to the arterial transit time (ATT) and (ii) loss of sensitivity because the signal strength decreases due to T1 decay of the spin label during the PLD. Furthermore, as the 2D acquisition occurs slice by slice from inferior to superior direction, the last slice was acquired ∼800 ms after the initial post labeling delay, making the effective PLD in the superior slices much longer than the nominal value, with the mean PLD across the brain approximately 1.9 s. Although 1.5 s might be lower than the ATT for watershed regions of the brain that are supplied by the most distal branches of their arteries, particularly in elderly patients, those are not the regions of interest for this study. Instead, the ROIs we considered were located in the medial temporal and parietal regions, which are characterized by relatively shorter ATT [MacIntosh et al., 2010]. We did not observe any visible transit artifacts when inspecting the mean CBF maps from each subject. Finally, we based our analyses on mean CBF values in relatively large ROIs and the mean CBF within an ROI would not change as long as the blood supplying the microvasculature has arrived within the ROI, even if intra‐arterial [Qiu et al., 2012].
The spatial distribution of CBF was different in the three methods. PASL showed higher CBF in the posterior circulation, cerebellum, and basal ganglia and lower CBF in the frontal lobe whereas BS 3D PCASL had an exactly opposite distribution. Although much of this difference can be attributed to the different labeling and acquisition schemes [Lovblad et al., 2015], differences in effective PLD and their mismatch with ATT also likely contributed. ATT can be measured experimentally, and would be highly beneficial for optimal accuracy in CBF quantification. Methods for rapidly measuring ATT have recently been described [Dai et al., 2013; von Samson‐Himmelstjerna et al., 2016].
The study had few limitations. First, the ASL sequences evaluated not only differed with respect to the labeling and the presence of background suppression, but the voxel sizes were also different, which could have confounded comparisons between methods. Our choice of voxel dimensions was based on the previous literature and our own experience in ASL. The PASL resolution was matched to ADNI ASL protocol to facilitate interpretation of our findings with respect to ADNI data. However, the 2D PCASL replicated a protocol we had successfully applied to Alzheimer's disease in prior work [Chen et al., 2011b; Musiek et al., 2012; Xie et al., 2016; Zhang et al., 2012]. For single shot 3D PCASL, the voxel size was chosen to be smaller than 2D PCASL in the z direction because of its characteristic blurring in that direction. As our analysis focused on CBF in relatively large ROIs and not at individual voxels, the SNR should be largely independent of voxel resolution as smaller voxels have lower SNR but a larger number is required to cover the same spatial extent. However, some residual effects of voxel size may still be expected due to differences in partial volume contamination and physiological noise.
A second limitation is that PET data of several subjects were separated from ASL acquisition by more than a year. Although this does not undermine the comparison between the concurrent ASL strategies, which was our primary goal, it could confound the comparison of ASL with PET. In particular, as the ASL data for 4 MCI patients were obtained later in the course of their disease than the PET data, group comparisons might be biased to show a greater effect size for ASL. We repeated the group analysis after excluding data from the 4 MCI patients whose ASL data was acquired one year later than the PET data. The restricted comparison did not appreciably change the findings.
Finally, no separate M0 image was acquired for the 2D acquisition techniques, so we used mean of the control images to quantify CBF from control–label perfusion difference; however, we corrected for this error using the method suggested by Alsop et al. [2015] to account for incomplete relaxation of tissue magnetization. In addition, CBF quantification was obtained with a fixed value of T1blood and its variability can introduce errors [Dolui et al., 2016; Hales et al., 2016; Wu et al., 2010], though any such errors are unlikely to affect comparison significantly between the ASL variants.
In conclusion, both PCASL and background suppression improve the sensitivity of ASL over PASL acquisitions for detecting group differences between MCI and elderly controls. In particular, despite increased blurring in the slice dimension, BS 3D PCASL showed the highest discrimination between MCI and control groups, and provided comparable discrimination to 18F‐FDG PET. Thus, BS 3D PCASL methodology may provide an alternative to 18F‐FDG PET that can be obtained during routine MRI in clinical practice and research, and accelerated 3D ASL acquisitions can be used to reduce readout times and blurring.
DISCLOSURE/CONFLICT OF INTEREST
There is no financial interest in relation to the work described in this paper.
REFERENCES
- Alsop DC, Casement M, de Bazelaire C, Fong T, Press DZ (2008): Hippocampal hyperperfusion in Alzheimer's disease. NeuroImage 42:1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez‐Garcia L, Lu H, Macintosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC, Zaharchuk G (2015): Recommended implementation of arterial spin‐labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med 73:102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop DC, Detre JA, Grossman M (2000): Assessment of cerebral blood flow in Alzheimer's disease by spin‐labeled magnetic resonance imaging. Ann Neurol 47:93–100. [PubMed] [Google Scholar]
- Ashburner J (2007): A fast diffeomorphic image registration algorithm. NeuroImage 38:95–113. [DOI] [PubMed] [Google Scholar]
- Asllani I, Habeck C, Scarmeas N, Borogovac A, Brown TR, Stern Y (2008): Multivariate and univariate analysis of continuous arterial spin labeling perfusion MRI in Alzheimer's disease. J Cereb Blood Flow Metab 28:725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavusoglu M, Pfeuffer J, Ugurbil K, Uludag K (2009): Comparison of pulsed arterial spin labeling encoding schemes and absolute perfusion quantification. Magn Reson Imag 27:1039–1045. [DOI] [PubMed] [Google Scholar]
- Cha YH, Jog MA, Kim YC, Chakrapani S, Kraman SM, Wang DJ (2013): Regional correlation between resting state FDG PET and pCASL perfusion MRI. J Cereb Blood Flow Metab 33:1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LL, Pa J, Duarte A, Schuff N, Weiner MW, Kramer JH, Miller BL, Freeman KM, Johnson JK (2009): Patterns of cerebral hypoperfusion in amnestic and dysexecutive MCI. Alzheimer Dis Assoc Disord 23:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Pike GB (2009): Human whole blood T2 relaxometry at 3 Tesla. Magn Reson Med 61:249–254. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang DJ, Detre JA (2011a): Test‐retest reliability of arterial spin labeling with common labeling strategies. J Magn Reson Imag 33:940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wolk DA, Reddin JS, Korczykowski M, Martinez PM, Musiek ES, Newberg AB, Julin P, Arnold SE, Greenberg JH, Detre JA (2011b): Voxel‐level comparison of arterial spin‐labeled perfusion MRI and FDG‐PET in Alzheimer disease. Neurology 77:1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Garcia D, de Bazelaire C, Alsop DC (2008): Continuous flow‐driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med 60:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Lopez OL, Carmichael OT, Becker JT, Kuller LH, Gach HM (2009): Mild cognitive impairment and Alzheimer disease: Patterns of altered cerebral blood flow at MR imaging. Radiology 250:856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Shankaranarayanan A, Alsop DC (2013): Volumetric measurement of perfusion and arterial transit delay using hadamard encoded continuous arterial spin labeling. Magn Reson Med 69:1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky AP (1992): Perfusion imaging. Magn Reson Med 23:37–45. [DOI] [PubMed] [Google Scholar]
- Dolui S, Wang Z, Wang DJ, Mattay R, Finkel M, Elliott M, Desiderio L, Inglis B, Mueller B, Stafford RB, Launer LJ, Jacobs DR, Jr ., Bryan RN, Detre JA (2016): Comparison of non‐invasive MRI measurements of cerebral blood flow in a large multisite cohort. J Cereb Blood Flow Metab 36:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Jahng GH, Hayasaka S, Kramer JH, Rosen HJ, Gorno‐Tempini ML, Rankin KP, Miller BL, Weiner MW, Schuff N (2006): Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology 67:1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukart J, Mueller K, Horstmann A, Vogt B, Frisch S, Barthel H, Becker G, Moller HE, Villringer A, Sabri O, Schroeter ML (2010): Differential effects of global and cerebellar normalization on detection and differentiation of dementia in FDG‐PET studies. NeuroImage 49:1490–1495. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Seara MA, Edlow BL, Hoang A, Wang J, Feinberg DA, Detre JA (2008): Minimizing acquisition time of arterial spin labeling at 3T. Magn Reson Med 59:1467–1471. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR (1975): "Mini‐mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12:189–198. [DOI] [PubMed] [Google Scholar]
- Garcia DM, Duhamel G, Alsop DC (2005): Efficiency of inversion pulses for background suppressed arterial spin labeling. Magn Reson Med 54:366–372. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B (2009): Accurate and robust brain image alignment using boundary‐based registration. NeuroImage 48:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther M, Oshio K, Feinberg DA (2005): Single‐shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med 54:491–498. [DOI] [PubMed] [Google Scholar]
- Hales PW, Kirkham FJ, Clark CA (2016): A general model to calculate the spin‐lattice (T1) relaxation time of blood, accounting for haematocrit, oxygen saturation and magnetic field strength. J Cereb Blood Flow Metab 36:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovitch P, Raichle ME (1985): What is the correct value for the brain–blood partition coefficient for water? J Cereb Blood Flow Metab 5:65–69. [DOI] [PubMed] [Google Scholar]
- Hu WT, Wang Z, Lee VM, Trojanowski JQ, Detre JA, Grossman M (2010): Distinct cerebral perfusion patterns in FTLD and AD. Neurology 75:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Jahng GH, Weiner MW, Miller BL, Chui HC, Jagust WJ, Gorno‐Tempini ML, Schuff N (2005): Pattern of cerebral hypoperfusion in Alzheimer disease and mild cognitive impairment measured with arterial spin‐labeling MR imaging: Initial experience. Radiology 234:851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E, Goodglass H, Weintraub S (1983): Boston Naming Test. Philadelphia: Lea & Febiger. [Google Scholar]
- Kilroy E, Apostolova L, Liu C, Yan L, Ringman J, Wang DJ (2014): Reliability of two‐dimensional and three‐dimensional pseudo‐continuous arterial spin labeling perfusion MRI in elderly populations: Comparison with 15O‐water positron emission tomography. J Magn Reson Imag 39:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S, Wang J, Jahng GH (2001): FAIR exempting separate T (1) measurement (FAIREST): A novel technique for online quantitative perfusion imaging and multi‐contrast fMRI. NMR Biomed 14:507–516. [DOI] [PubMed] [Google Scholar]
- Lee IA, Preacher KJ (2013): Calculation for the test of the difference between two dependent correlations with one variable in common [Computer software]. Available from http://quantpsy.org.
- Lovblad KO, Montandon ML, Viallon M, Rodriguez C, Toma S, Golay X, Giannakopoulos P, Haller S (2015): Arterial spin‐labeling parameters influence signal variability and estimated regional relative cerebral blood flow in normal aging and mild cognitive impairment: FAIR versus PICORE techniques. Am J Neuroradiol 36:1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh WM, Wong EC, Bandettini PA, Hyde JS (1999): QUIPSS II with thin‐slice TI1 periodic saturation: A method for improving accuracy of quantitative perfusion imaging using pulsed arterial spin labeling. Magn Reson Med 41:1246–1254. [DOI] [PubMed] [Google Scholar]
- MacIntosh BJ, Filippini N, Chappell MA, Woolrich MW, Mackay CE, Jezzard P (2010): Assessment of arterial arrival times derived from multiple inversion time pulsed arterial spin labeling MRI. Magn Reson Med 63:641–647. [DOI] [PubMed] [Google Scholar]
- Maleki N, Dai W, Alsop DC (2012): Optimization of background suppression for arterial spin labeling perfusion imaging. MAGMA 25:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson N, Tosun D, Insel PS, Simonson A, Jack CR, Jr. , Beckett LA, Donohue M, Jagust W, Schuff N, Weiner MW Alzheimer's Disease Neuroimaging I (2014): Association of brain amyloid‐beta with cerebral perfusion and structure in Alzheimer's disease and mild cognitive impairment. Brain 137:1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Heyman A, Mohs RC, Hughes JP, van Belle G, Fillenbaum G, Mellits ED, Clark C (1989): The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer's disease. Neurology 39:1159–1165. [DOI] [PubMed] [Google Scholar]
- Musiek ES, Chen Y, Korczykowski M, Saboury B, Martinez PM, Reddin JS, Alavi A, Kimberg DY, Wolk DA, Julin P, Newberg AB, Arnold SE, Detre JA (2012): Direct comparison of fluorodeoxyglucose positron emission tomography and arterial spin labeling magnetic resonance imaging in Alzheimer's disease. Alzheimer Dement 8:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsaerts HJ, van Osch MJ, Zelaya FO, Wang DJ, Nordhoy W, Wang Y, Wastling S, Fernandez‐Seara MA, Petersen ET, Pizzini FB, Fallatah S, Hendrikse J, Geier O, Gunther M, Golay X, Nederveen AJ, Bjornerud A, Groote IR (2015): Multi‐vendor reliability of arterial spin labeling perfusion MRI using a near‐identical sequence: Implications for multi‐center studies. NeuroImage 113:143–152. [DOI] [PubMed] [Google Scholar]
- Ordidge RJ, Wylezinska M, Hugg JW, Butterworth E, Franconi F (1996): Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magn Reson Med 36:562–566. [DOI] [PubMed] [Google Scholar]
- Petersen RC (2004): Mild cognitive impairment as a diagnostic entity. J Intern Med 256:183–194. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Roberts RO, Knopman DS, Boeve BF, Geda YE, Ivnik RJ, Smith GE, Jack CR Jr (2009): Mild cognitive impairment: Ten years later. Arch Neurol 66:1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Straka M, Zun Z, Bammer R, Moseley ME, Zaharchuk G (2012): CBF measurements using multidelay pseudocontinuous and velocity‐selective arterial spin labeling in patients with long arterial transit delays: Comparison with xenon CT CBF. J Magn Reson Imag 36:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME (1998): Behind the scenes of functional brain imaging: A historical and physiological perspective. Proc Natl Acad Sci USA 95:765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitan RM (1958): Validity of the trail making test as an indicator of organic brain damage. Percept Motor Skills 8:271–276. [Google Scholar]
- Spreen O, Strauss E (1998): A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary. New York: Oxford University Press; xvi, 736 p. [Google Scholar]
- Steiger JH (1980): Tests for comparing elements of a correlation matrix. Psychol Bull 87:245–251. [Google Scholar]
- Tukey JW (1977): Exploratory Data Analysis. Reading, Massachusetts: Addison‐Wesley Publishing; xvi, 688 p. [Google Scholar]
- Verfaillie SC, Adriaanse SM, Binnewijzend MA, Benedictus MR, Ossenkoppele R, Wattjes MP, Pijnenburg YA, van der Flier WM, Lammertsma AA, Kuijer JP, Boellaard R, Scheltens P, van Berckel BN, Barkhof F (2015): Cerebral perfusion and glucose metabolism in Alzheimer's disease and frontotemporal dementia: Two sides of the same coin? Eur Radiol 25:3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernandez‐Seara MA (2013): Comparison of 2D and 3D single‐shot ASL perfusion fMRI sequences. NeuroImage 66C:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidorreta M, Zhao L, Shankar S, Wolf DH, Alsop DC, Detre JA (2017): In‐Vivo Evaluation of PCASL Labeling Scheme and Position). In: Proceedings of the International Society of Magnetic Resonance in Medicine. Honolulu, USA.
- von Samson‐Himmelstjerna F, Madai VI, Sobesky J, Guenther M (2016): Walsh‐ordered hadamard time‐encoded pseudocontinuous ASL (WH pCASL). Magn Reson Med 76:1814–1824. [DOI] [PubMed] [Google Scholar]
- Wang Z (2012): Improving cerebral blood flow quantification for arterial spin labeled perfusion MRI by removing residual motion artifacts and global signal fluctuations. Magn Reson Imag 30:1409–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Aguirre GK, Rao H, Wang J, Fernandez‐Seara MA, Childress AR, Detre JA (2008): Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx. Magn Reson Imag 26:261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Das SR, Xie SX, Arnold SE, Detre JA, Wolk DA, Alzheimer's Disease Neuroimaging I (2013): Arterial spin labeled MRI in prodromal Alzheimer's disease: A multi‐site study. NeuroImage Clin 2:630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansapura JP, Holland SK, Dunn RS, Ball WS Jr (1999): NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imag 9:531–538. [DOI] [PubMed] [Google Scholar]
- Wechsler D (1987): WMS‐R: Wechsler Memory Scale–Revised: Manual. San Antonio: Psychological Corp.: Harcourt Brace Jovanovich; viii, 150 p. [Google Scholar]
- Williams DS, Detre JA, Leigh JS, Koretsky AP (1992): Magnetic resonance imaging of perfusion using spin inversion of arterial water. Proc Natl Acad Sci USA 89:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO, Nordberg A, Backman L, Albert M, Almkvist O, Arai H, Basun H, Blennow K, de Leon M, DeCarli C, Erkinjuntti T, Giacobini E, Graff C, Hardy J, Jack C, Jorm A, Ritchie K, van Duijn C, Visser P, Petersen RC (2004): Mild cognitive impairment–beyond controversies, towards a consensus: Report of the International Working Group on Mild Cognitive Impairment. J Intern Med 256:240–246. [DOI] [PubMed] [Google Scholar]
- Wolk DA, Detre JA (2012): Arterial spin labeling MRI: An emerging biomarker for Alzheimer's disease and other neurodegenerative conditions. Curr Opin Neurol 25:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EC, Buxton RB, Frank LR (1998): A theoretical and experimental comparison of continuous and pulsed arterial spin labeling techniques for quantitative perfusion imaging. Magn Reson Med 40:348–355. [DOI] [PubMed] [Google Scholar]
- Wu WC, Edlow BL, Elliot MA, Wang J, Detre JA (2009): Physiological modulations in arterial spin labeling perfusion magnetic resonance imaging. IEEE Trans Med Imag 28:703–709. [DOI] [PubMed] [Google Scholar]
- Wu WC, Fernandez‐Seara M, Detre JA, Wehrli FW, Wang J (2007): A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous arterial spin labeling. Magn Reson Med 58:1020–1027. [DOI] [PubMed] [Google Scholar]
- Wu WC, St Lawrence KS, Licht DJ, Wang DJ (2010): Quantification issues in arterial spin labeling perfusion magnetic resonance imaging. Top Magn Reson Imag 21:65–73. [DOI] [PubMed] [Google Scholar]
- Xekardaki A, Rodriguez C, Montandon ML, Toma S, Tombeur E, Herrmann FR, Zekry D, Lovblad KO, Barkhof F, Giannakopoulos P, Haller S (2015): Arterial spin labeling may contribute to the prediction of cognitive deterioration in healthy elderly individuals. Radiology 274:490–499. [DOI] [PubMed] [Google Scholar]
- Xie L, Dolui S, Das SR, Stockbower GE, Daffner M, Rao H, Yushkevich PA, Detre JA, Wolk DA (2016): A brain stress test: Cerebral perfusion during memory encoding in mild cognitive impairment. NeuroImage Clin 11:388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Antuono PG, Jones J, Xu Y, Wu G, Ward D, Li SJ (2007): Perfusion fMRI detects deficits in regional CBF during memory‐encoding tasks in MCI subjects. Neurology 69:1650–1656. [DOI] [PubMed] [Google Scholar]
- Ye FQ, Frank JA, Weinberger DR, McLaughlin AC (2000): Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST). Magn Reson Med 44:92–100. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Stafford RB, Wang Z, Arnold SE, Wolk DA, Detre JA (2012): Microvascular perfusion based on arterial spin labeled perfusion MRI as a measure of vascular risk in Alzheimer's disease. J Alzheimer Dis 32:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]


