Abstract
Sensory organ glia surround neuronal receptive endings (NREs), forming a specialized compartment important for neuronal activity, and reminiscent of glia-ensheathed synapses in the central nervous system. We previously showed that DAF-6, a Patched-related protein, is required in glia of the C. elegans amphid sensory organ to restrict sensory compartment size. LIT-1, a Nemo-like kinase, and SNX-1, a retromer component, antagonize DAF-6 and promote compartment expansion. To further explore the machinery underlying compartment size control, we sought genes whose inactivation restores normal compartment size to daf-6 mutants. We found that mutations in igdb-2, encoding a single-pass transmembrane protein containing Ig-like and fibronectin type III domains, suppress daf-6 mutant defects. IGDB-2 acts in glia, where it localizes to glial membranes surrounding NREs, and, together with LIT-1 and SNX-1, regulates compartment morphogenesis. Immunoprecipitation followed by mass spectrometry demonstrates that IGDB-2 binds to LGC-34, a predicted ligand-gated ion channel, and lgc-34 mutations inhibit igdb-2 suppression of daf-6. Our findings reveal a novel membrane protein complex and suggest possible mechanisms for how sensory compartment size is controlled.
1. Introduction
Animals require sensation to mount appropriate responses to environmental stimuli. Sensory organs detecting external cues are often comprised of glia, or glia-like cells, that form specialized compartments surrounding receptive endings of sensory neurons or neuron-like cells. Sustentacular cells in the olfactory epithelium, Deiters’ cells in the cochlea, and retinal pigment epithelial cells in the retina are well-known examples of such compartment-forming glia-like cells (Bok, 1993; Breipohl et al., 1974; Rio et al., 2002).
Sensory organ glia share several features with glial cells that surround excitatory synapses in vertebrate nervous systems. Both cell types express glial cell markers such as the intermediate filament protein GFAP, maintain extracellular ion and signaling molecule concentrations, engulf shed neuronal membranes, and exhibit complex intracellular calcium responses to upstream stimuli (Bok, 1993; Hansel et al., 2001; Hegg et al., 2009; Rio et al., 2002). Thus, understanding how sensory organ glia compartments arise and how they affect neuron receptive ending (NRE) functions, may shed light on activities of their central nervous system (CNS) counterparts (Shaham, 2010).
The nematode C. elegans provides a unique arena to study glia-neuron interactions, as developmental constraints allow neurons to survive even in the absence of glia (Bacaj et al., 2008). Glial loss or manipulation in this animal leads to profound sensory and synaptic deficits (Bacaj et al., 2008; Colón-Ramos et al., 2007; Felton and Johnson, 2011; Singhvi et al., 2016; Wallace et al., 2016). Importantly, interactions between single glia and neurons can be interrogated at the level of neuronal activity and animal behavior (Oikonomou and Shaham, 2011).
Each C. elegans possesses bilaterally symmetric anterior amphid sensory organs composed of 12 sensory neurons and two glial cells: a socket glial cell (AMso) and a sheath glial cell (AMsh) (Ward et al., 1975) (Fig. 1A). Amphid neurons extend ciliated dendrites anteriorly, passing through a matrix-filled tubular channel that opens to the environment (Perkins et al., 1986). AMso and AMsh glial membranes comprise the anterior and posterior sections of this channel, respectively, and are joined by tight junctions to form a continuous tube. Similar junctions attach glia to sensory neuron dendrites, sealing the posterior portion of the channel to form an isolated compartment surrounding ciliary NREs (Fig. 1B).
Fig. 1. ns122 is a daf-6 suppressor.
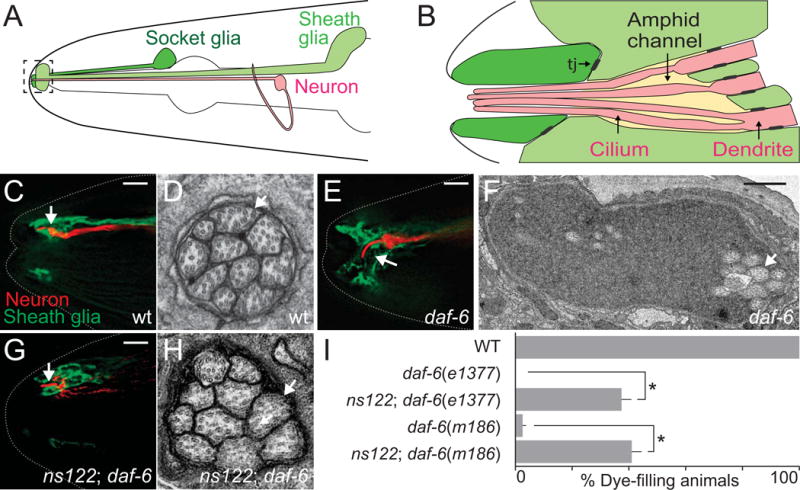
(A) The C. elegans amphid consists of 12 amphid neurons (one is shown in red) and the sheath (light green) and socket glia (dark green). The amphid channel is outlined and shown in greater detail in (B). Adapted from (Perkins et al., 1986). Tj, tight junctions. (C, E, G) The amphid channel (white arrow) in wild-type (C), daf-6(e1377) (E), and ns122; daf-6(e1377) (G) adult animals. The ASER neuron is visualized with mCherry (red, driven by the gcy-5 promoter) and the sheath glia with GFP (green, T02B11.3 promoter). Left is anterior. Scale bar, 5 μm.
(D, F, H) Transmission electron micrographs of cross-sections through the amphid channel in wild-type (D), daf-6(e1377) (F), and ns122; daf-6(e1377) (H) adult animals. White arrows point to cilia. Scale bar, 0.5 μm. Note the difference in scale between (C) and (E). (I) ns122 suppresses daf-6 dye-filling defects. n ≥ 100. Error bars, standard error of the mean (SEM), from ≥ 3 experiments. *p < 0.0001.
Amphid channel morphogenesis requires DAF-6, a Patched-related protein that restricts amphid channel expansion during development (Oikonomou et al., 2011; Perens and Shaham, 2005). Bloated channels in daf-6 mutants can be restored to their proper size by mutations in either lit-1 or snx-1 genes (Oikonomou et al., 2012; Oikonomou et al., 2011), suggesting that LIT-1, a Nemo-like kinase, and SNX-1, a retromer component, antagonize DAF-6 and promote channel growth. Indeed, lit-1 mutants alone exhibit abnormally constricted channels. daf-6, lit-1, and snx-1 also interact genetically with che-14, which encodes a Dispatched-related protein required for apical secretion from the AMsh glia (Michaux et al., 2000; Oikonomou et al., 2012; Oikonomou et al., 2011; Perens and Shaham, 2005). DAF-6, LIT-1, and SNX-1 proteins all function in amphid glia, and localize to the amphid channel surface (Oikonomou et al., 2012; Oikonomou et al., 2011; Perens and Shaham, 2005).
Here, we describe two novel regulators of amphid channel morphogenesis: IGDB-2, a single-pass transmembrane protein with Ig-like and fibronectin type III domains, and LGC-34, a predicted ligand-gated ion channel. We demonstrate that like lit-1 and snx-1 mutations, igdb-2 mutations restore inappropriately expanded channels in daf-6 mutants to their wild-type dimensions. IGDB-2 functions cell-autonomously in amphid glia, an activity requiring its N-terminal extracellular domain, and localizes to glial membranes forming the amphid compartment. igdb-2 mutations also genetically interact with lesions in other components driving compartment expansion. LGC-34 binds IGDB-2 in vivo and in cell culture, and lgc-34 loss suppresses igdb-2 mutant defects. We suggest, therefore, that IGDB-2 promotes amphid channel expansion, in part, by antagonizing LGC-34 activity. Our studies identify a novel complex controlling sensory compartment size, and suggest mechanisms for how compartment size may be regulated.
2. Materials and methods
2.1. C. elegans strains
Strains were cultured as previously described (Brenner, 1974). All animals were maintained and scored at 20°C for at least two generations without starvation. N2 animals were used as wild-type. Alleles in this study were: LGII lgc-34(gk532, gk751837), LGIII lit-1(ns132) (Oikonomou et al., 2011), LGIV igdb-2(ns122, gk214668), and LGX snx-1(ns133) (Oikonomou et al., 2012) and daf-6(e1377, m186) (Albert et al., 1981; Starich et al., 1995).
2.2. Identification of igdb-2(ns122)
SNP mapping was performed as described (Wicks et al., 2001). Briefly, CB4856 males were mated with ns122; daf-6 hermaphodites (N2 background). F2 recombinants were cloned, and F3 animals were scored for dye-filling as described in Section 2.6. SNP mapping placed ns122 in the region between +4.96 and +6.44 on LG IV. Fosmid rescue experiments narrowed the mapping interval to map units between LG IV +5.97 and +5.98. Whole genome sequencing of the ns122; daf-6 strain was performed, and the results analyzed using galign (Shaham, 2009). Mutations were found in two genes in the relevant interval: igdb-2 and T04A11.1. Rescue studies confirmed igdb-2 is the relevant gene.
2.3. Extrachromosomal arrays
The following arrays were used in this study: nsEx5869, nsEx5870, and nsEx5884 (PT02B11.3∷GFP and Pgcy-5∷mCherry); nsEx4695-nsEx4699 (WRM0629bG01 fosmid containing igdb-2 and pMH135 (Ppha-4∷GFP)); nsEx4743-nsEx4747 (pWW3 and pMH135); nsEx5663, nsEx5664, and nsEx5693 (pWW4 and Punc-122∷dsRed); nsEx5233-nsEx5236 (pWW5 and Punc-122∷dsRed); nsEx5436-nsEx5438 (pWW6 and Punc-122∷dsRed); nsEx5453-nsEx5456 (pWW7 and Punc-122∷dsRed); nsEx5373 (pWW8); Ex(pGT001(Pitr-1∷GFP) and pRF4(rol-6(su1006))) (Gower et al., 2001); nsEx5189, nsEx5190, nsEx5192, nsEx5195, and nsEx5232 (Pigdb-2∷IGDB-2(N-terminal deletion)∷GFP and Punc-122∷dsRed); nsEx5124, nsEx5125, nsEx5156, and nsEx5796 (Pigdb-2∷IGDB-2(middle deletion)∷GFP and Punc-122∷dsRed); and nsEx5155, nsEx5157, nsEx5158, and nsEx5201 (Pigdb-2∷IGDB-2(C-terminal deletion)∷GFP and Punc-122∷dsRed).
2.4. Integrated arrays
The following arrays were used: nsIs98 (Pmir-228∷GFP); nsIs521 (pWW2 and Punc-122∷dsRed); nsIs355 (pWW1 and pRF4); and sIs13244 (Plgc-34∷GFP and pCeh361(dpy-5(+))) (McKay et al., 2003).
2.5. Plasmid construction
pWW3 consists of Pigdb-2∷IGDB-2 (genomic SacI-XbaI fragment from 5.4kb upstream of ATG to 0.4kb downstream of TAA, cloned into pBS). pWW4 was pBS containing Pigdb-2∷IGDB-2(cDNA)∷GFP with: 1) the same promoter fragment as pWW3, 2) the igdb-2 cDNA amplified by polymerase chain reactions (PCR) from a mixed stage cDNA library, cloned into NheI-AscI sites, 3) GFP, and 4) unc-54 3′UTR. pWW5 consists of Pmir-228∷IGDB-2(cDNA)∷GFP (Pmir-228 (Miska et al., 2007) cloned into NotI-AscI and igdb-2 cDNA into NheI-KpnI sites in pSM-GFP). pWW6 and pWW7 are the same as pWW5 but with Pitr-1 (2.1kb) and Pdyf-7 (1.8kb) at NotI-XmaI, respectively (Gower et al., 2001; Heiman and Shaham, 2009). pWW8 is the same as pWW4, except with NLS-RFP fused to the unc-54 3′UTR, in place of IGDB-2(cDNA)∷GFP, cloned with BamHI-MluI. pWW9, pWW10, and pWW11 are the same as pWW4, but with the following cDNA deletion mutants: IGDB-2Δ25aa-784aa, IGDB-2Δ786aa-1412aa, and IGDB-2Δ1441aa-1526aa, respectively. Plasmids used for worm co-immunoprecipitation studies were pWW1 and pWW2. pWW1 consists of PT02B11.3∷3xFLAG∷GFP∷LIT-1 (inserted 3xFLAG into the Acc65I site in pGO38) (Oikonomou et al., 2011). pWW2 consists of Pigdb-2∷IGDB-2∷GFP∷3xFLAG (same as pWW3 but with GFP∷3xFLAG fused to the 3′ end of igdb-2, followed by the unc-54 3′UTR flanked by Acc65I sites). Plasmids used for cell culture co-immunoprecipitation studies were pWW12 (pLenti-CMV∷LGC-34∷GFP) and pWW13 (pLenti-CMV∷IGDB-2∷3xFLAG), which were made using the Gateway® cloning system (ThermoFisher).
2.6. Dye-filling assay
Animals were washed off NGM plates into M9 buffer containing 10 μg/ml diI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) (ThermoFisher D282) and rotated at 20°C in the dark for 2 hours. They were then transferred to new NGM plates and scored for dye-filling (animals with any subset of dye-filled amphid neurons were considered positive for dye-filling). For transgenic strains, at least three lines were scored and averaged. Statistical analysis of all dye-filling values was performed with the unpaired t-test (GraphPad), except for epistasis analysis which was performed with the one-way ANOVA test with Bonferroni’s multiple comparisons (GraphPad).
2.7. Fluorescence and transmission electron microscopy
Images were acquired with a DeltaVision Image Restoration microscope (Applied Precision) with an Olympus UPLSAPO (60X, 1.3NA) silicone oil objective for amphid channel morphology and with an inverted TCS SP8 laser scanning confocal microscope (Leica) with a PlanApo (either 63x, 1.40NA or 100x, 1.46NA) oil objective for expression pattern studies. Deltavision images were deconvolved with SoftWoRx (Applied Precision). All fluorescence images were analyzed with ImageJ (NIH). For transmission electron microscopy, animals were sectioned and imaged as previously described (Perens and Shaham, 2005; Wallace et al., 2016).
2.8. Co-immunoaffinity purification from worms and mass spectrometry
Mixed stage worms were grown on peptone-enriched plates with HB101 bacteria, resuspended in homogenization buffer (50 mM HEPES-NaOH (pH 7.6), 1 mM EDTA, 150 mM NaCl, and 10% glycerol), and frozen as previously described (Mets and Meyer, 2009). Frozen worm powder was resuspended in homogenization buffer (with the addition of 1% NP-40 and Halt protease inhibitor (ThermoFisher 78429)), sonicated, and centrifuged at 20,000 g for 15 minutes at 4°C. The supernatant (2–3 mg/ml, 5–10 ml total) was incubated with 200 μl anti-FLAG® M2 affinity gel (Sigma A2220) for 2 hours. Beads were washed 3 times with homogenization buffer and proteins eluted with SDS loading buffer. One third of the total eluate was resolved by SDS-PAGE (4–12% gradient) to one third the length of the gel, and stained with Coomassie Blue (BioRad 1610436). The protein was then in-gel trypsinized, and the extracted peptides were identified by MS/MS mass spectrometry (Rockefeller University Proteomics Resource Center). Western blot analysis was performed with anti-GFP (polyclonal rabbit, gift from Baolin Wang) and anti-FLAG® M2 (monoclonal mouse, Sigma F3165) at 1:1000, and HRP-conjugated donkey anti-rabbit and anti-mouse at 1:5000 (Jackson ImmunoResearch 711-035-152 and 711-035-150, respectively).
2.9. Cell culture and co-immunoaffinity purification
HEK293 cells were cultured in DMEM (ThermoFisher 11995065), supplemented with 10% heat-inactivated fetal bovine serum (Sigma F4135) and penicillin/streptomycin (ThermoFisher 15140-122). Transfections were performed using FuGENE HD (Promega E2311) according to the manufacturer protocol. Transfected cells were lysed with homogenization buffer and centrifuged at 20,000 g for 10 minutes. The supernatant was incubated with either anti-FLAG® M2 affinity gel or anti-GFP, as was done with worm lysate (see above).
3. Results
3.1. Mutations in igdb-2 suppress sensory compartment morphogenesis defects of daf-6 mutants
The amphid channel is a specialized sensory compartment, formed by the AMso and AMsh glial cells, through which sensory neurons extend cilia to access the external environment (Fig. 1B, C). These cilia are arranged in a stereotypical 3:4:3 pattern (Fig. 1D) and are tightly enveloped by the two glial cells. We previously showed that daf-6 mutants, harboring lesions in the DAF-6 Patched-related protein, exhibit a bloated channel containing excess extracellular material (Perens and Shaham, 2005). Within the bloated channel, neuronal cilia are disorganized and bent and fail to access the environment (Fig. 1E, F). The daf-6 mutant defects suggest that this gene normally blocks channel expansion.
To identify genes that drive channel growth, we mutagenized daf-6 mutants, and sought animals in which normal channel size is restored (see Oikonomou et al., 2011 for screen details). We previously described lesions in two genes, lit-1, encoding a Nemo-like kinase, and snx-1, encoding a retromer component, that suppress the daf-6 mutant defect (Oikonomou et al., 2012; Oikonomou et al., 2011). A third suppressor mutant, with allele designation ns122, was also found but not characterized.
To determine whether ns122 indeed restores normal channel morphology to daf-6 mutants, we examined ns122; daf-6 double mutants carrying transgenes driving GFP expression in AMsh glia and mCherry in the ASER amphid sensory neuron. We found that the ASER cilium in the double mutant extends through a channel as in the wild type in 20% of animals (n=10, compare Fig. 1G to 1C, and note difference from 1E). To confirm these observations, we analyzed double mutants by transmission electron microscopy (TEM). As shown in Fig. 1H, the amphid channel indeed appears largely normal, with cilia tightly bundled and exposed to the environment (n=3, compare Fig. 1H to 1F; Fig. S1). However, cilia are not arranged in a 3:4:3 pattern, suggesting partial suppression.
To assess channel defects more quantitatively, we used a third assay. Some amphid neurons in wild-type animals can take up dyes, such as DiI, from the environment. Such dye uptake is blocked in animals in which sensory cilia fail to access the environment (Perkins et al., 1986). Consistent with our imaging assays, while neurons of daf-6 mutants all fail to accumulate dye, 37% of ns122; daf-6(e1377) double mutants exhibit neuronal dye-filling in at least one of the two amphid sensory organs (Fig. 1I), confirming that ns122 suppresses daf-6 mutant defects. Crossing the ns122 allele to another daf-6 mutant, m186, results in a comparable dye-filling increase (Fig. 1I). Furthermore, three ns122; daf-6(e1377) animals that accumulated dye in only one amphid organ displayed only one amphid channel of more normal morphology by EM (Fig. S1), suggesting that dye-filling can serve to report on channel morphogenesis.
To identify the gene defective in ns122 mutants, we used single nucleotide polymorphism (SNP) mapping and whole-genome sequencing to localize the ns122 lesion to the igdb-2 gene (Materials and methods), which encodes a single-pass transmembrane protein with multiple Ig-like and fibronectin type III (FNIII) domains (Fig. 2A). We found a Q579ochre mutation in igdb-2(ns122) mutants, leading to a predicted truncation of the IGDB-2 protein, preserving the first Ig and FNIII domains. To determine whether this igdb-2 mutation is responsible for restoring wild-type channel shape to daf-6 mutants, we crossed another igdb-2 allele, gk214668, to daf-6 mutants. This allele harbors a Q78ochre mutation and leaves only the first Ig domain intact (Fig. 2A). 22% of igdb-2(gk214668); daf-6(e1377) double mutants exhibit dye-filling, suggesting that igdb-2 is indeed the relevant gene (Fig. 2B). Further supporting this conclusion, dye uptake in igdb-2(ns122); daf-6(e1377) mutants carrying an extrachromosomal array containing the igdb-2 fosmid is reduced from 37% (no transgene) to 3% (Fig. 2B). Similarly, igdb-2(ns122); daf-6(e1377) mutants carrying a transgene containing the igdb-2 genomic region (from 5.4 kb upstream of the start codon to 0.4 kb downstream of the stop codon) have reduced dye-filling (Fig. 2B); the same is true using the igdb-2 promoter region to drive expression of an igdb-2 cDNA∷GFP fusion.
Fig. 2. ns122 is an allele of igdb-2, which encodes an Ig/FNIII transmembrane protein.
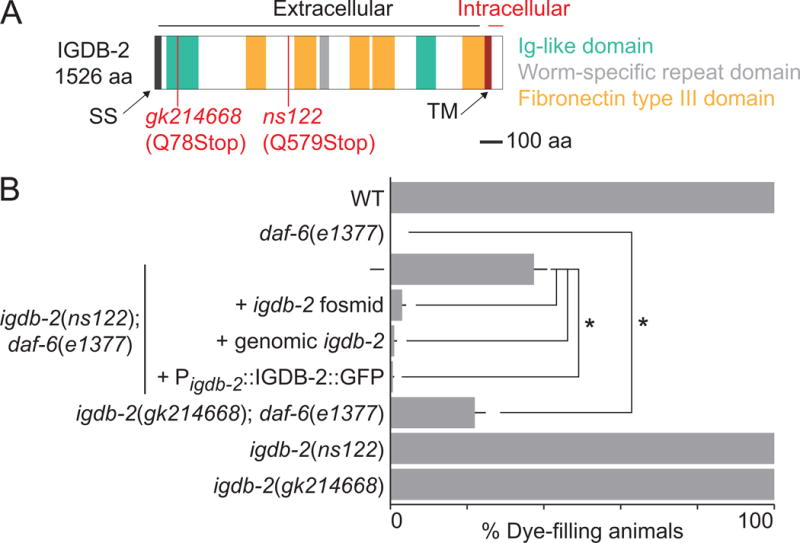
(A) Schematic of the IGDB-2 protein, marking truncations predicted in ns122 and gk214668. SS, signal sequence. TM, transmembrane domain. (B) Dye-filling assay for indicated genotypes. Alleles used are: daf-6(e1377) and igdb-2(ns122). (—) indicates no transgene. n ≥ 87. Error bars, SEM, from ≥ 3 experiments. *p < 0.0082.
Together, these data demonstrate that lesions in igdb-2 restore wild-type channel morphology to daf-6 mutants.
3.2. igdb-2 can act in AMso glia and localizes to sensory compartment membranes
To determine whether igdb-2 functions in neurons or glia to regulate channel morphogenesis, we performed cell-specific rescue experiments. As shown in Fig. 3A, igdb-2(ns122); daf-6(e1377) mutants expressing an igdb-2 cDNA∷GFP fusion transgene under control of the glia-specific mir-228 promoter, expressed during channel morphogenesis (Miska et al., 2007), have significantly reduced dye accumulation compared to non-transgenic animals. igdb-2 cDNA∷GFP expression in AMso glia alone, using an itr-1 promoter fragment (Gower et al., 2001), also significantly reduces dye uptake. Importantly, igdb-2 cDNA∷GFP expression using the ciliated sensory neuron-specific dyf-7 promoter, also active at the time of compartment morphogenesis (Heiman and Shaham, 2009), does not affect dye filling of igdb-2(ns122); daf-6(e1377) mutants. Thus, igdb-2 functions at least in part in AMso glia and not in neurons to regulate compartment shape.
Fig. 3. igdb-2 acts and is expressed in glia to regulate amphid channel morphogenesis.
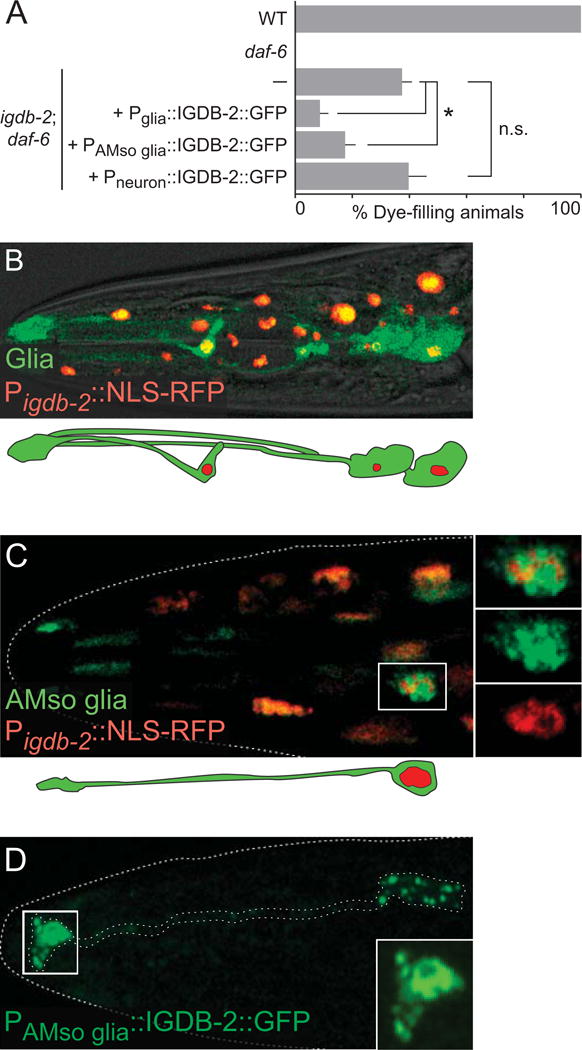
(A) Dye-filling assay for indicated genotypes. Alleles used are: daf-6(e1377) and igdb-2(ns122). (—) indicates no transgene. Promoters used for expression in glia, AMso glia, and neurons are Pmir-228 (Miska et al., 2007), Pitr-1 (Gower et al., 2001), and Pdyf-7 (Heiman and Shaham, 2009), respectively. n ≥ 100. Error bars, SEM, from ≥ 3 experiments. *p < 0.0048, n.s. not significant. (B, C) Image of an adult (head) expressing Pigdb-2∷NLS-RFP (red) and either (B) Pmir-228∷GFP (in all glia, green) or (C) Pitr-1∷GFP (in socket glia, green). Schematic of glia is shown below (B) and (C). (D) Adult expressing Pitr-1∷IGDB-2∷GFP (in socket glia, green). Outlined area in (C) and (D) are shown in greater detail in inset. Scale bars, 5 μm. Left is anterior.
The igdb-2 gene expression pattern also supports this conclusion. Animals co-expressing igdb-2 promoter∷nuclear-RFP and glial mir-228 promoter∷GFP exhibit signal co-localization in glia, among other cells (Fig. 3B). A similar experiment using itr-1 promoter∷GFP to mark AMso glia, revealed co-localization with igdb-2 promoter∷GFP in these cells (Fig. 3C), consistent with our rescue studies.
To examine where IGDB-2 protein localizes within AMso glia, we generated animals carrying a transgene driving igdb-2 cDNA∷GFP expression in these cells. We found an enrichment of IGDB-2 near the amphid channel (Fig. 3D) and in vesicular structures in the cell body. This supports the notion that IGDB-2 is trafficked on vesicles to the amphid channel, where it participates in channel morphogenesis. These observations are consistent with previous findings demonstrating that DAF-6, LIT-1, and SNX-1 also localize to the amphid channel (Oikonomou et al., 2012; Oikonomou et al., 2011; Perens and Shaham, 2005).
3.3. The IGDB-2 membrane-distal extracellular domain is required for compartment morphogenesis
The IGDB-2 protein consists of a large extracellular domain containing Ig-like and FNIII protein-protein interaction domains, as well as a short intracellular domain, which could be used for signal transduction. To uncover the functional significance of these domains, we generated igdb-2(ns122); daf-6(e1377) animals carrying transgenes in which the igdb-2 promoter drives expression of truncated igdb-2 cDNA∷GFP proteins. As shown in Fig. 4, animals expressing IGDB-2ΔN∷GFP, lacking membrane-distal extracellular sequences, exhibit dye accumulation comparable to non-transgenic mutants. However, in mutants expressing either IGDB-2Δmiddle∷GFP or IGDB-2ΔC∷GFP, only 6% and 16% of animals take up dye, respectively, indicating that these transgenes remain functional. These results, therefore, indicate that the N-terminal membrane-distal domain of IGDB-2, containing one Ig domain, two FNIII domains, and a single nematode-specific repeat domain, is required for IGDB-2 channel morphogenesis functions, and that the membrane-proximal and intracellular domains are dispensable for activity. Thus, IGDB-2 is unlikely to function through intracellular signaling, and more likely influences compartment shape by interacting with extracellular moieties.
Fig. 4. The N-terminal membrane-distal domain of IGDB-2 is required for IGDB-2 function in amphid channel morphogenesis.
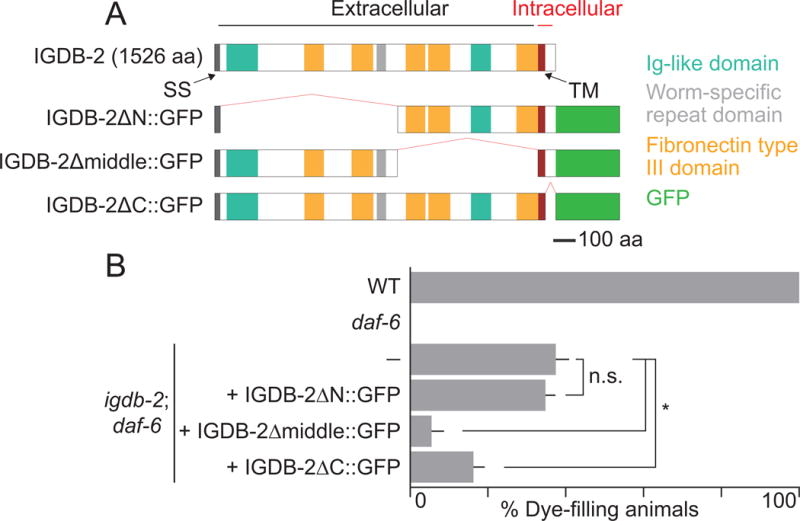
(A) Schematic of wild-type and truncated IGDB-2∷GFP proteins. (B) Dye-filling assay for indicated genotypes. Alleles used are: daf-6(e1377) and igdb-2(ns122). (—) indicates no transgene. n ≥ 100. Error bars, SEM, from ≥ 3 experiments. *p < 0.0024, n.s. not significant.
3.4. igdb-2 genetically interacts with other genes controlling compartment size
Like igdb-2 mutations, lesions in lit-1 and snx-1 only partially suppress daf-6 mutant defects. One explanation for this is that more than one pathway drives compartment expansion; thus, disruption of a single pathway may not be sufficient to restore wild-type sensory channel morphology to daf-6 mutants. We therefore sought to determine whether igdb-2, lit-1, and snx-1 function in independent pathways. To this end, we generated the following triple mutants: 1) lit-1; igdb-2; daf-6, 2) igdb-2; snx-1 daf-6, and 3) lit-1; snx-1 daf-6. We then assessed their ability to take up dye, and compared them with the relevant daf-6 double mutants. We found that 39%, 26%, and 45% of triple mutants accumulate dye in amphid sensory neurons, respectively (Fig. 5). Neither lit-1; igdb-2; daf-6 nor igdb-2; snx-1 daf-6 mutants were statistically different from the corresponding double mutants, suggesting that igdb-2 acts with lit-1 and snx-1. However, lit-1; snx-1 daf-6 mutants showed a significantly higher suppression (45%) than lit-1; daf-6 (25%) mutants, suggesting parallel functions for lit-1 and snx-1, as previously reported (Oikonomou et al., 2012). However, this synergistic effect was much weaker than observed earlier (Oikonomou et al., 2012), and not even seen when comparing lit-1; snx-1 daf-6 mutants to snx-1 daf-6 mutants (34%). We speculate, therefore, that the differences seen in earlier findings may be due to background mutations in the double or triple mutants. Given the weakness of this synergy, and the lack of synergy between either lit-1 or snx-1 and igdb-2, these values together do not support parallel pathways between lit-1, snx-1, and igdb-2.
Fig. 5. igdb-2 genetically interacts with other genes controlling compartment size.
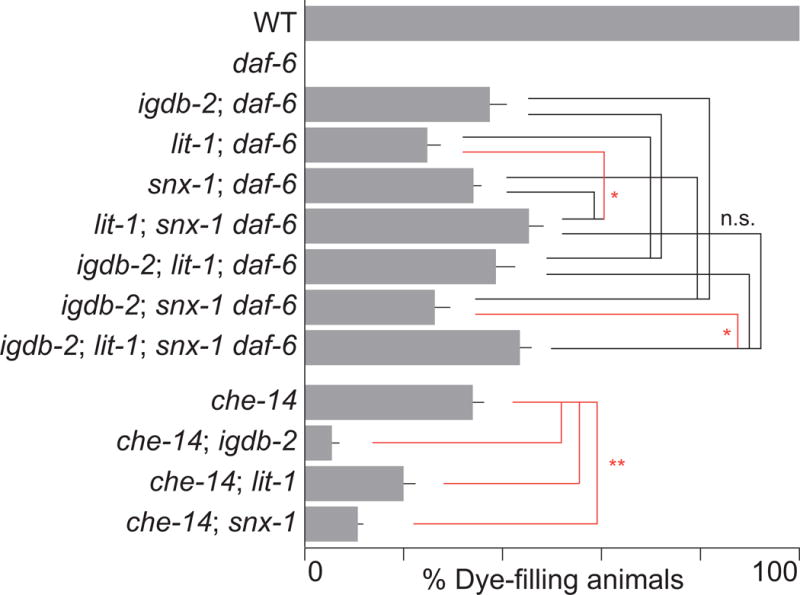
Dye-filling assay for indicated genotypes. Alleles used are: daf-6(e1377), igdb-2(ns122), lit-1 (ns132), snx-1(ns133), and che-14(ok193). n ≥ 100. Error bars, SEM, from ≥ 3 experiments. n.s. not significant (black lines), *p < 0.019 (red lines), by one-way ANOVA analysis with Bonferroni’s multiple comparisons. **p < 0.0044 (red lines), by unpaired t-test.
To verify this epistasis analysis, we generated lit-1; igdb-2; snx-1 daf-6 quadruple mutants. 44% of animals could take up dye, statistically comparable to the lit-1; igdb-2; daf-6 and igdb-2; snx-1 daf-6 triple mutants. This further supports a common pathway model. While there is a slight difference between the quadruple mutant and igdb-2; snx-1 daf-6, this trend is not supported by the other results.
Taken together, these results suggest either that IGDB-2, LIT-1, and SNX-1 function in the same process, or that daf-6 mutant defects cannot be suppressed much beyond ~45%.
To distinguish between these models, we tested whether igdb-2 interacts genetically with che-14/Dispatched, which also regulates amphid channel morphogenesis (Michaux et al., 2000). We previously showed that lit-1 and snx-1 mutations enhance dye-filling defects in che-14 mutants, in contrast to their suppression of daf-6 defects (Oikonomou et al., 2012; Oikonomou et al., 2011). To determine if igdb-2 acts similarly, we crossed igdb-2(ns122) to che-14 mutants. 6% of double mutants took up dye, significantly less than the 34% in che-14 single mutants, demonstrating that igdb-2 does enhance the che-14 dye-filling defect. We thus favor a model in which IGDB-2 acts in the same process as LIT-1 and SNX-1.
3.5. LGC-34, a predicted ligand-gated ion channel, binds to IGDB-2
To identify IGDB-2 interactors that may also direct sensory compartment morphogenesis, we generated animals expressing a FLAG-tagged IGDB-2 protein, detectable by Western blot (Fig. 6A), and performed co-immunoaffinity purification of this fusion protein using anti-FLAG beads (Fig. 6A, B). As a control, we purified LIT-1 from a strain expressing FLAG-tagged LIT-1. Precipitates were subjected to MS/MS mass spectrometry to identify interacting proteins (Table S1). Peptides derived from LGC-34, a ligand-gated ion channel, were found to precipitate with IGDB-2∷FLAG but not LIT-1∷FLAG. Importantly, peptides derived from WRM-1/β-catenin, a known LIT-1-interacting protein, were found in the LIT-1, but not in the IGDB-2 precipitate, demonstrating both the success and specificity of our method. To confirm the IGDB-2-LGC-34 interaction, we co-expressed IGDB-2-FLAG and LGC-34-GFP in HEK293 cells. Immunoprecipitation using anti-GFP beads specifically pulled down IGDB-2-FLAG only when co-expressed with LGC-34-GFP (Fig. 6C, compare right to left lane in top IP panel). In the converse experiment, while anti-FLAG beads nonspecifically bind a residual amount of LGC-34-GFP (Fig. 6C, left lane of bottom IP panel), co-expression with IGDB-2-FLAG, precipitated much more LGC-34-GFP (Fig. 6C, right lane of bottom IP panel), suggesting that LGC-34 indeed co-immunoprecipitates with IGDB-2. Taken together, these co-immunoprecipitation experiments reveal that these two proteins specifically interact in this setting as well.
Fig. 6. LGC-34, a predicted ligand-gated ion channel, binds to IGDB-2.
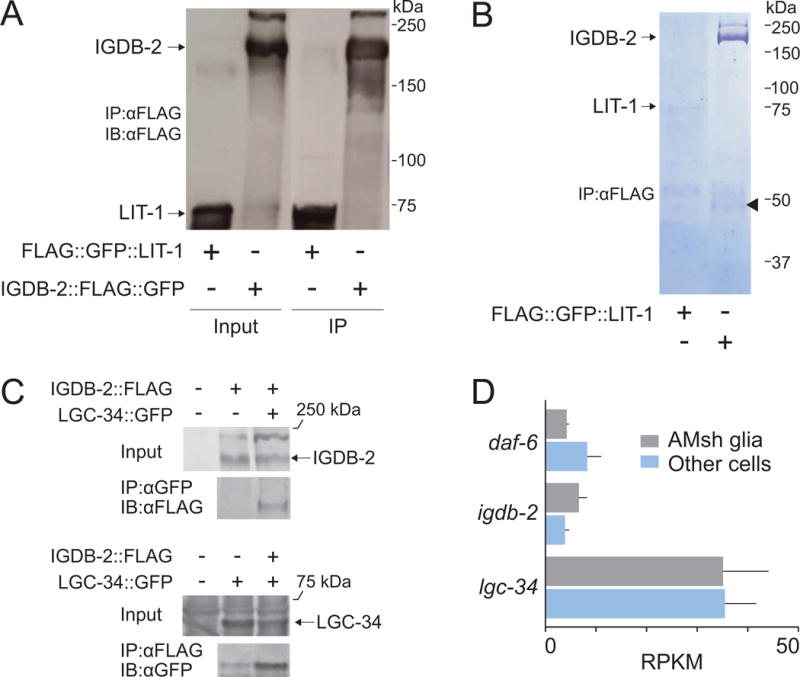
(A) IGDB-2∷FLAG was purified from animals with anti-FLAG beads and detected by Western blot. FLAG∷LIT-1 was expressed and purified as a control. IP, immunoprecipitated. (B) Proteins co-immunoprecipitated with IGDB-2∷FLAG were resolved by SDS-PAGE and the gel stained with Coomassie Blue. The arrowhead marks LGC-34. (C) IGDB-2-FLAG and LGC-34-GFP were co-expressed in HEK293 cells. IGDB-2 co-immunoprecipitates with LGC-34, purified with anti-GFP beads (top), and vice versa, with anti-FLAG beads (bottom). (D) Gene expression profiling of daf-6, igdb-2, and lgc-34 in amphid sheath glia and other cells of the worm (Based on Wallace et al., 2016; and S.W. Wallace, unpublished results). RPKM, Reads Per Kilobase of transcript per Million mapped reads. Error bars, SEM, from 3 experiments.
To determine whether LGC-34 and IGDB-2 are co-expressed in C. elegans glia, we analyzed previous AMsh glia expression profiling and found reads corresponding to both lgc-34 and igdb-2 (Fig. 6D; Wallace et al., 2016; and S.W. Wallace, unpublished results). Both mRNAs are detected at higher levels than daf-6, a known glial gene (Perens and Shaham, 2005), suggesting that LGC-34 and IGDB-2 are likely both expressed in glia.
3.6. LGC-34 restricts sensory compartment expansion and is inhibited by IGDB-2
To uncover possible lgc-34 functions, we assessed whether lgc-34 mutations affect amphid compartment morphology. lgc-34(gk532) contains a deletion spanning the lgc-34 5′ UTR and the first 2 exons (Fig. 7A). We found that lgc-34(gk532); igdb-2; daf-6 triple mutants had significantly reduced dye-filling, compared to igdb-2; daf-6 double mutants (Fig. 7B). To confirm this, we crossed a second lgc-34 allele, gk751837, predicted to generate a truncated protein with no transmembrane domains, into igdb-2; daf-6 double mutants. The resulting triple mutant also displayed a reduction in igdb-2-dependent daf-6 suppression, with only 12% of triple mutants taking up dye. Neither lgc-34 allele had a dye-filling defect on its own, and did not significantly restore dye filling to daf-6 mutants (Fig. S2). Thus, lgc-34 appears to participate in amphid compartment morphogenesis downstream of igdb-2.
Fig. 7. lgc-34 restricts sensory compartment expansion and is inhibited by igdb-2.
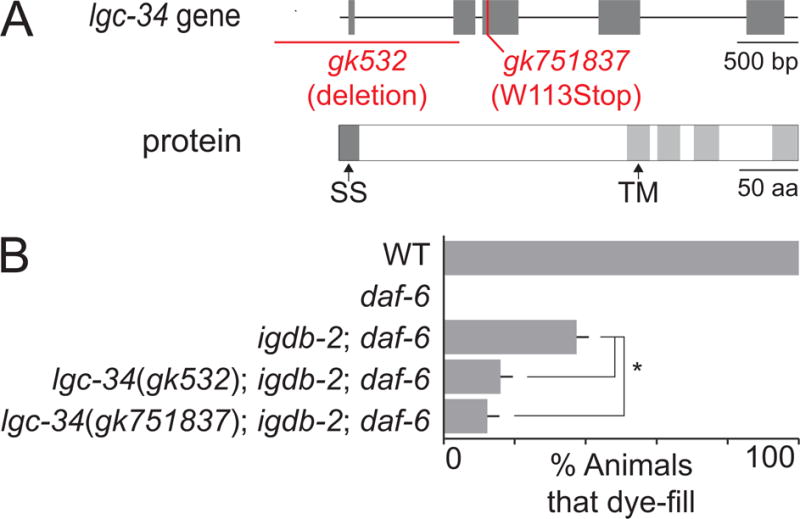
(A) Schematic of the lgc-34 gene (top), marked with regions affected in the gk532 and gk751837 alleles, and LGC-34 protein (bottom). SS, signal sequence, and TM, transmembrane domain. (B) Dye-filling assay for indicated genotypes. Alleles used are: daf-6(e1377), igdb-2(ns122), lgc-34(gk532), and lgc-34(gk751837). n ≥ 100. Error bars, SEM, from ≥ 3 experiments. *p < 0.006.
4. Discussion
4.1. A model for amphid compartment morphogenesis
The results presented here are consistent with a model in which igdb-2 normally inhibits lgc-34, which in turn restricts sensory compartment growth (Fig. 8). Since lgc-34 mutations do not fully suppress igdb-2 mutant defects, igdb-2 also likely functions independently, and perhaps together with lit-1 and snx-1 to influence amphid channel morphogenesis.
Fig. 8. A model for sensory compartment morphogenesis.
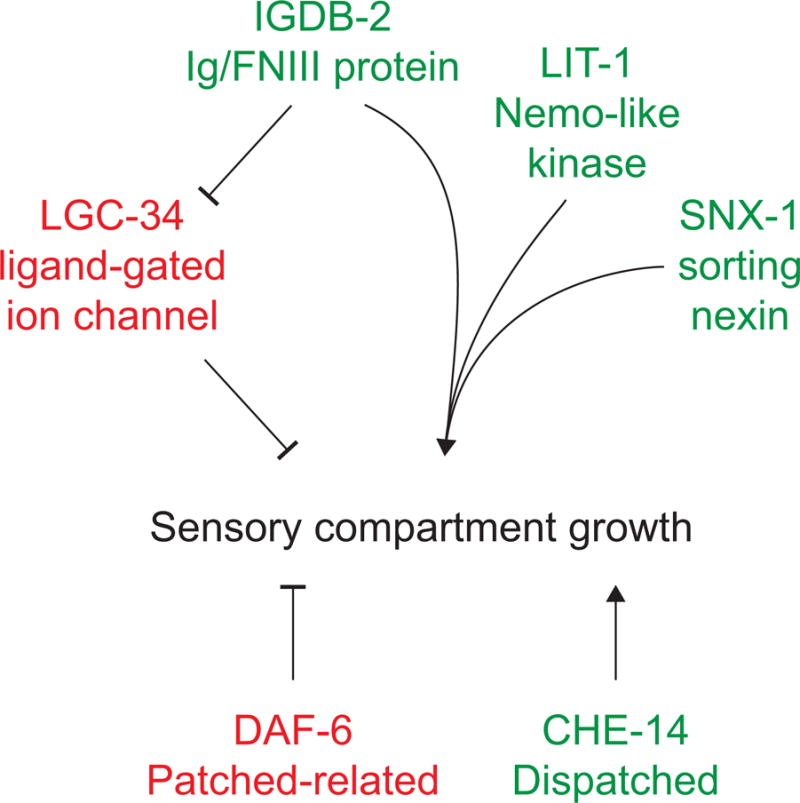
IGDB-2 inhibits LGC-34, which in turn restricts sensory compartment expansion. IGDB-2 also independently promotes expansion, possibly with LIT-1 and SNX-1. DAF-6 restricts sensory compartment growth (Oikonomou et al., 2011), while CHE-14 promotes it (Michaux et al., 2000).
We also demonstrated that igdb-2 functions, at least in part, in the AMso glia. This was somewhat surprising, given that daf-6 is required in the adjacent AMsh glia to restrict amphid channel expansion (Perens and Shaham, 2005). Thus, igdb-2 could be opposing daf-6 in a paracrine manner, or both could be acting from both the AMso and AMsh glia. We favor the latter model based on the following observations. First, both igdb-2 and daf-6 are expressed in AMso and AMsh glia (Wallace et al., 2016; S.W. Wallace, unpublished results; and Perens and Shaham, 2005). Second, AMso glia-specific expression of igdb-2 rescues ns122 defects, but to a lesser degree than expression under the igdb-2 or pan-glial mir-228 promoter. This suggests that igdb-2 may be acting from other cells, like the AMsh glia. Third, infrared laser-induced expression of daf-6 in the AMsh glia in daf-6 mutants restores dye-filling, but only in 30% of animals (Singhal and Shaham, 2017). Thus, daf-6 function may similarly be required in the AMso glia, where it may function with IGDB-2.
4.2. IGDB-2 protein domains are found in other lumen-forming proteins
IGDB-2 contains Ig-like and FNIII domains, possibly providing insight into how the amphid channel forms. Ig/FNIII proteins are classified as cell adhesion molecule (CAM) members of the immunoglobulin superfamily, and play prominent roles in neurodevelopment (Sytnyk et al., 2017). A number of proteins with similar domains are specifically involved in lumen formation: Neogenin, a Netrin receptor in the zebrafish, promotes neural tube formation (Mawdsley et al., 2004); Neuroglian, the Drosophila L1-CAM homolog, promotes formation of glial canals of the antennal lobe (Chen and Hing, 2008); and Robo, the Slit receptor, promotes formation of astrocytic tunnels leading to the olfactory bulb in mice (Kaneko et al., 2010). Given that Neogenin, Neuroglian, and Robo all function as membrane receptors for cell-cell communication, IGDB-2 may function in a similar manner, directing communication between the AMso and AMsh glia, or between the glia and amphid neurons.
4.3. IGDB-2 binds LGC-34
Our findings suggest that IGDB-2 functions through LGC-34 to control amphid channel morphogenesis. What could be the functional nature of this interaction? One hypothesis is that IGDB-2 may alter membrane localization or stability of LGC-34. This scenario is reminiscent of how another Ig/FNIII transmembrane protein and Hedgehog coreceptor, Cdo, regulates activity of the Kir2.1 K+ channel: Cdo forms a complex with Kir2.1 and increases cell surface expression of Kir2.1 in differentiating myoblasts (Leem et al., 2016). Notably, the Drosophila homolog of Cdo, Ihog (Interference hedgehog), forms a complex with Patched and a related Ig/FNIII protein, Boi (Brother of Ihog) (Zheng et al., 2010). This is intriguing given the genetic interactions we uncovered between igdb-2 and daf-6, encoding a Patched-related protein. Moreover, Ihog interacts with Hedgehog in vitro and with Patched through its first and second FNIII domains (Zheng et al., 2010), respectively. Similarly, IGDB-2 structure-function analysis showed that the N-terminal membrane-distal domain, which contains its first two FNIII domains, is necessary for IGDB-2 function.
Alternatively, IGDB-2 may regulate the open/closed state of LGC-34, thus mediating ion flux across amphid glia, and possibly glial cell osmolarity. Consistent with this model, igdb-2 lesions appear to enhance dye-filling defects of mutants carrying mutations in CHE-14, a Dispatched-related protein involved in osmoregulation and amphid channel morphogenesis (Michaux et al., 2000). Indeed, the expansion and contraction of the amphid channel could be consistent with changes in fluid accumulation and osmoregulation.
LGC-34 may also be gated by a specific ligand, since other LGC (ligand-gated ion channels of the cys loop superfamily) proteins in C. elegans are neurotransmitter receptors. For example, LGC-55 is a tyramine-gated chloride channel expressed in head neurons and GLR glia, and likely functions in neurons to regulate head movements (Ringstad et al., 2009). In vertebrate glia, ligand-gated ion channels are known to activate downstream signaling cascades (Verkhratsky and Steinhäuser, 2000), providing possible clues for processes activated by LGC-34 and inhibited by IGDB-2.
Based on sequence analysis, LGC-34 is highly divergent from other LGCs and has only an atypical cysteine loop; this is normally involved in ligand-binding and present in most other LGCs (Jones and Sattelle, 2008). Thus, it is also possible that LGC-34, along with IGDB-2, may be acting through a novel mechanism to regulate sensory compartment growth.
Supplementary Material
Highlights.
-
–
IGDB2, a C. elegans Ig/FNIII protein, participates in sensory organ formation.
-
–
IGDB2 functions in sensory organ glia.
-
–
IGDB2 binds the putative ion channel LGC-34.
-
–
IGDB2 antagonizes the function of DAF-6/Patched-related.
Acknowledgments
We thank Shaham lab members for discussions and comments. We thank the Rockefeller University Proteomics and Bio-Imaging Resource Centers for technical support, and Baolin Wang and Howard Baylis for reagents. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). W.W. was supported by NIH Medical Scientist Training Program grant T32GM07739. S.S. was supported by NIH grants HD078703, NS064273 and NS081490.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert PS, Brown SJ, Riddle DL. Sensory control of dauer larva formation in Caenorhabditis elegans. The Journal of comparative neurology. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D. The retinal pigment epithelium: a versatile partner in vision. Journal of cell science. 1993;(Supplement 17):189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- Breipohl W, Laugwitz HJ, Bornfeld N. Topological relations between the dendrites of olfactory sensory cells and sustentacular cells in different vertebrates. An ultrastructural study. Journal of Anatomy. 1974;117:89–94. [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Hing H. The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Developmental Neurobiology. 2008;68:1029–1045. doi: 10.1002/dneu.20644. [DOI] [PubMed] [Google Scholar]
- Colón-Ramos DA, Margeta MA, Shen K. Glia Promote Local Synaptogenesis Through UNC-6 (Netrin) Signaling in C. elegans. Science (New York, NY) 2007;318:103–106. doi: 10.1126/science.1143762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton CM, Johnson CM. Modulation of dopamine-dependent behaviors by the Caenorhabditis elegans Olig homolog HLH-17. Journal of neuroscience research. 2011;89:1627–1636. doi: 10.1002/jnr.22694. [DOI] [PubMed] [Google Scholar]
- Gower NJD, Temple GR, Schein JE, Marra M, Walker DS, Baylis HA. Dissection of the promoter region of the inositol 1,4,5-trisphosphate receptor gene, itr-1, in C. elegans: a molecular basis for cell-specific expression of IP3R isoforms1. Journal of molecular biology. 2001;306:145–157. doi: 10.1006/jmbi.2000.4388. [DOI] [PubMed] [Google Scholar]
- Hansel DE, Eipper BA, Ronnett GV. Neuropeptide Y functions as a neuroproliferative factor. Nature. 2001;410:940–944. doi: 10.1038/35073601. [DOI] [PubMed] [Google Scholar]
- Hegg CC, Irwin M, Lucero MT. Calcium store-mediated signaling in sustentacular cells of the mouse olfactory epithelium. Glia. 2009;57:634–644. doi: 10.1002/glia.20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, Shaham S. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell. 2009;137:344–355. doi: 10.1016/j.cell.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Sattelle DB. The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invertebrate Neuroscience. 2008;8:41–47. doi: 10.1007/s10158-008-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N, Marín O, Koike M, Hirota Y, Uchiyama Y, Wu JY, Lu Q, Tessier-Lavigne M, Alvarez-Buylla A, Okano H, Rubenstein JLR, Sawamoto K. New Neurons Clear the Path of Astrocytic Processes for Their Rapid Migration in the Adult Brain. Neuron. 2010;67:213–223. doi: 10.1016/j.neuron.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem YE, Jeong HJ, Kim HJ, Koh J, Kang K, Bae GU, Cho H, Kang JS. Cdo Regulates Surface Expression of Kir2.1 K+ Channel in Myoblast Differentiation. PloS one. 2016;11:e0158707. doi: 10.1371/journal.pone.0158707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawdsley DJ, Cooper HM, Hogan BM, Cody SH, Lieschke GJ, Heath JK. The Netrin receptor Neogenin is required for neural tube formation and somitogenesis in zebrafish. Developmental biology. 2004;269:302–315. doi: 10.1016/j.ydbio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJM, Kai H, Li D, Mah A, Marra M, Mcghee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Gene Expression Profiling of Cells, Tissues, and Developmental Stages of the Nematode C. elegans. Cold Spring Harbor symposia on quantitative biology. 2003;68:159–170. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G, Gansmuller A, Hindelang C, Labouesse M. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Current Biology. 2000;10:1098–1107. doi: 10.1016/s0960-9822(00)00695-3. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLOS Genetics. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Shaham S. Some, but not all, retromer components promote morphogenesis of C. elegans sensory compartments. Developmental biology. 2012;362:42–49. doi: 10.1016/j.ydbio.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Perens EA, Lu Y, Watanabe S, Jorgensen EM, Shaham S. Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS biology. 2011;9:e1001121. doi: 10.1371/journal.pbio.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Shaham S. The glia of Caenorhabditis elegans. Glia. 2011;59:1253–1263. doi: 10.1002/glia.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perens EA, Shaham S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Developmental cell. 2005;8:893–906. doi: 10.1016/j.devcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Developmental biology. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Abe N, Horvitz HR. Ligand-Gated Chloride Channels Are Receptors for Biogenic Amines in C. elegans. Science. 2009;325:96–100. doi: 10.1126/science.1169243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C, Dikkes P, Liberman MC, Corfas G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. The Journal of comparative neurology. 2002;442:156–162. doi: 10.1002/cne.10085. [DOI] [PubMed] [Google Scholar]
- Shaham S. galign: a tool for rapid genome polymorphism discovery. PloS one. 2009;4:e7188. doi: 10.1371/journal.pone.0007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. Chemosensory organs as models of neuronal synapses. Nature reviews. Neuroscience. 2010;11:212–217. doi: 10.1038/nrn2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nature Communications. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, Liu B, Friedman Christine J, Fong J, Lu Y, Huang XY, Shaham S. A Glial K/Cl Transporter Controls Neuronal Receptive Ending Shape by Chloride Inhibition of an rGC. Cell. 2016;165:936–948. doi: 10.1016/j.cell.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnyk V, Leshchyns’ka I, Schachner M. Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends in neurosciences. 2017;40:295–308. doi: 10.1016/j.tins.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A, Steinhäuser C. Ion channels in glial cells. Brain research reviews. 2000;32:380–412. doi: 10.1016/s0165-0173(99)00093-4. [DOI] [PubMed] [Google Scholar]
- Wallace Sean W, Singhvi A, Liang Y, Lu Y, Shaham S. PROS-1/Prospero Is a Major Regulator of the Glia-Specific Secretome Controlling Sensory-Neuron Shape and Function in C. elegans. Cell Reports. 2016;15:550–562. doi: 10.1016/j.celrep.2016.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode caenorhabditis elegans. The Journal of comparative neurology. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RHA. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature genetics. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zheng X, Mann RK, Sever N, Beachy PA. Genetic and biochemical definition of the Hedgehog receptor. Genes & development. 2010;24:57–71. doi: 10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


