Abstract
Aroclor 1254 (A1254) is the most toxic commercial PCB mixture produced, primarily due to its relatively high concentrations of dioxin-like congeners. This study demonstrates a comparative evaluation of dechlorination of A1254 and PCB-118 by indigenous organohalide respiring bacteria enriched from three PCB impacted sites: Grasse River (GR), NY; Fox River (FR), WI; and Baltimore Harbor (BH), MD. PCB-118 dechlorination rates in GR, BH, and FR was 0.0308, 0.015, and 0.0006 Cl−/biphenyl/day, respectively. A1254 dechlorination rates in GR, FR, and BH were 0.0153, 0.0144, and 0.0048 Cl−/biphenyl/day, respectively. A1254 dechlorination was achieved through the removal of doubly-/singly-flanked chlorines in meta and para positions of mostly penta- followed by hexa- and hepta-chlorinated congeners by 88%, 69%, and 51% in GR, and 88%, 87%, and 83% in FR, respectively, while in BH mostly hepta- (70%) followed by hexa-chlorinated congeners (66%) were dechlorinated. A previously developed Anaerobic Dechlorination Model (ADM) quantified a total of 17 toxicity-related dechlorination pathways in all three sediment microcosms. The toxic equivalency of A1254 based on seven dioxin-like congeners decreased by about 53%, 45% and 21%, in GR, FR and BH microcosms, respectively. The dechlorination products were generally tetra- and tri-chlorinated congeners with unflanked chlorines, all of which is susceptible to further degradation by aerobic bacteria. Concerning the toxic congeners, ADM can be useful to initiate further research focusing on the stimulation of the toxicity reducing pathways for risk assessment and effective remediation strategies.
Keywords: Anaerobic dechlorination, pathway modeling, Aroclor 1254, Polychlorinated biphenyls, dioxin TEF
Graphical abstract
1. Introduction
Polychlorinated biphenyls (PCBs) are ubiquitous environmental contaminants which are persistent, lipophilic and strongly hydrophobic with a high potential for bioaccumulation in living organisms. Environmental PCB exposure is a major health concern due to the toxic, carcinogenic, and endocrine disruptive effects of PCBs [1]. Among the commercial PCB mixtures manufactured in the USA, A1254 had one of the widest applications ranging from coolants for transformer and capacitor oils, ink solvents, pesticide extenders, plasticizers to a variety of adhesives [2]. A1254 is the most toxic of the commercial mixtures with a greater proportion of non- and mono-ortho chlorinated congeners with dioxin-like properties since the total weight percent of toxic compounds in A1254 is between 11.99% (Lot G4 production) and 23.8% by weight (Lot A4 production or Late production). A1254 is 8–16 times more toxic than Aroclor 1248, and about 3–6 times more toxic when compared to Aroclor 1242 and 1260 [3]. Among the PCB congeners of A1254, PCB-118 is the most abundant dioxin-like congener accounting for an average 7% to 13.5% by weight depending on the production lot [3]. Sales record of world biggest PCB producer, the Monsanto U.S., between 1957 and 1974 pointed out that Aroclor 1254 was the second most produced/sold Aroclor mixtures after Aroclor 1242 [4].
One potential mechanism for reducing the risks associated with A1254 is through microbial degradation (bioremediation) since anaerobic and aerobic microorganisms were shown to transform PCBs under a variety of laboratory and environmental conditions [5–8]. Several microorganisms were previously isolated that can degrade PCBs aerobically [9–12], although the aerobic degradation of PCBs is only effective for lightly chlorinated congeners limiting aerobic degradation of A1254 [9, 13]. Typically, only the top few millimeters of sediments are aerobic and the largest reservoirs of PCBs in rivers and lakes are in the anaerobic zones of sediments. Complete degradation of heavily chlorinated PCBs such as A1254 can therefore only occur after anaerobic dechlorination by organohalide respiring bacteria [14]. There are a limited number of studies on dechlorination of A1254 [9, 13, 15, 16] where the impact of use of various inoculum on transformation of this mixture is investigated. In this study, we compared the dechlorination and detoxification of A1254 by using inoculum from sites e.g. GR, FR, and BH with three different PCB contamination histories. An in-depth analysis of biotransformation pathways is essential to better compare and understand the fate of these compounds in the three sets. Individual quantification of anaerobic dechlorination pathways was revealed through a previously developed model ADM [17], which enabled the investigation of toxicity change in microcosms representing each site. Rates and pathways of PCB dechlorination can vary greatly between PCB-impacted sites due to the different populations of indigenous organohalide respiring bacteria [15]. In addition to the dechlorination potential of A1254 and PCB-118 by the indigenous microorganisms, changes in the dechlorinating communities were evaluated by denaturing high pressure liquid chromatography (DHPLC).
2. Materials and Methods
2.1. Chemicals
All PCBs (99–100% purity) were purchased from AccuStandard. PCE was purchased from Sigma-Aldrich. All other chemicals were reagent grade.
2.2. Sediment sampling
Sediments collected with a Ponar grab sampler were stored anaerobically in glass jars sealed with Teflon lined tops at 4°C in the dark prior to use. GR sediment was collected during Spring 2008 from the lower GR in the Village of Massena, NY, US, as described previously [18]. GR was contaminated primarily with A1248 from aluminum production since the 1930s [19]. FR sediment was collected from the Lower Fox River site located in central and northeastern Wisconsin, US, during dredging in Fall 2008 as described previously [20]. FR was contaminated primarily with A1242 from a number of carbonless paper plants along the river [21]. BH sediment was collected in late Spring 2009 from the Northwest Branch of BH a coastal embayment located in a highly urbanized watershed of the Chesapeake Bay, US, as described previously [22]. BH was primarily contaminated with A1260 with smaller amounts of A1254 [22]. Sediments were black in color and had a sulfide odor indicative of reduced anoxic conditions.
2.3. Sediment Microcosm
Low-sulfate (<0.3 mM) estuarine medium [23] prepared without Na2S was anaerobically dispensed as 50-mL aliquots into 160-mL serum bottles and autoclaved at 121°C for 20 minutes. The final pH of the medium was 6.8. All subsequent additions were performed in an anaerobic glove box (Coy Laboratory Products, Ann Arbor, Michigan, USA) containing N2:CO2:H2 (75:20:5). Microcosms were prepared as described previously [24] by adding 10 mL of sediments (GR, FR, and BH) and a fatty acid mixture (acetate, propionate, and butyrate) at a final concentration of 2.5 mM. PCB-118 or A1254 solubilized in acetone were added (0.2%, v/v) to the microcosms at a final concentration of 100 ppm, or 50 ppm, respectively. Microcosms were sealed with 20-mm Teflon-coated butyl stoppers (West Pharmaceutical, Inc.) secured with aluminum crimp seals and incubated statically at 30°C in the dark and sampled immediately after inoculation and subsequently every 30 days.
2.4. PCB extractions and analytical procedures
Microcosms were sampled every 30 days. Triplicate samples for PCBs (AccuStandard, Inc., New Haven, CT) were analyzed by the extraction of 1 mL culture with 5 mL of hexane (Fisher Scientific, PA) on a wrist action shaker (Burrell Corp., PA) overnight according to previously explained method [24]. PCBs 30 and 204 were added as internal standards. Recovery of PCB-166 added as a surrogate was 88±2%. No surrogate recovery correction was performed. Calibration table consisted of 132 congener groups with co-elution, and a total of 172 individual congeners prepared and analyzed as explained before [24]. The PCB concentrations were measured as µg PCB/mL of microcosm slurry and converted to mol%. Total chlorines per biphenyl was calculated as the product of the average number of chlorines and molar concentration of each congener divided by the sum of the total molar concentration of all congeners [24]. The dechlorination rate was calculated within the linear slope of the dechlorination curve by dividing total chlorine removed per biphenyl with the time elapsed in days [11]. Reduction in the total toxic equivalent (TEQ) was calculated based on the toxic equivalent factor for each dioxin-like congener relative to 2,3,7,8-tetrachlorinated dibenzo-p-dioxin as defined by the World Health Organization [25]. The TCDD (2,3,7,8-Tetrachlorodibenzo-p-dioxin) equivalency, i.e. dioxin-like toxicity of each sediment microcosm for time 0 and time final was calculated by multiplying the concentrations of toxic congeners by the TEF (Toxicity Equivalency Factors) values [25].
2.5. DNA extraction and enumeration of PCB dehalorespiring bacteria by qPCR
Triplicate DNA samples was extracted from 0.25 mL of sediment slurry with a 96-well bead-beating plate (MOBIO Laboratories, Inc.) according to the manufacturer’s directions. DNA was eluted in 100 µL of Tris buffer and quantified with a NanoDrop 1000 Spectrophotometer (ThermoScientific). Extracted DNA samples had an A260/280 ratio of ≥1.6 and an A260/230 ratio of ≥2.0.
Putative PCB dechlorinating microorganisms within the organohalide respiring Chloroflexi were enumerated with primers Chl-348F and Dehal-884R targeting the16S rRNA genes [11] as described previously [22]. Amplifications efficiencies of standards were 98±5% with R2=0.999.
2.6. Community analysis of PCB dechlorinating bacteria
PCR amplified 16S rRNA gene analysis was conducted on denaturing high pressure liquid chromatography (DHPLC) using a WAVE 3500 HT system (Transgenomic, Omaha, NE) equipped with a florescence detector (excitation 490 nm, emission 520 nm). Primers 348F/884 were used both for DHPLC following PCR or qPCR products were used where indicated. PCR reactions were performed as described previously [18]. PCR products of the correct length (ca. 500 bp) were confirmed by electrophoresis using a 1.2% (w/v) high-melt agarose gel prior to analysis by DHPLC. The 16S rRNA gene fragments were analyzed according to a previous method [18].
2.7. Anaerobic dechlorination model (ADM)
ADM [17] was utilized to investigate and compare dechlorination pathways occurring in three sediment microcosms. ADM systematically analyzes changes in a contaminant profile that result from microbial reductive dechlorination according to empirically determined dechlorination pathways, while a mol balance between the parent congener being dechlorinated and the product congener being formed is maintained. No aerobic PCB degradation was assumed, and the total mol% of parent congeners at the beginning of dechlorination is equal to the total mol% of all dechlorination by-products. Success of model fit is evaluated by comparing predicted profile to the measured profile with the help of the following parameters: Q, percent improvement in similarity [21], cosine θ, coefficient of proportional similarity [26] and the coefficient of determination, R2 [27]. A good model fit is indicated by cosine θ and R2 values close to 1 and a Q value close to 100% [17]. Relative standard deviation (RSD) values associated with the pathways indicates likeliness of occurrence of a pathway; such that lower RSD values indicate higher likeliness. As a result, any pathway with a quantification value above the median and RSD value lower than 100 was arbitrarily selected as a cut-off criteria during the determination of major dechlorination pathways of each sediment microcosm.
3. Results
3.1. PCB-118 dechlorination
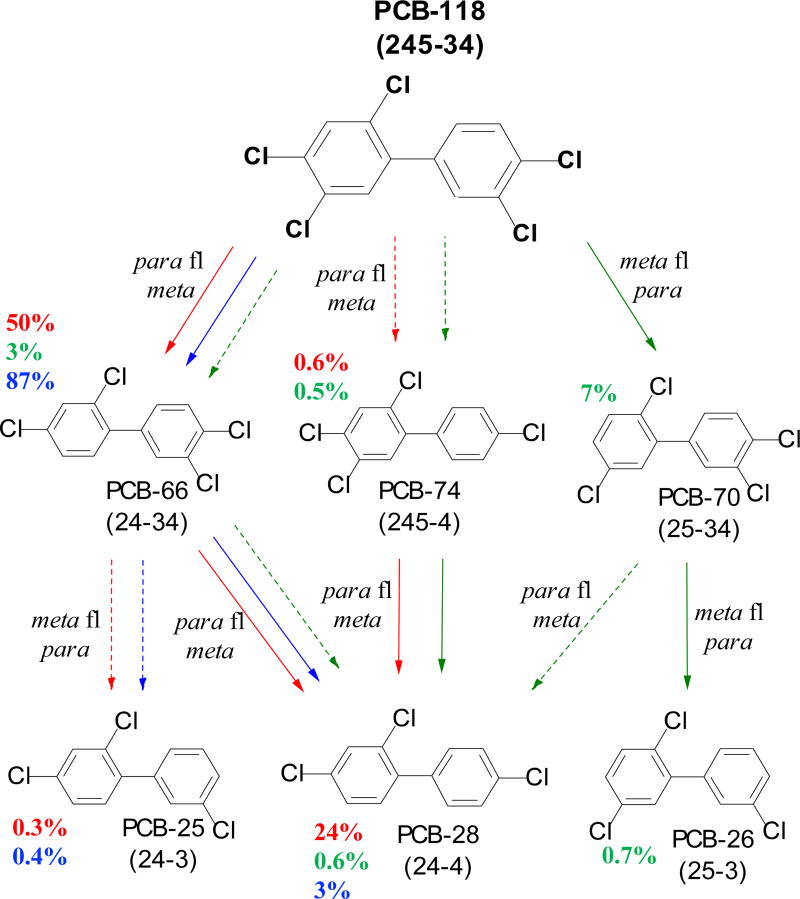
In the GR sediment microcosm, an average 74 mol% PCB-118 was dechlorinated in 30 days primarily to PCB-66 (2,3’,4,4’-CB) by 50 mol% and PCB-28 (2,4,4’-CB) by about 24 mol% via two sequential removal of para-flanked chlorines in the meta position. In contrast, FR sediment culture dechlorinated an average 11 mol% of PCB-118 to mostly PCB-70 (7 mol%) and to lesser extent to PCB-66 (3 mol%) in 180 days, which were subsequently dechlorinated to PCB-26 and PCB-28. The results indicate that the FR culture preferentially targeted meta-flanked para chlorines, but also had some ability to remove para-flanked meta chlorines. After a 120-day lag time in BH microcosms, PCB-118 (90 mol%) was rapidly dechlorinated to PCB-66 by 87 mol% and eventually to PCB-28 (3 mol%) following a similar dechlorination pathway as GR (Figure 1). The dechlorination rates achieved in GR, BH, and FR for PCB-118 were 30.8×10−3, 15×10−3and 0.6×10−3 total Cl− removed per biphenyl per day (Cl−/bp day), respectively.
Figure 1.
Dechlorination pathways of PCB-118 (245-34) in GR (red), FR (green), and BH (blue). Solid arrows indicate main pathways, while dashed arrows show minor pathways. End mol% of each PCB congener are shown for individual sediment microcosms. Chlorine removal mechanisms observed for each sediment microcosm during the dechlorination of PCB-118 and its daughter products are shown next to the arrows.
3.2. Aroclor 1254 dechlorination
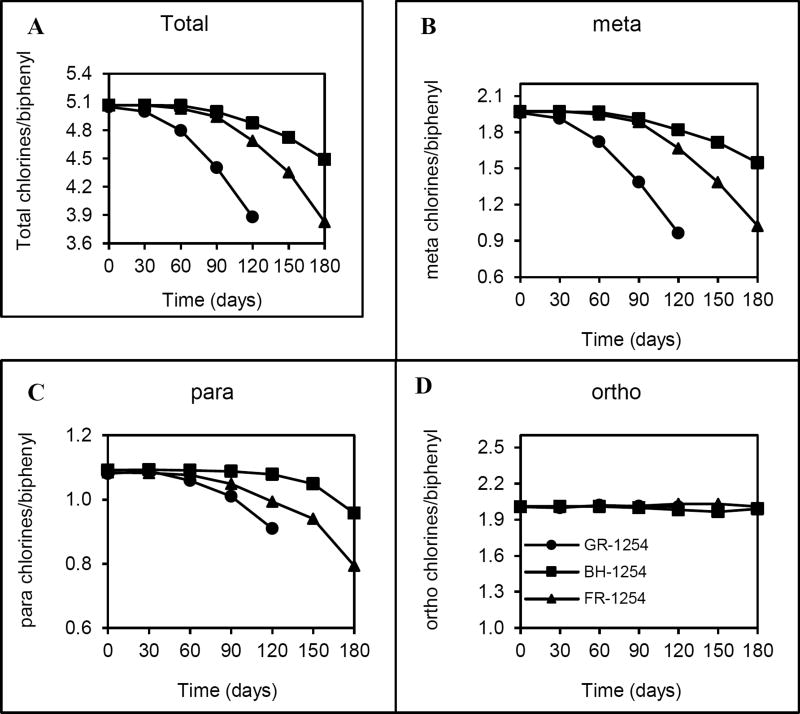
The dechlorination of A1254 resulted in removal of mostly doubly plus singly flanked meta and para chlorines. A1254 dechlorination rates of GR, FR, and BH were 15.3×10−3 Cl−/day, 14.4×10−3 Cl−/day, and 4.8×10−3 Cl−/day (Table 2). Chlorine removal from meta position in GR was higher than the others (Figure 2-A), while more para chlorine was removed in FR microcosms, 26.8% compared to the others (Figure 2-C). No ortho dechlorination (Figure 2-D) and unflanked chlorine removal was observed during the A1254 dechlorination.
Table 2.
Dechlorination characteristics of three sediment microcosms
| Sediment Microcosm |
Dechlorination %
|
Maximum Rate (Cl−/bp day) |
Average Rate (Cl−/bp day) |
|||||
|---|---|---|---|---|---|---|---|---|
| Total chlorine |
ortho chlorine |
meta chlorine |
para chlorine |
Doubly flanked chlorine |
Single flanked chlorine |
|||
| GR | 23.1 | - | 50.8 | 15.8 | 71.4 | 75.1 | 15.3 | 9.7 |
| FR | 24.5 | - | 48.1 | 26.7 | 86.9 | 77.3 | 14.4 | 6.9 |
| BH | 11.4 | - | 21.4 | 12.2 | 66.4 | 35.6 | 4.8 | 3.2 |
-: either not observed or not statistically significant (p>0.05)
Figure 2.
Dechlorination of A1254 with three sediment microcosms (GR, FR, and BH) as total (A), meta (B), para (C), and ortho (D) chlorines/biphenyl over time. Each data point is the mean of three replicate microcosms.
An analysis of homolog distributions observed in GR and FR microcosms revealed that dechlorination of A1254 proceeded mostly through removal of chlorine from penta- (5-CB) followed by hexa- (6-CB) and hepta- (7-CB) chlorinated congeners ranging between 88%, 69%, and 51% for GR, and 88%, 87%, and 83% for FR, respectively (Figure S1). On the other hand, BH microcosms, which contain a much higher percentage of higher chlorinated homologs (Table 1), dechlorinated mostly 7-CB congeners (70%), followed by 6-CB (66%), and 5-CB congeners (40%). The dechlorination products of highly chlorinated congeners resulted in unflanked 4-CB and 3-CB in all three sediment microcosms at varying levels (Table S1).
Table 1.
Analysis of PCB homologues in GR, FR, and BH sediment samples and commercial Aroclor mixtures
3.3. Detailed pathway analysis of Aroclor 1254 dechlorination by ADM
Since all AD pathways identified in the literature depend on the evaluation of initial PCB profiles (e.g. Aroclors) and a final dechlorinated profile, intermediate time points are not evaluated for the identification of AD pathways in this study.
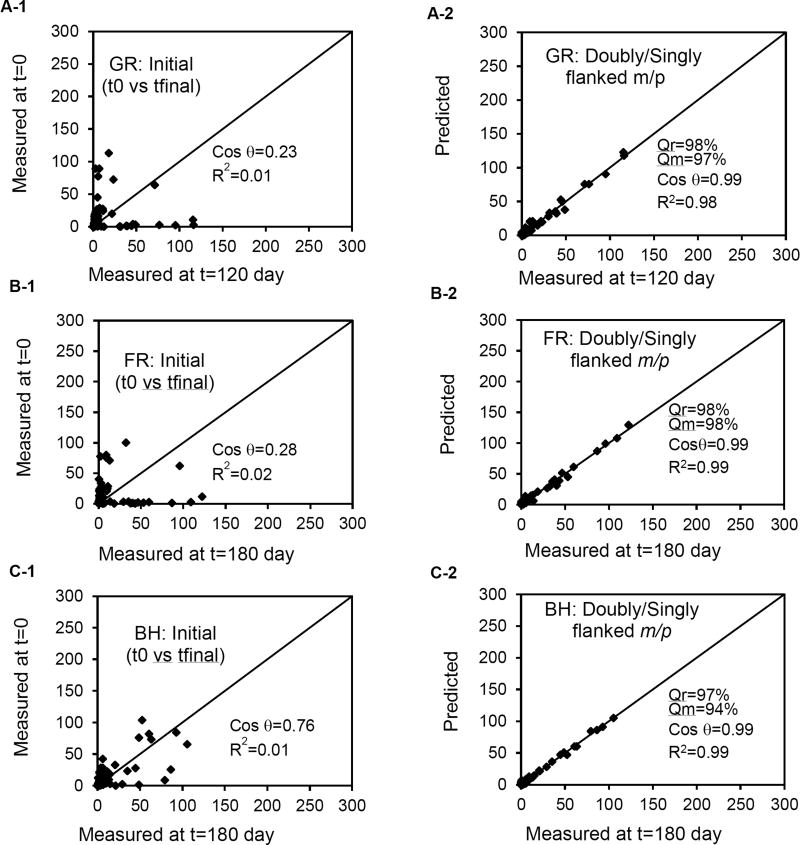
A comparison of the PCB profiles of day 0 and day final of GR (120), FR (180) and BH (180) sediment microcosms indicated that the day 0 PCB profiles of GR and FR were very different from their day final profiles as indicated with very low R2 and Cos θ values (Figure 3A-1, and B-1). In contrast, the PCB profile of BH sediment microcosm at day 0 was much closer to its profile after dechlorination (Figure 3C-1). This is because PCBs in BH sediment microcosms were dechlorinated much less when compared to the others. The goodness of fit results (Figure 3) show that ADM satisfactorily predicts dechlorination pathways undergoing in each sediment microcosm. Accordingly, a detailed pathway analysis is presented here.
Figure 3.
The scatter plot of measured PCB profiles at day final and time 0 for GR (A-1), FR (B-1), and BH (C-1), and the scatter plot of predicted vs measured PCB profiles for doubly and singly flanked meta and para for GR (A-2) FR (B-2) and BH (C-2) by the ADM.
ADM predicted a total of 170 possible pathways for all three microcosms, of which no more than 9 were quantified as zero. The complete list of major dechlorination pathways quantified in the three sediment microcosms is given in Table S2. A comparative evaluation of pathways quantified by ADM yield the following: (1) 44, 59, and 48 pathways were identified as major pathways by the ADM for GR, FR and BH, respectively. (2) 26 out of 70 major pathways quantified by the ADM are common for all three sediments (Table S2). (3) Common pathways indicate doubly/singly flanked meta and singly flanked para Cl removal from the biphenyl, indicating the significance of this dechlorination route. On the other hand, FR microcosms also have a high number of para removal pathways among its major pathways, 21 out of 53. (4) When the three are compared in a pairwise fashion, GR and FR microcosms are observed to share much more common pathways, such as 40 out of 70 major pathways, while only 9 of them were observed in BH microcosms in which para Cl was primarily removed from the biphenyl. As biologically shown (Table 2) and accurately identified by ADM, doubly or singly flanked meta/para chlorines were preferentially dechlorinated in all microcosms (Table S2).
3.4. Analysis of PCB toxicity
Among the toxic PCB congeners [25], PCB-81 (co-elutes with 87), 126, and 169 are not present in A1254 and the production of these congeners would only be possible through an ortho dechlorination which was not observed in any microcosm (Table 2). Therefore, during toxicity assessment, these congeners were not considered. ADM quantified a total of 17 toxicity-related dechlorination pathways in all three sediment microcosms, while 10 of them were identified as major pathway in BH, 8 of these pathways were also the major pathway in GR and FR (shown in bold in Table 3). The pathways resulting in the conversion of a toxic congener to a non-toxic or less toxic congener considered for toxicity reduction. The dioxin like toxicity of GR, FR and BH microcosms decreased significantly in time by about 53%, 45% and 21%, respectively.
Table 3.
Dechlorination pathways of toxic congenersa identified by WHO [25] quantified by ADM in each sediment microcosms together with their quantification values (mole ‰)
| Dechlorination pathway | Average ± Standard deviation (mole‰)
|
||
|---|---|---|---|
| GR | FR | BH | |
| PCB 77 (34-34) --> PCB 35 (34-3) | 14.47±14.25 | 13.29±13.34 | 4.06±4.06 |
| PCB 77 (34-34) --> PCB 37 (34-4) | 18.22±17.51 | 10.22±10.27 | 5.45±4.43 |
| PCB 105 (234-34) --> PCB 66 (24-34) | 10.86±4.41 | 10.2±4.43 | 16.58±6.53 |
| PCB 114 (2345-4) --> PCB 63 (235-4) | 1.53±1.24 | 1.74±1.35 | 0.44±0.51 |
| PCB 114 (2345-4) --> PCB 74 (245-4) | 1.18±1.2 | 0.85±1.29 | 1.49±1.19 |
| PCB 118 (245-34) --> PCB 66 (24-34) | 30.57±16.47 | 16.64±11.75 | 31.26±15.94 |
| PCB 118 (245-34) --> PCB 67 (245-3) | 21.02±16.5 | 17.71±10.78 | 8.1±10.46 |
| PCB 118 (245-34) --> PCB 70 (25-34) | 24.68±15.5 | 22.42±10.58 | 12.96±11.58 |
| PCB 118 (245-34) --> PCB 74 (245-4) | 28.31±18.03 | 19.4±11.77 | 13.6±10.1 |
| PCB 123 (345-24) --> PCB 66 (24-34) b, g | 10.01±6.36 | 4.28±4.37 | 12.66±10.01 |
| PCB 123 (345-24) --> PCB 68 (24-35) | 3.71±3.04 | 4.16±3.37 | 3.72±4.26 |
| PCB 156 (2345-34) --> PCB 107 (235-34) b, f | 6.4±1.42 | 9.78±1.43 | 5.12±1.84 |
| PCB 156 (2345-34) --> PCB 118 (245-34) b, c | 4.81±2.43 | 3.6±1.44 | 6.93±1.86 |
| PCB 157 (234-345) --> PCB 105 (234-34) c | 0.2±0.38 | 0.07±0.22 | 0.24±0.43 |
| PCB 157 (234-345) --> PCB 108 (234-35) | 0.26±0.44 | 0.57±0.66 | 0.27±0.44 |
| PCB 157 (234-345) --> PCB 122 (345-23) | 0.32±0.48 | 0.49±0.62 | 0.2±0.35 |
| PCB 157 (234-345) --> PCB 123 (345-24) | 0.4±1.28 | 0.4±0.51 | 0.43±0.49 |
Pathways in bold are the major pathways in all microcosms.
Among the toxic congeners other than listed here, 81, 126 and 169 are not present in Aroclor 1254, and also 169 was not in the analysis method, therefore, they are not quantified by the model.
Major pathways seen in BH, GR, and FR microcosms, respectively.
Pathways resulting in no change in toxicity.
3.5. Enumeration and community analysis of dechlorinating phylotypes
In all sediment microcosms, the number of putative dechlorinators increased (Figure S2A). The highest increase, 38-fold from (1.58±0.38) ×10+5 to (7.63±0.25) ×10+6, was seen in GR sediment microcosms amended with PCB-118, while the increase was about 25-fold in GR microcosms with A1254. The increase in the number of putative organohalide respiring bacteria in FR and BH microcosms during PCB-118 dechlorination was lower than GR microcosm with 16.3 and 24-folds, respectively.
DHPLC analysis of day zero and day final samples of all three sediment microcosms spiked with A1254 and PCB-118 are given in Figure S3. Each peak theoretically is the 16s rRNA gene amplified from a unique microorganism among the population; the peak height semi-quantitatively shows the abundance of that 16s rRNA gene in the population. In all of the microcosms the initial community diversity was high (about 6–10 apparent different organisms), and the community was distinct for each sediment. Microbial diversity of GR sediment microcosms (PCB-118 or A1254) changed from day 0 to day 120 (final day) and some of the phylotypes present initially were enriched, but the enriched phylotypes in GR A1254 and PCB-118 microcosms were different from each other (Figure S3A and B). Similarly, in FR microcosms, the same organisms were apparently enriched in both PCB-118 and A1254 (Figure S3C and D). However, in BH microcosms, the presence of A1254 did not enhance an enrichment of any particular members of the BH community (Figure S3E and F).
4. Discussion
PCB-118 was rapidly dechlorinated in GR while there was a 120 day of lag in BH microcosms. Similar results were reported by Fagervold et al. [28] for BH culture in initial microcosms. Accordingly, PCB-118 was one of the top congeners dechlorinated in A1254 (Table S1). On the other hand, dechlorination of PCB-118 in FR microcosms was comparatively lower and slower, with 11% dechlorination in 180 days. Interestingly, PCB-118 as a congener in A1254 mixture in FR microcosms dechlorinated very highly indicating the stimulatory effect of the presence of multiple congeners [15, 28–30]. Similarly, as reported in that study [28], the presence of specific or multiple congeners in Aroclor 1260 promoted the reductive dechlorination of PCB 194 by the BH culture.
None of the studied sediments indicated ortho dechlorination and unflanked chlorine removal during the A1254 dechlorination. Different sediments and sediment conditions exhibit distinct dechlorination preferred patterns, or processes [31–34]. The processes involve stepwise dechlorination that removes para- and meta-chlorines and leaves predominantly lightly chlorinated ortho compounds as a result [35]. Based on dechlorination processes, some of the factors that may influence which chlorine on a biphenyl will be subjected to dechlorination include: 1) the position of the chlorine (ortho, meta or para), 2) the surrounding configuration of chlorines (unflanked, single-flanked or double-flanked), 3) the chlorine configuration of the opposite ring, 4) environmental conditions and 5) the microbial populations present [34]. Double-flanked chlorines are generally dechlorinated first even though these reactions produce the least amount of energy. The preference for double and then single flanked chlorines might be explained based on the chemistry of chlorinated biphenyls. Microbial reductive dechlorination has been anticipated as a two-step process, first, an electron is transferred to the chlorinated biphenyl and a carbanion intermediate is produced [36]. The negative charge is stabilized by resonance throughout the biphenyl molecule and the surrounding chlorine atoms. The capability of the molecule to stabilize through resonance also effects the overall reactivity, or standard potential (E°), of different PCB congeners. Generally, higher chlorinated congeners have higher E° values and reactivity in environments with low redox potential. Additionally, PCB molecules with ortho chlorines are less planar, have lower E° values, and are chemically less reactive [37, 38]. The reactivity of a specific PCB chlorine is dependent upon both the chemical properties of the congener and catalytic properties of the microbes [39, 40].
Ortho dechlorination capable of targeting PCB congener with two and more ortho-chlorines has been reported previously [41–44]. Rare ortho dechlorination had been reported in Woods Pond and Baltimore Harbor sediments; the Baltimore Harbor estuarine sediment contains the dechlorinator ortho-17 (o-17) which is capable of targeted ortho dechlorination [41, 43–45]. In a recent study, in marine sediment collected from Hunters Point California, PCB 116 which have both meta- and para-chlorines dechlorination was reported via a stepwise removal of the two ortho-chlorines resulting in the accumulation of only one product, PCB 14 [46]. Ortho dechlorination was also observed in freshwater sediments from Woods Pond and Silver Lake where unflanked ortho-chlorine from PCB 30 (2,4,6-CB) was removed [40]. Recently, a rare ortho dechlorination, targeting mono-ortho PCB congeners was observed in Grasse sediment preferentially targeting congeners with a single unflanked ortho-chlorine atom and unflanked meta- and/or para-chlorines [47].
ADM successfully predicted terminal products of dechlorination of A1254. Typically, congeners having less than four chlorines and mostly unflanked, namely, PCB 53, 16/32, 17, 51, 49, 52/43, 47, 25, 26, 27, 28 were observed to be accumulated mostly in FR and GR microcosms at day 180 and 120, respectively. In addition to these terminal products, accumulation of several other PCB congeners such as PCB 66, 44, 70/76 and 95 were also observed in BH microcosms, which might have resulted from the lower dechlorination rate. The dechlorination patterns obtained suggested that the FR microorganisms were more capable than the GR and BH organisms of removing the para chlorine. Previously, Rhee et al. [16] reported about 38% of total chlorine removal during A1254 dechlorination resulting in PCB 8/5, 31/28, and 47/48 as major end products after 24 months of incubation. In the study of Quensen et al. [15], after more than 6 months of incubation, about 63% meta plus para chlorines were removed and the major products reported were PCB 1 and 4/10. In both studies, inoculum obtained from Hudson River but from different locations on Hudson River. These results suggest that there were possibly different PCB-dechlorinating microorganisms with distinctive specificities for PCB dechlorination at different sites.
The activity differences of the sediment cultures might be related to contamination history of the sediments. The findings indicated that active indigenous PCB dechlorinating communities were present at all three PCB-impacted sites and that a significantly higher dechlorination rate detected in GR was linked with an enriched dechlorinating population. Surprisingly, enumeration of the putative organohalide respiring bacteria in sediments of the three sites showed that initial high numbers of putative dechlorinating bacteria do not always lead to a higher dechlorination activity (i.e. dechlorination rate). Specifically, the initial number of putative dechlorinating bacteria in BH and FR was about 10-fold higher than that of GR sediment. As was explained in the previous sections, GR microcosms had the highest dechlorination activity with a shorter lag time and a greater extent of dechlorination when compared to the others. The number of putative organohalide respiring bacteria did not appear to influence the observed differences in the activities at these three sites which were in accordance with the findings of Kjellerup et al. [18].
As stated by May et al. [29] individual congeners can influence (either by stimulation or inhibition) the activity of individual PCB dechlorinating bacteria. Kjellerrup et al. [18] suggested that different congener profiles could lead to the selection of different dechlorinating communities. Moreover, Quensen et al. [15] also showed that Silver Lake sediment inoculum which was contaminated primarily with Aroclor 1260 exhibited shorter lag time and more rapid dechlorination of Aroclor 1260 than Hudson River. Accordingly, different dechlorination activities might be selected depending on the particular Aroclor present at a site. For example, GR was contaminated with Aroclor 1248 [19], while the major PCB source to Fox River sediments was Aroclor 1242 [21], and Aroclor 1260 for BH [22]. As seen from Table 1, analysis of PCBs in GR and FR sediments showed that their homologue profiles were similar, whereas BH was significantly different. This might explain why the preference of BH sediment culture dechlorinating selectively highly chlorinated (7-CB and 6-CB) PCB congeners that were different than GR and FR as well as its lower dechlorination rate. Historical contamination of the BH sediments with Aroclor 1260 most probably enabled the enrichment of bacterial culture capable with a preference towards highly chlorinated congeners. Since FR and GR had similar site and PCB contamination profiles, PCB congener preference of the cultures from these sites was also similar. It is likely that PCB congener profiles and properties of contaminated sites promoted the diversity, selection and enrichment of dechlorinating culture in these sediment microcosms.
Although the total A1254 dechlorination of FR and GR is close to each other (about 23–24%), their Cl removal patterns showed differences. For example, para- and doubly flanked-chlorine removal is a bigger portion for FR when compared to GR. On the other hand, the maximum and average dechlorination rate was higher in GR than that of FR. Higher intrinsic total PCB concentration of GR (Table 1) possibly had an influence on the rate. The concentration of PCB congeners accessible for dechlorination and the concentration of potential electron donors were possibly higher in GR sediments favoring enrichment of the PCB dechlorinating phylotypes as higher concentrations of inherent PCBs was associated with selection of a more active PCB dechlorinating population in GR [18]. Moreover, the lag time exerted by BH sediment culture during PCB-118 dechlorination was very long compared to the other sites. This possibly indicated the presence of slow and possibly different microbial communities as microbial populations have an important effect on dechlorination capability. Also, a 3 to 6-month lag phase was observed in dechlorination of Aroclor 1260 in initial microcosms inoculated with BH sediment and the lag time decreased to less than 50 days only after the fourth transfer [28]. This observation shows that BH culture is not very active and needed to be enriched by transferring several times. Similarly, PCB-53, 51, 52, 49, 47, and 15/17 were reported as end products by Fagervold et al. [28] which are among the top terminal products of this study (Table S1). Also, similar to BH microcosm results of this study, an increase in PCB 66/95/93/102/88, 79/99/113 and 67/100 was observed in the previous study, indicating preference of BH culture towards higher chlorinated congeners that dechlorinates to these relatively lower chlorinated ones.
ADM provided an in-depth analysis of all dechlorination pathways observed in each sediment microcosm by identifying and quantifying a large number of possible dechlorination pathways. ADM is a helpful tool to identify toxicity reducing pathways, especially, those concerning toxic congeners 77, 105, 118, 156, 157, 167. Regarding the toxic congeners, model can be benefited to initiate further biological studies focusing on the stimulation of these specific pathways by this way, the toxicity reducing pathways could be promoted in situ for effective bioremediation.
According to DHPLC chromatograms, one phylotype was responsible for all of the activity in GR and FR microcosms. Similar to GR, in FR sediment microcosm, the initial community was enriched for one or more dominant phylotypes by PCB-118 or A1254 (Figure S3). This suggests that the organisms enriched were the organisms responsible for the dechlorination observed. In BH microcosms, one or more phylotypes were enriched by amendment with PCB-118 (Figure S3F) suggesting that the enriched microorganisms were responsible for the dechlorination of PCB-118 and its intermediates. However, the presence of A1254 did not necessarily enrich any particular members of the BH community (Figure S3F). This observation confirms the low dechlorination activity of BH sediment microcosms amended with A1254 when compared to those of in GR and FR sediment microcosms. Each sediment sample was isolated from different sites with their own contamination histories and site characteristics, therefore, the existence of different dechlorination patterns suggests that there were different species or strains of microorganisms dechlorinating PCBs in these three sediments, each with its own specificity. The complementary nature of the specificities observed for these three sediments implies that greater overall dechlorination might be achieved by combining these cultures.
Supplementary Material
HIGHLIGHTS.
Anaerobic dechlorination model determined Aroclor 1254 dechlorination pathways
Major toxicity reducing pathways were determined in three sediment cultures
Dioxin-like toxicity was reduced by 53%, 45%, 21% in GR, FR, BH, respectively
Dechlorination end products were unflanked tri- & tetra-chlorinated biphenyls
Acknowledgments
We thank U. Ghosh for providing the sediments, R. Payne for his help with PCB congener analysis. This work was supported by in part by the National Institute of Environmental Health Science Superfund Research Program (5R01ES16197-2) and US Department of Defense Strategic Environmental Research & Development Program (ER-1492). Devrim Kaya was supported by the Scientific and Technical Research Council of Turkey (TUBITAK- BIDEB: 2214).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting information available
Supplementary data related to this article can be found. Figure S1 shows homolog chlorine distributions of A1254 in GR (A), FR (B), and BH (C) microcosms during the incubation period, Table S1 shows the summary of highly dechlorinated and accumulated PCB congeners during A1254 dechlorination in GR, FR, and BH sediment microcosms, Table S2 shows major dechlorination pathways quantified by the ADM in GR, FR, and BH microcosms and their quantification values (mole ‰), Figure S2 shows changes in number of 16S gene copies over time in GR, FR, and BH microcosms (A) and Agarose gel showing the qPCR performed for PCB dechlorinating bacterium DF-1 and slurry samples of each sediment microcosms (B), and Figure S3 shows Results of DHPLC community analysis of putative dechlorinating Chloroflexi 16S rRNA genes in GR microcosms spiked with A) PCB-118 or B) A1254, in FR microcosms spiked with C) PCB-118 or D) A1254, and in BH microcosms spiked with E) PCB-118 or F) Aroclor1254 at day 0 and day final. The possible phylotypes are labeled as P1, P2, P3, P4, P5, and P6 in a) GR-118.
References
- 1.USEPA. Methodology for evaluating potential carcinogenicity in support of reportable quantity adjustments pursuant to CERCLA section 102. Washington, DC: 1988. [Google Scholar]
- 2.Erickson MD. Analytical Chemistry of PCBs. CRC-Lewis Publishers; Boca Raton, New York: 1997. [Google Scholar]
- 3.Frame GM, Wagner RE, Carnahan JC, Brown JF, May RJ, Smullen LA, Bedard DL. Comprehensive, quantitative, congener-specific analyses of eight aroclors and complete PCB congener assignments on DB-1 capillary GC columns. Chemosphere. 1996;33:603–623. [Google Scholar]
- 4.Johnson GW, Quensen JF, III, Chiarenzelli J, Hamilton C. Environmental Forensics: A Contaminant Specific Guide. Elsevier; Amsterdam: 2006. Polychlorinated Biphenyls; pp. 187–225. [Google Scholar]
- 5.Wu Q, Sowers KR, May HD. Microbial reductive dechlorination of aroclor 1260 in anaerobic slurries of estuarine sediments. Appl. Environ. Microbiol. 1998;64:1052–1058. doi: 10.1128/aem.64.3.1052-1058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tartakovsky B, Michotte A, Cadieux JCA, Lau PCK, Hawari J, Guiot SR. Degradation of Aroclor 1242 in a single-stage coupled anaerobic/aerobic bioreactor. Wat. Res. 2001;35:4323–4330. doi: 10.1016/s0043-1354(01)00175-0. [DOI] [PubMed] [Google Scholar]
- 7.Fava F, Zanaroli G, Young LY. Microbial reductive dechlorination of pre-existing PCBs and spiked 2,3,4,5,6-pentachlorobiphenyl in anaerobic slurries of a contaminated sediment of Venice Lagoon (Italy) FEMS Microbiol. Ecol. 2003;44:309–318. doi: 10.1016/S0168-6496(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 8.Tharakan J, Tomlinson D, Addagada A, Shafagati A. Biotransformation of PCBs in Contaminated Sludge: Potential for Novel Biological Technologies. Engineering in Life Sciences. 2006;6:43–50. [Google Scholar]
- 9.Yadav JS, Quensen JF, 3rd, Tiedje JM, Reddy CA. Degradation of polychlorinated biphenyl mixtures (Aroclors 1242, 1254, and 1260) by the white rot fungus Phanerochaete chrysosporium as evidenced by congener-specific analysis. Appl. Environ. Microbiol. 1995;61:2560–2565. doi: 10.1128/aem.61.7.2560-2565.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohn WW, Tiedje JM. Microbial reductive dehalogenation. Microbiol. Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagervold SK, Watts JE, May HD, Sowers KR. Sequential reductive dechlorination of meta-chlorinated polychlorinated biphenyl congeners in sediment microcosms by two different Chloroflexi phylotypes. Appl. Environ. Microbiol. 2005;71:8085–8090. doi: 10.1128/AEM.71.12.8085-8090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba D, Yoshida N, Katayama A. Effects of inhibitors on anaerobic microbial consortium with enhanced dechlorination activity in polychlorinated biphenyl mixture. J Biosci Bioeng. 2007;104:268–274. doi: 10.1263/jbb.104.268. [DOI] [PubMed] [Google Scholar]
- 13.Bedard DL, Wagner RE, Brennan MJ, Haberl ML, Brown JF., Jr Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 1987;53:1094–1102. doi: 10.1128/aem.53.5.1094-1102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegel J, Wu Q. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol Ecol. 2000;32:1–15. doi: 10.1111/j.1574-6941.2000.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 15.Quensen JF, Boyd SA, Tiedje JM. Dechlorination of Four Commercial Polychlorinated Biphenyl Mixtures (Aroclors) by Anaerobic Microorganisms from Sediments. Appl. Environ. Microbiol. 1990;56:2360–2369. doi: 10.1128/aem.56.8.2360-2369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee GY, Sokol RC, Bethoney CM, Bush B. A Long-Term Study of Anaerobic Dechlorination of Pcb Congeners by Sediment Microorganisms - Pathways and Mass-Balance. Environ. Toxicol. Chem. 1993;12:1829–1834. [Google Scholar]
- 17.Demirtepe H, Kjellerup B, Sowers KR, Imamoglu I. Evaluation of PCB dechlorination pathways in anaerobic sediment microcosms using an anaerobic dechlorination model. J Hazard Mater. 2015;296:120–127. doi: 10.1016/j.jhazmat.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Kjellerup BV, Sun X, Ghosh U, May HD, Sowers KR. Site-specific microbial communities in three PCB-impacted sediments are associated with different in situ dechlorinating activities. Environ Microbiol. 2008;10:1296–1309. doi: 10.1111/j.1462-2920.2007.01543.x. [DOI] [PubMed] [Google Scholar]
- 19.USEPA. USEPA. NY: 2014. Grasse River Superfund Site Community Involvement Plan for Remedial Design and Remedial Action. 2014. [Google Scholar]
- 20.Chun CL, Payne RB, Sowers KR, May HD. Electrical stimulation of microbial PCB degradation in sediment. Wat. Res. 2013;47:141–152. doi: 10.1016/j.watres.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imamoglu I, Li K, Christensen ER, McMullin JK. Sources and dechlorination of polychlorinated biphenyl congeners in the sediments of Fox River, Wisconsin. Environ. Sci. Technol. 2004;38:2574–2583. doi: 10.1021/es035165x. [DOI] [PubMed] [Google Scholar]
- 22.Payne RB, May HD, Sowers KR. Enhanced reductive dechlorination of polychlorinated biphenyl impacted sediment by bioaugmentation with a dehalorespiring bacterium. Environ Sci Technol. 2011;45:8772–8779. doi: 10.1021/es201553c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkaw M, Sowers KR, May HD. Anaerobic ortho Dechlorination of Polychlorinated Biphenyls by Estuarine Sediments from Baltimore Harbor. Appl. Environ. Microbiol. 1996;62:2534–2539. doi: 10.1128/aem.62.7.2534-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaya D, Imamoglu I, Sanin FD, Payne RB, Sowers KR. Potential risk reduction of Aroclor 1254 by microbial dechlorination in anaerobic Grasse River sediment microcosms. J. Haz. Mat. 2017;321:879–887. doi: 10.1016/j.jhazmat.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE. The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicological sciences: an official journal of the Society of Toxicology. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis JC. Statistics and Data Analysis in Geology. John Wiley & Sons; New York: 2002. [Google Scholar]
- 27.Manly BFJ. Statistics for environmental science and management. Boca Raton: Chapman & Hall/CRC; 2009. [Google Scholar]
- 28.Fagervold SK, May HD, Sowers KR. Microbial reductive dechlorination of aroclor 1260 in Baltimore harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl. Environ. Microbiol. 2007;73:3009–3018. doi: 10.1128/AEM.02958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May HD, Cutter LA, Miller GS, Milliken CE, Watts JE, Sowers KR. Stimulatory and inhibitory effects of organohalides on the dehalogenating activities of PCB-dechlorinating bacterium o-17. Environ Sci Technol. 2006;40:5704–5709. doi: 10.1021/es052521y. [DOI] [PubMed] [Google Scholar]
- 30.Bedard DL, Van Dort H, Deweerd KA. Brominated Biphenyls Prime Extensive Microbial Reductive Dehalogenation of Aroclor 1260 in Housatonic River Sediment. Appl. Environ. Microbiol. 1998;64:1786–1795. doi: 10.1128/aem.64.5.1786-1795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudkova V, Demnerova K, Bedard DL. Sediment-free anaerobic microbial enrichments with novel dechlorinating activity against highly chlorinated commercial PCBs. J. Chem. Technol. Biotechnol. 2012;87:1254. [Google Scholar]
- 32.Hughes AS, Vanbriesen JM, Small MJ. Identification of structural properties associated with polychlorinated biphenyl dechlorination processes. Environ. Sci. Technol. 2010;44:2842. doi: 10.1021/es902109w. [DOI] [PubMed] [Google Scholar]
- 33.Bedard DL, Pohl EA, Bailey JJ, Murphy A. Characterization of the PCB substrate range of microbial dechlorination process LP. Environ. Sci. Technol. 2005;39:6831. doi: 10.1021/es050255i. [DOI] [PubMed] [Google Scholar]
- 34.Bedard DL, Quensen JF. Microbial reductive dechlorination of polychlorinated biphenyls. In: Young LY, Cerniglia CE, editors. Microbial transformation and degradation of toxic organic chemicals. A. John Wiley; New York: 1995. pp. 127–216. [Google Scholar]
- 35.Wiegel J, Wu QZ. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol. 2000;32:1. doi: 10.1111/j.1574-6941.2000.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 36.Nies L, Vogel TM. Identification of the proton source for the microbial reductive dechlorination of 2,3,4,5,6-pentachlorobiphenyl. Appl. Environ. Microbiol. 1991;57:2771–2774. doi: 10.1128/aem.57.9.2771-2774.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connors TF, Rusling JF, Owlia A. Determination of standard potentials and electron-transfer rates for halobiphenyls from electrocatalytic data. Anal. Chem. 1985;57:170–174. doi: 10.1021/ac00279a042. [DOI] [PubMed] [Google Scholar]
- 38.Rusling JF, Miaw CL. Kinetic estimation of standard reduction potential of polyhalogenated biphenyls. Environ. Sci. Technol. 1989;23:476–479. [Google Scholar]
- 39.Brown JJF, Wagner RE, Feng H, Bedard DL, Brennan MJ, Carnahan JC, May RJ. Environmental dechlorination of PCBs. Environ. Toxicol. Chem. 1987;6:579–593. [Google Scholar]
- 40.Williams WA. Microbial reductive dechlorination of trichlorobiphenyls in anaerobic sediment slurries. Environ. Sci. Technol. 1994;28:630. doi: 10.1021/es00053a015. [DOI] [PubMed] [Google Scholar]
- 41.Cutter LA, Watts JEM, Sowers KR, May HD. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ. Microbiol. 2001;3:699. doi: 10.1046/j.1462-2920.2001.00246.x. [DOI] [PubMed] [Google Scholar]
- 42.Pulliam Holoman TR, Elberson MA, Cutter LA, May HD, Sowers KR. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 1998;64:3359. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutter L, Sowers KR, May HD. Microbial dechlorination of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl. Environ. Microbiol. 1998;64:2966. doi: 10.1128/aem.64.8.2966-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandort HM, Bedard DL. Reductive ortho-dechlorination and meta-dechlorination of a polychlorinated biphenyl congener by anaerobic microorganisms. Appl. Environ. Microbiol. 1991;57:1576. doi: 10.1128/aem.57.5.1576-1578.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkaw M, Sowers KR, May HD. Anaerobic ortho dechlorination of polychlorinated biphenyls by estuarine sediments from Baltimore Harbor. Appl. Environ. Microbiol. 1996;62:2534. doi: 10.1128/aem.62.7.2534-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kjellerup BV, Naff C, Edwards SJ, Ghosh U, Baker JE, Sowers KR. Effects of activated carbon on reductive dechlorination of PCBs by organohalide respiring bacteria indigenous to sediments. Wat. Res. 2014;52:1. doi: 10.1016/j.watres.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, Gregory KB, VanBriesen JM. Microbial-Catalyzed Reductive Dechlorination of Polychlorinated Biphenyls in Hudson and Grasse River Sediment Microcosms: Determination of Dechlorination Preferences and Identification of Rare Ortho Removal Pathways. Environ. Sci. Technol. 2016;50:12767–12778. doi: 10.1021/acs.est.6b03892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.