Abstract
Identification of the primary allele(s) in HLA class II associated diseases remains challenging because of a tight linkage between alleles of HLA-DR and -DQ loci. In the present study, we determined the genotypes of seven HLA loci (HLA-A, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1) for 1200 Japanese patients with primary biliary cholangitis and 1196 controls. Observation of recombination derivatives facilitated an evaluation of the effects of individual HLA alleles consisting of disease-prone/disease-resistant HLA haplotypes. Consequently, a primary contribution of DQB1*06:04 (odds ratio: 0.19, p = 1.91 × 10−22), DQB1*03:01 (odds ratio: 0.50, p = 6.76 × 10−10), DRB1*08:03 (odds ratio: 1.75, p = 1.01 × 10−7) and DQB1*04:01 (odds ratio: 1.50, p = 9.20 × 10−6) was suggested. Epistasis of the protective DQB1*06:04 to risk conferred by DRB1*08:03 was demonstrated by subpopulation analysis, implicating the presence of an active immunological mechanism that alleviates pathogenic autoimmune reactions. Further, the contribution of the aforementioned HLA alleles as well as an HLA-DP allele, DPB1*02:01 to the association signals of 304 loci among 4103 SNPs in the HLA region at the genome-wide level of significance (p values less than 5 × 10−8) was demonstrated by the stepwise exclusion of the individuals possessing these HLA alleles from the comparison.
Introduction
Primary biliary cholangitis (PBC) is a relatively rare disease that is predominantly observed in middle-age women. It is characterized by chronic and progressive destruction of intra-hepatic bile ducts and cholestasis1. Several lines of evidence have demonstrated that autoimmunity contributes to the development of PBC, as a consequence of the breakdown of immunological tolerance to autologous antigens. These include pathognomonic antibodies against mitochondrial components (anti-mitochondrial antibodies, AMA) produced very early in the disease process2, the involvement of cell-mediated immunity as suggested by histological findings showing the accumulation and activation of immune competent cells in the portal area of the liver and cellular responses to autologous antigens in in vitro studies3–5, and the fact that patients often experience a wide spectrum of autoimmune disorders such as Sjögren’s syndrome, autoimmune hepatitis, Hashimoto’s thyroiditis, rheumatoid arthritis, and systemic sclerosis (including limited cutaneous systemic sclerosis, formerly known as CREST syndrome)6, 7. As is the case in other autoimmune disorders, PBC has been associated with HLA polymorphisms8–13, and in most of these conditions, the impact of specific HLA alleles on the antigenic repertoire of effector cells was suggested as a mechanism underlying autoimmunity.
Genome-wide association studies (GWAS) of PBC in different populations have revealed the involvement of genetically determined alterations in certain immunological pathways, such as those related to IL12 signal transduction, TNF/TLR signal transduction, and B cell differentiation to plasma cells. However, most of these genes have not been universally identified in studies thus far, with the exception of polymorphic markers in the HLA region14–16. One of the outstanding features of HLA genes is that they exhibit the highest degree of polymorphism among human functional genes. Hundreds to thousands of alleles have been identified at the loci encoding HLA class I (HLA-A, -B, and -C) and class II (HLA-DR, -DQ, and -DP) molecules, some of which exist in particular preferential combinations known as “common HLA haplotypes” in a relatively ethnicity-specific manner. In the present study, we examined the effects of HLA polymorphisms on the development of PBC and demonstrated that multiple HLA alleles show highly significant genome wide-association signals for single-nucleotide polymorphisms (SNPs) in the HLA region.
Results
Clinical characteristics of the study population
This study enrolled 1200 Japanese patients with PBC (Table 1). A female predominance was observed, with a female to male ratio of 7.63. The majority of patients (71.6%) did not progress beyond clinical stage I by the time of their latest clinical evaluation. Patients in the clinical stage III group included 112 cases who had undergone liver transplantation (9.4%) and 8 cases who died of progression to hepatic failure (0.7%). Clinical and histological staging did not differ between genders.
Table 1.
Basic characteristics of the patients with PBC.
| All patients | Female | Male | Female vs. Male | |
|---|---|---|---|---|
| Total number | n = 1200a | n = 1060 | n = 139 | female: male = 7.63:1 |
| Age of onset (mean years ± SD) | 57 ± 12 | 57 ± 12 | 60 ± 11 | ns |
| Liver biopsy | ||||
| Scheuer 0 | 9/808 (1.1%) | 8/714 (1.1%) | 1/94 (1.1%) | ns |
| Scheuer 1 | 424/808 (52.5%) | 376/714 (52.7%) | 48/94 (51.1%) | ns |
| Scheuer 2 | 223/808 (27.6%) | 197/714 (27.6%) | 26/94 (27.7%) | ns |
| Scheuer 3 | 80/808 (9.9%) | 68/714 (9.5%) | 12/94 (12.8%) | ns |
| Scheuer 4 | 72/808 (8.9%) | 65/714 (9.1%) | 7/94 (7.4%) | ns |
| Outcome | ||||
| hepatocellular carcinoma | 22/1043 (2.1%) | 17/922 (1.8%) | 5/121 (4.1%) | ns |
| liver transplantation | 112/1199 (9.4%) | 101/1060 (9.5%) | 12/139 (8.6%) | ns |
| fatal hepatic failure | 8/1199 (0.7%) | 5/1060 (0.5%) | 3/139 (2.2%) | ns |
| Clinical stage at latest evaluation | ||||
| I | 813/1136 (71.6%) | 725/1006 (72.1%) | 88/130 (67.7%) | ns |
| II | 188/1136 (16.5%) | 163/1006 (16.2%) | 25/130 (19.2%) | ns |
| III | 135/1136 (11.9%) | 118/1006 (11.7%) | 17/130 (13.1%) | ns |
| Concomitant diseases | ||||
| Sjögren’s syndrome | 177/1043 (17.0%) | 169/922 (18.3%) | 8/121 (6.6%) | OR = 3.17, p = 0.0013 |
| systemic sclerosisb | 49/1043 (4.7%) | 47/922 (5.1%) | 2/121 (1.7%) | ns |
| Hashimoto’s thyroiditis | 102/1043 (9.8%) | 96/922 (10.4%) | 6/121 (5.0%) | ns (OR = 2.23, p = 0.058) |
| autoimmune hepatitis | 82/1043 (7.9%) | 76/922 (8.2%) | 6/121 (5.0%) | ns |
| Raynaud’s phenomenon | 32/1043 (3.1%) | 31/922 (3.4%) | 1/121 (0.8%) | ns |
| rheumatoid arthritis | 44/1043 (4.2%) | 43/922 (4.7%) | 1/121 (0.8%) | OR = 5.87, p = 0.0485 |
| Autoantibodies | ||||
| AMA | 956/1085 (88.1%) | 839/960 (87.4%) | 117/125 (93.6%) | OR = 0.47, p = 0.0439 |
| ANA | 752/1042 (72.2%) | 693/926 (74.8%) | 59/116 (50.9%) | OR = 2.87, p = 5.65 × 10−8 |
| gp210 | 331/1134 (29.2%) | 290/1003 (28.9%) | 41/131 (31.3%) | ns |
| CENP-B | 324/1166 (27.8%) | 302/1031 (29.3%) | 22/135 (16.3%) | OR = 2.13, p = 0.00153 |
| SS-A | 178/1166 (15.3%) | 166/1031 (16.1%) | 12/135 (8.9%) | OR = 1.97, p = 0.0285 |
The prevalence of complications and autoantibodies was compared between female and male patients with PBC; significantly increased number and frequency were highlighted in bold. ns: not significant.
aThere are three patients whose information about sex, age and clinical symptoms was not available.
bIncluding limited cutaneous systemic sclerosis (also known as CREST).
Concomitant autoimmune disorders were generally more prevalent in female patients. Among them, Sjögren’s syndrome, systemic sclerosis, and rheumatoid arthritis differed significantly in prevalence between female and male patients. The AMA-positive rate was higher in male than female patients (93.6% vs. 87.4%), but other autoantibodies that may be accompanied by autoimmune complications were more prevalent in female patients. Aside from the higher prevalence of PBC and autoimmune diseases in females, there were no gender-based differences in clinical or pathological features, the prevalence of hepatocellular carcinoma, or the levels of antibodies against the nuclear pore complex gp210.
HLA allele carrier status
In the present study, 158 HLA alleles (18 A, 42 B, 38 DRB1, 13 DQA1, 22 DQB1, 5 DPA1, and 20 DPB1) or allele groups were identified. Among them, 87 alleles (11 A, 21 B, 20 DRB1, 10 DQA1, 11 DQB1, 3 DPA1, and 11 DPB1) that were carried by more than 1% of individuals in either patients or controls were identified and the carrier frequencies compared between the two groups (Supplementary Table 1). Forty-four alleles (3 A, 7 B, 14 DRB1, 5 DQA1, 9 DQB1, 2 DPA1, and 4 DPB1) demonstrated positive associations with p values less than 0.05 (Supplementary Table 1). The associations of the 22 remained significant (p < 5.75 × 10−4) after correction for multiple-testing (Table 2), and 10 (A*33:03, B*44:03, DRB1*13:02, DRB1*08:03, DQA1*01:02, DQB1*06:04, DQB1*03:01, DQB1*06:01, DPA1*01:03, and DPB1*04:01) reached nominal genome-wide significance (p < 5 × 10−8). Although this highly significant association was detected for all six HLA loci, the HLA- DR and -DQ alleles exhibited the most significant effects in terms of both disease-promoting (DRB1*08:03 and DQB1*06:01) and disease-suppressive (DRB1*13:02, DQB1*06:04, and DQB1*03:01) activity.
Table 2.
Carrier frequencies for selected HLA alleles in PBC patients and controls.
| HLA allele | Carriers in PBC | Carriers in controls | Odds ratio (95% CI) | p value | ||
|---|---|---|---|---|---|---|
| HLA-A | n = 1200 | n = 1196 | ||||
| A*33:03 † | 88 | (7.3%) | 199 | (16.6%) | 0.40 (0.30–0.52) | 2.35 × 10−12 |
| A*02:01/07/18 | 395 | (32.9%) | 293 | (24.5%) | 1.51 (1.26–1.81) | 5.29 × 10−6 |
| HLA-B | n = 1200 | n = 1196 | ||||
| B*44:03 a | 66 | (5.5%) | 187 | (15.6%) | 0.31 (0.23–0.42) | 7.01 × 10−16 |
| B*07:02 | 90 | (7.5%) | 146 | (12.2%) | 0.58 (0.44–0.77) | 1.11 × 10−4 |
| HLA-DRB1 | n = 1200 | n = 1194 | ||||
| DRB1*13:02 a | 47 | (3.9%) | 175 | (14.7%) | 0.24 (0.17–0.33) | 1.35 × 10−19 |
| DRB1*08:03 a | 283 | (23.6%) | 179 | (15.0%) | 1.75 (1.42–2.16) | 1.01 × 10−7 |
| DRB1*14:03 | 7 | (0.6%) | 32 | (2.7%) | 0.21 (0.09–0.49) | 5.10 × 10−5 |
| DRB1*04:05 | 390 | (32.5%) | 292 | (24.5%) | 1.49 (1.24–1.78) | 1.31 × 10−5 |
| HLA-DQA1 | n = 1198 | n = 783 | ||||
| DQA1*01:02 a | 173 | (14.4%) | 208 | (26.6%) | 0.47 (0.37–0.59) | 2.20 × 10−11 |
| HLA-DQB1 | n = 1199 | n = 1195 | ||||
| DQB1*06:04 a | 37 | (3.1%) | 171 | (14.3%) | 0.19 (0.13–0.28) | 1.91 × 10−22 |
| DQB1*03:01 a | 144 | (12.0%) | 256 | (21.4%) | 0.50 (0.40–0.63) | 6.76 × 10−10 |
| DQB1*06:01 a | 520 | (43.4%) | 403 | (33.7%) | 1.51 (1.27–1.78) | 1.25 × 10−6 |
| DQB1*04:01 | 378 | (31.5%) | 280 | (23.4%) | 1.50 (1.25–1.80) | 9.20 × 10−6 |
| DQB1*04:02 | 137 | (11.4%) | 87 | (7.3%) | 1.64 (1.24–2.18) | 4.99 × 10−4 |
| HLA-DPA1 | n = 1200 | n = 783 | ||||
| DPA1*01:03 a | 585 | (48.8%) | 495 | (63.2%) | 0.55 (0.46–0.67) | 2.58 × 10−10 |
| HLA-DPB1 | n = 1200 | n = 1196 | ||||
| DPB1*04:01 a | 35 | (2.9%) | 131 | (11.0%) | 0.24 (0.17–0.36) | 9.63 × 10−15 |
| DPB1*02:01 | 378 | (31.5%) | 485 | (40.6%) | 0.67 (0.57–0.80) | 3.95 × 10−6 |
| DPB1*05:01 | 815 | (67.9%) | 729 | (61.0%) | 1.36 (1.15–1.60) | 3.72 × 10−4 |
The statistical tests of HLA alleles were listed for p values less than the significance levels corrected by Bonferroni’s procedure based on the number of the observed alleles in greater than 1% either patients or controls: 11 A, 21 B, 20 DRB1, 10 DQA1, 11 DQB1, 3 DPA1 and 11 DPB1 alleles; total 87 HLA alleles; p < 0.05/87 = 5.75 × 10−4.
aThese alleles reached genome-wide significance, p < 5 × 10−8.
HLA haplotype analysis
Because the linkage between certain HLA alleles is so tight, high level of linkage disequilibrium (LD) occurs, carrying a certain portion of the significant difference in allele or carrier frequencies at the loci of interest observed in patients-control comparisons to potentially be attributable to over- or under-representation of the alleles of other loci, which are more likely to be causative variants. We tried to identify such primary associations among the significant DRB1 and DQB1 alleles by haplotype analysis of the four most significant DRB1-DQB1 combinations (Table 3). The risk conferred by the DRB1*08:03-DQB1*06:01 haplotype (OR = 1.86, p = 1.98 × 10−9), but not by haplotypes composed of the other DRB1 alleles and DQB1*06:01 (OR = 1.11, p = 0.28) indicated that DQB1*06:01 itself had a nominal practical effect on the disease development of PBC (Table 3(A)). Instead, DRB1*08:03 appeared to be primarily associated with the risk of PBC, although the effect of DRB1*08:03 alone could not be evaluated sufficiently because the haplotypes without DQB1*06:01 were observed in only a small number (four persons each) of patients and controls (Table 3(A)). In the case of the combination of DRB1*13:02 and DQB1*06:04, it is likely that DQB1*06:04 is the principal contributor to the disease resistance because the DRB1*13:02-DQB1*06:04 haplotype effect (OR = 0.19, p = 1.15 × 10−23, Table 3(B)) was equivalent to the effect of DQB1*06:04 (OR = 0.19, p = 1.15 × 10−23, Table 2) and was stronger than that of DRB1*13:02 (OR = 0.24, p = 6.19 × 10−21, Table 2). The frequencies of haplotypes including DRB1*13:02 but not DQB1*06:04 were not decreased in the patient group (OR = 1.50, p = 0.40, Table 3(B)). Indeed, carriers of the second most prevalent haplotype consisting of DRB1*13:02, DRB1*13:02-DQB1*06:09 17, were more represented in the patient group (OR = 1.59, p = 0.37, Supplementary Table 2). Furthermore, the protective alleles of the HLA-A, -B, and -DP loci that reached the genome-wide significance, A*33:03, B*44:03, DPA1*01:03, and DPB1*04:01, were all at a high level of LD with DQB1*06:04, and their protective effects were thus considered to be secondary to those of DQB1*06:04, which appeared to be a principal contributor. Similarly, the protective effect of DQB1*03:01 and the risk effect of DQB1*04:01 were suspected based on a comparison of the effects of haplotype carrier status. Specifically, DQB1*03:01 was shared by several protective haplotypes, including DRB1*14:03-DQB1*03:01 (although we could not evaluate the effect of DRB1*14:03 because all chromosomes with DRB1*14:03 also carry DQB1*03:01 (Table 3(C))). DQB1*04:01 elevated disease risk regardless of the presence or absence of DRB1*04:05 in the case of the combination of DRB1*04:05 and DQB1*04:01, but not vice versa (Table 3(D)).
Table 3.
Carrier status for HLA-DRB1-DQB1 haplotypes consisting of risk/protective alleles in PBC patients and controls.
| DRB1-DQB1 haplotype | PBC (n = 1199) | Controls (n = 1193) | Odds ratio (95% CI) | p value | ||
|---|---|---|---|---|---|---|
| (A) DRB1*08:03 and/or DQB1*06:01 | ||||||
| DRB1*08:03-DQB1*06:01 | 278 | (23.2%) | 174 | (14.6%) | 1.77 (1.43–2.18) | 7.82 × 10 −8 |
| DRB1*08:03-not (DQB1*06:01) | 4 | (0.3%) | 4 | (0.3%) | 1.33 (0.42–4.21) | 0.62 |
| not (DRB1*08:03)-DQB1*06:01 | 242 | (20.2%) | 228 | (19.1%) | 1.11 (0.92–1.34) | 0.28 |
| (B) DRB1*13:02 and/or DQB1*06:04 | ||||||
| DRB1*13:02-DQB1*06:04 | 36 | (3.0%) | 169 | (14.2%) | 0.19 (0.13–0.27) | 1.84 × 10 −22 |
| DRB1*13:02- not (DQB1*06:04) | 11 | (0.9%) | 6 | (0.6%) | 1.50 (0.58–3.88) | 0.40 |
| not (DRB1*13:02)-DQB1*06:04 | 1 | (0.1%) | 2 | (0.2%) | 0.47 (0.04–5.24) | 0.53 |
| (C) DRB1*14:03 and/or DQB1*03:01 | ||||||
| DRB1*14:03-DQB1*03:01 | 7 | (0.6%) | 32 | (2.7%) | 0.21 (0.09–0.49) | 5.10 × 10 −5 |
| DRB1*14:03- not (DQB1*03:01) | 0 | (0.0%) | 0 | (0.0%) | — | — |
| not (DRB1*14:03)-DQB1*03:01 | 137 | (11.4%) | 224 | (18.8%) | 0.54 (0.43–0.68) | 8.54 × 10 −8 |
| (D) DRB1*04:05 and/or DQB1*04:01 | ||||||
| DRB1*04:05-DQB1*04:01 | 373 | (31.1%) | 277 | (23.2%) | 1.49 (1.24–1.79) | 1.44 × 10 −5 |
| DRB1*04:05- not (DQB1*04:01) | 17 | (1.4%) | 15 | (1.3%) | 0.99 (0.55–1.79) | 0.98 |
| not (DRB1*04:05)-DQB1*04:01 | 5 | (0.4%) | 2 | (0.2%) | 1.59 (0.38–6.65) | 0.53 |
Haplotypes composed of given DRB1 and/or DQB1 alleles were compared. The statistical tests reaching significance (p < 0.05) were highlighted in bold.
Interaction between DR-DQ risk/protective factors
The effect of a given risk/protective factor may enhance or attenuate the action of a second factor beyond the extent anticipated by an independent additive effect model. The interactions between the four most significant primary risk/protective factors identified above were analyzed. For this purpose, augmentation or attenuation of the effect was evaluated by comparing the frequencies of carriers of the factor of interest in subpopulations stratified by the presence or absence of each of the other three factors (Table 4); for example, the effect of DRB1*08:03 was not influenced by the existence of DQB1*03:01 or DQB1*04:01, but was profoundly affected by the presence of DQB1*06:04 (Table 4). It is of noteworthy that the interaction between DRB1*08:03 and DQB1*06:04 was asymmetrical; the protective effect of DQB1*06:04 was not influenced by the disease-promoting effect of DRB1*08:03 (OR = 0.14, p = 0.00417), while the disease-promoting effect of DRB1*08:03 was almost completely negated by the presence of DQB1*06:04 (OR = 1.08, p = 0.92). A similar but inverse asymmetric interaction between DRB1*08:03 and DQB1*03:01 was also demonstrated by stratification analysis; the risk of DRB1*08:03 was evident in the presence of DQB1*03:01 (OR = 2.71, p = 0.000882) but the protective effect conferred by DQB1*03:01 was not observed in the presence of DRB1*08:03 (OR = 0.78, p = 0.41).
Table 4.
Effect of the risk/protective DRB1/DQB1 alleles on the presence/absence of another risk/protective DRB1/DQB1 allele.
| Effect of HLA allele | HLA background | |
|---|---|---|
| Present | Absent | |
| Effect of DRB1*08:03 in the presence/absence of DQB1*06:04 | ||
| Frequencies in subset of patients/controls | 5.4%/4.7% | 24.1%/16.6% |
| Odds ratio (95% CI), p | 1.16 (0.24–5.74), p = 0.85 | 1.59 (1.29–1.97), p = 1.70 × 10 −5 |
| Effect of DQB1*06:04 in the presence/absence of DRB1*08:03 | ||
| Frequencies in subset of patients/controls | 0.7%/4.5% | 3.8%/16.1% |
| Odds ratio (95% CI), p | 0.15 (0.03–0.73), p = 0.0068 | 0.21 (0.14–0.30), p = 8.16 × 10 −19 |
The effect of the most significant risk/protective DRB1 and DQB1 alleles was evaluated by comparing the frequencies between patients with PBC and control population in the presence/absence of another risk/protective allele. Significant differences (p < 0.05) in the comparisons are highlighted in bold.
SNP association
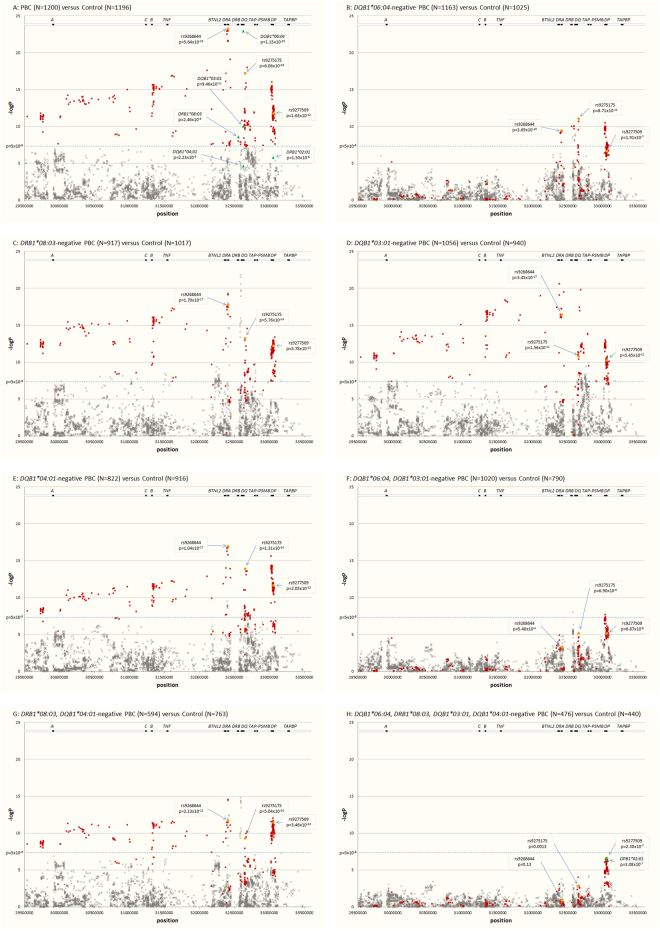
For all patients and controls, 4103 SNPs in the HLA region (bound by rs446198 at position chr 6:29507426 of GRCh37 assembly and rs367408 at position chr 6:33505746) were genotyped16, 18 and the data were further analyzed for the 1200 patients with PBC and 1196 controls whose HLA data were available. When the smaller of two p values which were obtained by applying the dominant effect model of either the predominant allele or the less frequent allele was taken as the effect of each SNP locus, 305 SNPs of them showed p values less than 5 × 10−8 (Fig. 1). Among them, rs9268644 near the HLA-DRA locus gave the minimal p value (p = 5.64 × 10−24) with an odds ratio of 0.39 (Table 5). In our first round of GWAS, rs9275175 in the HLA-DQB1 locus was identified as the most significant SNP16, but it was not as significant as rs9268644 in this setting (OR = 0.41, p = 6.06 × 10−18, Table 5). Three hundred and five nominally significant association signals were distributed from the HLA-A to HLA-DP loci in accordance with the results of the HLA association analysis (Fig. 1A).
Figure 1.
Association signals of genetic markers in the HLA region. For each genetic marker -log p was plotted, selected HLA class II alleles (green triangle) and 4103 SNPs (x) in the HLA region (bound by rs446198 at position chr 6:29507426 of GRCh37 assembly and rs367408 at position chr 6:33505746). Three hundred and one SNPs with a p value less than 5 × 10−8 (−log p greater than 7.301, the level is shown by blue dotted line) in the comparison between all patients (N = 1200) and controls (N = 1196) are shown by red x symbols throughout the panels. Three SNPs presented in Table 5 are highlighted by orange circles. (A) Comparison between all patients and controls; (B) DQB1*06:04-negative patients and controls; (C) DRB1*08:03-negative patients and controls; (D) DQB1*03:01-negative patients and controls; (E) DQB1*04:01-negative patients and controls; (F) DQB1*06:04-negative, DQB1*03:01-negative patients and controls; (G) DRB1*08:03-negative, DQB1*04:01-negative patients and controls; (H) all four allele-negative patients and controls. The location of genes encoded in the region, HLA-A (A), -B (B), -C (C), -DRA1 (DRA), -DRB1 (DRB), -DQA1/-DQB1 (DQ), -DPA1/-DPB1 (DP), TNF, BTNL2, TAP1/TAP2/PSMB8/PSMB9 (TAP-PSMB) and TAPBP, is presented at the top of each panel.
Table 5.
Association of SNPs in the HLA region with PBC in the absence of major HLA factors identified in this study.
| Population | Number of individuals | SNPs with p < 5 × 10−8 | Odds ratio (95% CI), p | |||
|---|---|---|---|---|---|---|
| Patients | Controls | rs9268644* | rs9275175† | rs9277509§ | ||
| All | ||||||
| 1200 | 1196 | 305/4103 | 0.39 (0.33–0.47), p = 5.64 × 10 −24 | 0.41 (0.33–0.50), p = 6.06 × 10−18 | 0.55 (0.46–0.65), p = 1.63 × 10−12 | |
| DQB1*06:04 –negative | ||||||
| 1163 | 1025 | 62/4103 | 0.52 (0.43–0.64), p = 3.69 × 10−10 | 0.46 (0.36–0.58), p = 8.71 × 10−12 | 0.63 (0.53–0.75), p = 1.91 × 10−7 | |
| DRB1*08:03 –negative | ||||||
| 917 | 1017 | 361/4103 | 0.42 (0.34–0.51), p = 1.70 × 10−18 | 0.44 (0.36–0.55), p = 5.76 × 10−14 | 0.51 (0.42–0.61), p = 5.78 × 10−13 | |
| DQB1*03:01 –negative | ||||||
| 1056 | 940 | 333/4103 | 0.41 (0.33–0.51), p = 3.45 × 10−17 | 0.43 (0.34–0.56), p = 1.56 × 10−11 | 0.54 (0.45–0.65), p = 5.45 × 10−11 | |
| DQB1*04:01 –negative | ||||||
| 822 | 916 | 251/4103 | 0.41 (0.33–0.50), p = 1.04 × 10−17 | 0.43 (0.34–0.53), p = 1.31 × 10−14 | 0.49 (0.40–0.60), p = 2.05 × 10−12 | |
| DQB1*06:04 -negative, DQB1*03:01 –negative | ||||||
| 1020 | 790 | 3/4103 | 0.65 (0.51–0.83), p = 0.00054 | 0.54 (0.41–0.71), p = 6.90 × 10−6 | 0.65 (0.53–0.78), p = 6.87 × 10−6 | |
| DRB1*08:03 -negative, DQB1*04:01 –negative | ||||||
| 594 | 763 | 225/4103 | 0.45 (0.35–0.56), p = 2.13 × 10−12 | 0.48 (0.38–0.61), p = 5.04 × 10−10 | 0.45 (0.36–0.57), p = 3.46 × 10−12 | |
| DQB1*06:04 -negative, DRB1*08:03 -negative, DQB1*03:01 -negative, DQB1*04:01 -negative | ||||||
| 476 | 440 | 0/4103 | 0.79 (0.59–1.07), p = 0.13 | 0.62 (0.46–0.83), p = 0.0013 | 0.49 (0.37–0.65), p = 2.30 × 10 −7 | |
*A SNP located in the DR locus (the first intron of DRA, chr 6:32408044–32408044), which showed the minimal p value for the dominant model of variant allele in the comparison of all patients and controls (shown in bold).
†A SNP located near the DQ locus (20 kb upstream of DQB1, chr 6: 32654147–32654147), which showed the minimal p value in our previous GWAS (16).
§A SNP located in the DP locus (the fifth intron of DPB1, chr 6: 33054207–33054207), which showed the minimal p value for the dominant model of a variant allele in the absence of four major HLA factors (shown in bold).
To evaluate the contribution of HLA alleles on the association signal of these SNPs in the HLA region, individuals carrying the HLA of interest were excluded from the dominant effect models. We then calculated odds ratio and p value as described above and compared the p values before and after the stepwise exclusion of HLA alleles of interest (Table 5 and Fig. 1B–H). Among the four most significantly associated HLA alleles, DQB1*06:04 shows strongest impact as revealed by the comparison between 1163 DQB1*06:04-negative patients and 1025 DQB1*06:04-negative controls in which only 62 SNPs remained with a p value less than 5 × 10−8 (Table 5 and Fig. 1B), although the other three alleles did as well but to a reduced extent (Fig. 1C–E). Furthermore, when the two protective alleles DQB1*06:04 and DQB1*03:01 were combined, 302 signals became less significant than the threshold (Fig. 1F), while the combination of two disease-promoting alleles, DRB1*08:03 and DQB1*04:01 had a weaker impact (Fig. 1G) even though the number of patients and controls remained larger than the case that for the protective alleles. In the absence of the four HLA-DR/DQ alleles, no SNP reached a nominal genome-wide significance level, but a peak association signal was found in the HLA-DPB1 locus, rs9277509 with p = 2.30 × 10−7 and an odds ratio of 0.49 in the comparison of the four HLA-DR/DQ-negative subpopulation of patients (N = 476) and controls (N = 440) (Table 5 and Fig. 1H). The fact that the SNP association signal remained after excluding the carriers of these four HLA-DR/DQ alleles could be explained by DPB1*02:01 exhibiting a similar p value (3.08 × 10−7) to the same subpopulation analysis (Fig. 1H).
Discussion
The association of certain SNP alleles in the HLA class II region with the development of PBC has been consistently reported as a major finding in several GWAS studies across different ethnic groups, including Japanese14–16. These results suggest that one or more HLA class II–linked genetic factors influence susceptibility to PBC, a theory that has been strongly supported by a number of HLA association studies8–13. In the present study, we recruited a large number of patients and healthy control individuals and could therefore obtain confirmatory results at genome-wide significance levels (p < 5 × 10−8) for the strongest disease-promoting effect of DRB1*08:03 and the strongest protective effect of DQB1*06:04 in Japanese individuals. The effects of both haplotypes were previously reported by us10 and others12. Further, 22 out of the 87 HLA factors examined in the present study were significant after Bonferroni’s correction for multiple testing (p < 5.75 × 10−4), which is known to be very conservative. This was the case even when we applied the statistical test for the comparison of carrier frequencies, which is generally less sensitive than the test for allele frequencies but is more relevant for the dominant model of inheritance. Because of LD between the alleles carried by common HLA haplotypes in the ethnic population of interest, some of our findings result from secondary associations due to LD with the primary allele. Therefore, we performed haplotype association analysis to discern possible interdependencies among them. We identified four major risk/protective factors in the HLA-DR-DQ region for PBC in a Japanese population, DQB1*06:04, DRB1*08:03, DQB1*03:01, and DQB1*04:01. As discussed in previous reports8, 9, 11, some of these risk/protective HLA alleles share similarities with clinically important alleles in other populations, such as the risk-increasing DRB1*08:01 allele and the protective DRB1*11 and DRB1*13 alleles in individuals of European descent. As shown in previous studies, the molecular basis underlying the effects of alleles, whether they are risk-promoting or protective, may involve common amino acid residues exclusive to each group8, 12. However, false association errors hindered the results of these studies, and therefore determining the principally associated allele is critical for this type of analysis.
The identification of multiple independent risk/protective factors within the HLA region prompted us to evaluate the effects of an interaction between them, since this has not previously been performed except for a study examining the genotype effect of the HLA-DRB1 locus9, in which the risk-promoting effect of DRB1*08 and the protective effect of DRB1*11 were independent and competed with each other. Some risk/protective factors behaved differently, being either enhanced or attenuated, depending on the presence or absence of another factor. For example, the major disease-promoting effect associated with DRB1*08:03 disappeared almost completely in the presence of the protective effect of DQB1*06:04. A similar but opposite relationship between the disease-promoting HLA-DR and the protective HLA-DQ factors was observed between DRB1*08:03 and DQB1*03:01. Recently, DQB1*06:04 (and DRB1*13:02) was also reported to be a protective allele against autoimmune thyroid diseases19; this allele exhibited dominant epistatic effects on HLA risk factors in a similar fashion to that observed in our study. Elucidating the immunological implications of the unrivaled protective effect conferred by DQB1*06:04 may lead to the identification of an active suppressive mechanism for the development of PBC, and could thus identify potential targets for disease prevention.
Epigenetic control of gene expression is another factor that could further elucidate the genetic contribution for PBC disease susceptibility which was not covered by studies of genetic polymorphisms such as our HLA analysis or GWAS. Indeed, alterations in DNA methylation patterns in immune cells were found in patients with PBC20, 21. Furthermore, somatic changes in the genetic material such as sex chromosome loss leading to monosomy X were also reported and may elucidate the underlying mechanism for the female predominance of PBC22, 23.
In summary, this study analyzed a large population of patients with PBC and an equivalently sized control group to confirm the presence of multiple disease-promoting and -protective genetic factors in the HLA region. Interactions between these genetic factors will provide a better understanding of the complicated pathogenic mechanisms of PBC.
Methods
Study design
Case-control study: patient samples were collected at 60 medical institutions in Japan, 32 of which belong to the National Hospital Organization Study Group for Liver Disease in Japan (NHOSLJ). After obtaining written informed consent, we collected patient blood samples for serum and DNA analysis. All study protocols were approved by the institutional review boards of Nagasaki University, NHOSLJ and the other participating institutions according to the Declaration of Helsinki issued by the World Medical Association.
Subject population
All patients met at least two of the following three criteria for the definitive diagnosis of PBC: (i) persistent elevation of serum alkaline phosphatase, an enzyme indicative of cholestasis; (ii) positive AMA test; and (iii) liver biopsy showing non-suppurative inflammation and destruction of the interlobular bile ducts (florid duct lesions), which are characteristics of PBC3. Patients with positive serological markers for persistent hepatitis B or C virus infection were excluded from this study. Liver biopsy data were available for 857 of the 1280 patients (67.0%). Histological diagnosis and staging was performed according to Scheuer’s classification3. Patients were categorized into three different clinical stages based on liver biopsy results and clinical manifestations: clinical stage I, Scheuer’s stage 1 or 2 in liver biopsy or unknown histological stage without signs of portal hypertension or liver cirrhosis; clinical stage II, Scheuer’s stage 3 or 4 in liver biopsy or any histological stage with signs of portal hypertension or liver cirrhosis but without jaundice (total bilirubin less than 2 mg/dL); clinical stage III, any Scheuer’s stage with persistent jaundice (total bilirubin 2 mg/dL or above). Data for clinical staging were provided by patients’ primary caregivers via the collection of fixed case record form.
HLA genotyping
The HLA-A, -B, -DQA1, and -DQB1 genotypes were determined by HLA-DNA typing kits based on reverse SSO hybridization using Luminex xMAP technology, LABType SSO® (One Lambda, Canoga Park, CA, USA) and WakFlow HLA (Wakunaga Pharmaceutical, Osaka, Japan) according to the manufacturers’ instructions. The HLA-DRB1, -DPA1, and -DPB1 genotypes were determined by direct sequencing of the PCR products with 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), followed by matching to an allele database implemented in ASSIGN ATF ver. 1.0.2.45. as described elsewhere20. Most HLA alleles were genotyped at the four-digit level, which correspond to the unique amino acid sequences of the precursor polypeptides. However, because of ambiguous matching of probe reaction patterns in HLA genotyping, the following groups of alleles could not be distinguished in certain individuals and the four-digit designation could therefore not be achieved: a group of A*02 alleles with Phe at position 9 (A*02:01, A*02:07, and A*02:18), another A*02 allele group with Tyr at position 9 (A*02:06 and A*02:10), A*11 alleles (A*11:01 and A*11:02), A*24 alleles (A*24:02 and A*24:20), B*13 alleles (B*13:01 and B*13:02), B*15 alleles with B62 antigen specificity (B*15:01, B*15:07, B*15:27, and B*15:28), B*15 alleles with B75 antigen specificity (B*15:02 and B*15:11), and DQA1*03 alleles (DQA1*03:01, DQA1*03:02, and DQA1*03:03). These alleles were designated A*02:01/07/18, A*02:06/10, A*11, A*24, B*13, B*15:01/07/27/28, B*15:02/11, and DQA1*03, respectively. Certain DRB1, DPA1, and DPB1 alleles were not distinguished from others if the nucleotide sequences of the second exon were indistinguishable, e.g. DRB1*12:01, DRB1*12:06, and DRB1*12:10. In this case, the most probable allele, DRB1*12:01, was assigned.
SNP genotyping
The study population in the present study includes 487 patients and 476 controls who were analyzed in our first round of GWAS16. Other patients and controls were collected for the second phase of GWAS18. After collection and cleaning of SNP genotyping data as previously described16, the genotype data for 4103 SNPs in the HLA region (bounded by rs446198 at position chr 6:29507426 of GRCh37 assembly and rs367408 at position chr 6:33505746) were evaluated. The subjects’ first, second and third-degree relatives (parent-offspring, siblings, uncle/aunt-nephew/niece) were excluded from this study based on a test of identity-by-descent using SNP data collected in the GWAS.
Statistical analysis
The association of disease phenotype and HLA carrier status or SNP genotype was evaluated by the odds ratio as calculated by Woolf’s formula and examined by the chi-square test with 2 × 2 contingency tables, unless otherwise indicated. HLA-DRB1-DQB1 haplotypes were empirically determined for all genotyped individuals with reference to publically accessible HLA haplotype frequency data (HLA Laboratory, Kyoto Japan, http://hla.or.jp/haplo/haplodl.php?lang = en). Haplotype association analysis was applied to compare the relative effect size between HLA-DRB1 and -DQB1 alleles consisting of significant haplotypes. In order to examine the interaction between selected HLA alleles, the effect of HLA carrier status was evaluated in subpopulations stratified by another HLA carrier status of interest. Statistical tests, including those mentioned above, were performed using STATA Release 12 (StataCorp, College Station, TX, USA).
Electronic supplementary material
Acknowledgements
We are indebted to all volunteers who participated in our PBC project. We also thank Drs Toshiki Nikami, Hajime Ota, Hiroshi Kouno, Hirotaka Kouno, Makoto Nakamuta, Nobuyoshi Fukushima, Tatsuji Komatsu, Toshiki Komeda, Yukio Ohara, Toyokichi Muro, Tsutomu Yamashita, Kaname Yoshizawa, Yoko Nakamura, Masaaki Shimada, Noboru Hirashima, Kazuhiro Sugi, Keisuke Ario, Eiichi Takesaki, Atsushi Naganuma, Hiroshi Mano, Haruhiro Yamashita, Kouki Matsushita, Fujio Makita, Hideo Nishimura, Kiyoshi Furuta, Naohiro Takahashi, Masahiro Kikuchi, Naohiko Masaki, Hitoshi Takaki, Takeaki Sato, Masahiko Takahashi, Tetsuo Yamamoto, Hironori Sakai, Michio Kato, Iwao Yabuuchi, Yuko Nagaoki, Noriaki Naeshiro, Shigeki Hayashi, Koichi Honda, Jinya Ishida, Yukio Watanabe, Masakazu Kobayashi, Michiaki Koga, Takeo Saoshiro, Michiyasu Yagura, Yuji Kamitsukasa, Keisuke Hirata (Members of PBC Research in the NHO Study Group for Liver Disease in Japan (NHOSLJ)), Atsushi Tanaka, Hajime Takikawa (Department of Medicine, Teikyo University School of Medicine, Tokyo, Japan), Mikio Zeniya (Department of Gastroenterology and Hepatology, Tokyo Jikei University School of Medicine, Tokyo, Japan), Etsuko Hashimoto, Makiko Taniai (Department of Medicine and Gastroenterology, Tokyo Women’s Medical University, Tokyo, Japan), Hiromasa Ohira (Department of Gastroenterology and Rheumatic Diseases, Fukushima Medical University of Medicine, Fukushima, Japan), Kazuhide Yamamoto, (Department of Gastroenterology and Hepatology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan), Masanori Abe, Morikazu Onji (Department of Gastroenterology and Metabology, Ehime University Graduate School of Medicine, Matsuyama, Japan), Kazuhiko Nakao, Tatsuki Ichikawa, Hidetaka Shibata (Department of Gastroenterology and Hepatology, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan), Satoshi Yamagiwa (Division of Gastroenterology and Hepatology, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan), Shuichi Kaneko, Masao Honda, Kuniaki Arai (Department of Gastroenterology, Kanazawa University Graduate School of Medicine, Kanazawa, Japan), Takafumi Ichida, Katsuji Hirano (Department of Gastroenterology and Hepatology, Juntendo University Shizuoka Hospital, Shizuoka, Japan), Masataka Seike (Faculty of Medicine, Oita University, Oita, Japan), Shotaro Sakisaka, Yasuaki Takeyama (Department of Gastroenterology and Medicine, Fukuoka, University School of Medicine, Fukuoka, Japan), Masaru Harada, Michio Senju (The Third Department of Internal Medicine, School of Medicine, University of Occupational and Environmental Health, Kitakyushu, Japan), Osamu Yokosuka, Tatsuo Kanda (Department of Medicine and Clinical Oncology, Graduate School of Medicine, Chiba University, Chiba, Japan), Yoshiyuki Ueno (Department of Gastroenterology, Yamagata University Faculty of Medicine, Yamagata, Japan), Hirotoshi Ebinuma (Division of Gastroenterology and Hepatology, Department of Internal Medicine, Keio Graduate School of Medicine, Tokyo, Japan), Tomohiro Tanaka, Noriyo Yamashiki, (Organ Transplantation Service, The University of Tokyo, Tokyo, Japan), Sumito Tamura, Yasuhiko Sugawara, Norihiro Kokudo (Hepatobiliary-Pancreatic Surgery Division, Artificial Organ and Transplantation Division, Department of Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan), Akira Mori, Shintaro Yagi, Shinji Uemoto (Division of Hepato-Biliary-Pancreatic and Transplant Surgery, Department of Surgery, Graduate School of Medicine, Kyoto University, Kyoto, Japan), Ken Shirabe, Akinobu Taketomi, Yoshihiko Maehara (Department of Surgery and Science, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan), Atsumasa Komori, Kiyoshi Migita, Masahiro Ito, Shinya Nagaoka, Seigo Abiru, and Hiroshi Yatsuhashi (Clinical Research Center, NHO Nagasaki Medical Center, Omura, Japan), for collecting clinical data and blood samples, and for obtaining informed consent from PBC cases. This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (JSPS) to MY (#26293076) and MN (#20590800, #23591006, #26293181), a Grant-in-Aid for Clinical Research from the National Hospital Organization (NHO) to MN, and a Grant-in-Aid for Scientific Research on Innovative Areas by the Ministry of Education, Culture, Sports, Science and Technology of Japan to KT (#22133008).
Author Contributions
M.Y., K.T. and M.N. conceived and designed the experiments. H.N. and M.N. managed the clinical data collection and sample preparation and storage. M.Y., H.N., M.K., N.N. and Y.H. analyzed the samples and data. M.Y. and H.N. wrote the main manuscript text and prepared figure 1. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11148-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michio Yasunami, Email: yasunami-michio@koseikan.jp.
Minoru Nakamura, Email: nakamuram@nagasaki-mc.com.
References
- 1.Selmi C, Bowlus CL, Gershwin ME, Coppel RL. Primary biliary cirrhosis. Lancet. 2011;377:1600–1609. doi: 10.1016/S0140-6736(10)61965-4. [DOI] [PubMed] [Google Scholar]
- 2.Van de Water J, et al. Detection of autoantibodies to recombinant mitochondrial proteins in patients with primary biliary cirrhosis. N Engl J Med. 1989;320:1377–1380. doi: 10.1056/NEJM198905253202104. [DOI] [PubMed] [Google Scholar]
- 3.Scheuer P. Primary biliary cirrhosis. Proc R Soc Med. 1967;60:1257–1260. [PMC free article] [PubMed] [Google Scholar]
- 4.Van de Water J, et al. Evidence for the targeting by 2-oxo-dehydrogenase enzymes in the T cell response of primary biliary cirrhosis. J Immunol. 1991;146:89–94. [PubMed] [Google Scholar]
- 5.Björkland A, et al. Blood and liver-infiltrating lymphocytes in primary biliary cirrhosis: increase in activated T and natural killer cells and recruitment of primed memory T cells. Hepatology. 1991;13:1106–1111. doi: 10.1002/hep.1840130617. [DOI] [PubMed] [Google Scholar]
- 6.Gershwin ME, et al. Risk factors and comorbidities in primary biliary cirrhosis: a controlled interview-based study of 1032 patients. Hepatology. 2005;42:1194–1202. doi: 10.1002/hep.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chazouillères O, et al. Primary biliary cirrhosis-autoimmune hepatitis overlap syndrome: clinical features and response to therapy. Hepatology. 1998;28:296–301. doi: 10.1002/hep.510280203. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson PT, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology. 2006;44:667–674. doi: 10.1002/hep.21316. [DOI] [PubMed] [Google Scholar]
- 9.Invernizzi P, et al. Human leukocyte antigen polymorphisms in Italian primary biliary cirrhosis: a multicenter study of 664 patients and 1992 healthy controls. Hepatology. 2008;48:1906–1912. doi: 10.1002/hep.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura M, et al. Analysis of HLA-DRB1 polymorphisms in Japanese patients with primary biliary cirrhosis (PBC): The HLA-DRB1polymorphism determines the relative risk of antinuclear antibodies for disease progression in PBC. Hepatol Res. 2010;40:494–504. doi: 10.1111/j.1872-034X.2010.00631.x. [DOI] [PubMed] [Google Scholar]
- 11.Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology. 2011;54:714–723. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umemura T, et al. Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology. 2012;55:506–511. doi: 10.1002/hep.24705. [DOI] [PubMed] [Google Scholar]
- 13.Invernizzi P, et al. Classical HLA-DRB1 and DPB1 alleles account for HLA associations with primary biliary cirrhosis. Genes Immun. 2012;13:461–468. doi: 10.1038/gene.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschfield GM, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mells GF, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet. 2012;91:721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga K, et al. Sequence-based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes. Immunogenetics. 1997;46:199–205. doi: 10.1007/s002510050262. [DOI] [PubMed] [Google Scholar]
- 18.Kawashima M, et al. Genome-wide association studies identify PRKCB as a novel genetic susceptibility locus for primary biliary cholangitis in the Japanese population. Hum Mol Genet. 2017;26:650–659. doi: 10.1093/hmg/ddw406. [DOI] [PubMed] [Google Scholar]
- 19.Ueda S, et al. Identification of independent susceptible and protective HLA alleles in Japanese autoimmune thyroid disease and their epistasis. J Clin Endocrinol Metab. 2014;99:E379–383. doi: 10.1210/jc.2013-2841. [DOI] [PubMed] [Google Scholar]
- 20.Lleo A, et al. Immunoglobulin M levels inversely correlate with CD40 ligand promoter methylation in patients with primary biliary cirrhosis. Hepatology. 2012;55:153–160. doi: 10.1002/hep.24630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lleo A, et al. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin Epigenetics. 2015;7:61. doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Invernizzi P, et al. Frequency of monosomy X in women with primary biliary cirrhosis. Lancet. 2004;363:533–535. doi: 10.1016/S0140-6736(04)15541-4. [DOI] [PubMed] [Google Scholar]
- 23.Lleo A, et al. Y chromosome loss in male patients with primary biliary cirrhosis. J Autoimmun. 2013;41:87–91. doi: 10.1016/j.jaut.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki A, et al. Human leukocyte antigen class I polymorphisms influence the mild clinical manifestation of Plasmodium falciparum infection in Ghanaian children. Hum Immunol. 2011;72:881–888. doi: 10.1016/j.humimm.2011.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.