Abstract
Hsp104 is a ring-forming protein disaggregase that rescues stress-damaged proteins from an aggregated state. To facilitate protein disaggregation, Hsp104 cooperates with Hsp70 and Hsp40 chaperones (Hsp70/40) to form a bi-chaperone system. How Hsp104 recognizes its substrates, particularly the importance of the N domain, remains poorly understood and multiple, seemingly conflicting mechanisms have been proposed. Although the N domain is dispensable for protein disaggregation, it is sensitive to point mutations that abolish the function of the bacterial Hsp104 homolog in vitro, and is essential for curing yeast prions by Hsp104 overexpression in vivo. Here, we present the crystal structure of an N-terminal fragment of Saccharomyces cerevisiae Hsp104 with the N domain of one molecule bound to the C-terminal helix of the neighboring D1 domain. Consistent with mimicking substrate interaction, mutating the putative substrate-binding site in a constitutively active Hsp104 variant impairs the recovery of functional protein from aggregates. We find that the observed substrate-binding defect can be rescued by Hsp70/40 chaperones, providing a molecular explanation as to why the N domain is dispensable for protein disaggregation when Hsp70/40 is present, yet essential for the dissolution of Hsp104-specific substrates, such as yeast prions, which likely depends on a direct N domain interaction.
Introduction
The capacity of cells to tolerate a variety of stress conditions is essential for organismal health. To protect cells from damage, molecular chaperones assist in the folding of other proteins, thereby providing the first line of defense against protein misfolding and aggregation1. However, the majority of stress-inducible molecular chaperones does not recognize or remodel protein aggregates. Instead, cells have evolved powerful ATP-driven protein disaggregases that have the remarkable ability to rescue stress-damaged proteins from an aggregated state2–4.
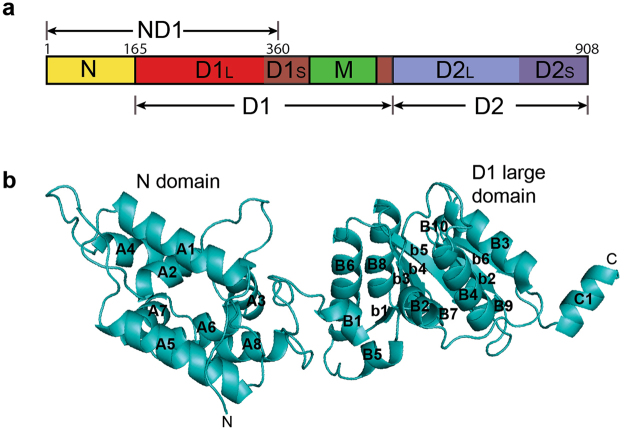
Members of the ring-forming Hsp104 family are the principal protein disaggregases in fungi (Hsp104), plants (Hsp101), and eubacteria (ClpB), but are absent in animal cells5. Hsp104 chaperones are multi-domain proteins consisting of an N domain, an M domain, and two AAA+ type nucleotide-binding domains, termed D1 and D2, which can be subdivided into a larger α/β domain (D1- and D2-large) and a smaller α-helical domain (D1- and D2-small) (Fig. 1a). The M domain serves an important regulatory function6 and mediates the physical interaction with Hsp707,8. The 3D structures of the isolated Hsp104 N domain9 and of full-length Hsp10410 have been determined, including that of their hexamer assembly in different nucleotide states11–13.
Figure 1.
Crystal structure of Hsp104ND1. (a) Schematic diagram of the Hsp104 domain organization. (b) Ribbon diagram of Hsp104ND1. Secondary structural elements are labeled according to their domain assignment: N domain (A), D1-large domain (B), and D1-small domain (C). An upper case letter designates an α-helix and a lower case letter a β-strand.
It is now widely accepted that Hsp104 recognizes amorphous and ordered protein aggregates14–16, and extracts polypeptides that are then threaded through the Hsp104 hexamer by pore loops in the D1 and D2 domains17,18, thereby coupling the energy of ATP binding and hydrolysis to drive protein unfolding. Although Hsp104 is a functional ATPase, efficient protein disaggregation depends on the functional cooperation and physical interaction with the Hsp70 system (Hsp70/40), which directs the Hsp104 hexamer to amorphous aggregates and yeast prions in vivo 14,15,19, activates the Hsp104 motor in a species-specific manner7,8,20,21, and assists in the recovery of Hsp104 substrates that depend on Hsp70/40 for folding11,21,22. Although the Hsp104:Hsp70/40 bi-chaperone system represents the functionally active form in vivo, the Hsp104 protein-remodeling activity can also be elicited in vitro in the absence of Hsp70/40 by mutation or under chemically defined nucleotide conditions that asymmetrically decelerate the ATPase activity of Hsp10423. Additionally, modification of the M domain by point mutation24,25 or by insertion of T4 lysozyme within the M domain helix 2 (Hsp104T4L)7,11 renders Hsp104 constitutively active for protein disaggregation and independent of Hsp70/40 chaperones in a substrate-dependent manner.
Despite major advances in our mechanistic understanding of Hsp104 function, it remains poorly understood how Hsp104 recognizes and binds protein substrates of diverse sequence, shape, and size. In this context, the importance of the N domain to Hsp104 function is perhaps most controversial. Initially proposed to be essential for ClpB function26,27 by mediating substrate interaction26,28–31, it was later shown that the N domain is dispensable for protein disaggregation in vitro 32,33 and thermotolerance development in vivo 33,34. Although the N domain appears to be dispensable for protein disaggregation, it was shown more recently that the N domain orientation can affect protein disaggregation35 and abolishes ClpB-dependent protein disaggregation when mutated36. Consistent with an important but non-essential role in eubacteria, the N domain of Saccharomyces cerevisiae Hsp104 is dispensable for thermotolerance development and yeast prion replication14,37, as well as for protein binding and recovery in vitro and in vivo 38. However, the Hsp104 N domain enhances protein disaggregation in vitro 7, mediates the interaction with the prion form of Sup35p15, and is essential for yeast prion dissolution16 and curing by Hsp104 overexpression37.
Delineating the functional role of the non-essential N domain is difficult because of the close physical and synergistic interaction between Hsp104 and the cognate Hsp70 chaperone system. Like Hsp104, Hsp70 is a functional ATPase that chaperones misfolded and aggregation-prone proteins. Because Hsp70 binding to the M domain is essential to unleash the protein disaggregating activity of Hsp104, Hsp104 mutants that abolish protein disaggregation could be impaired in substrate interaction, in Hsp70 cooperation, or both. Here, we present the crystal structure of an S. cerevisiae Hsp104 N-terminal fragment (residues 1–360) consisting of the N domain and an extended D1-large domain. We find that the N domain is bound to a C-terminal helical segment of the neighboring D1 domain, mimicking substrate interaction. We show by structure-guided mutagenesis that the pseudo two-fold symmetrical N domain features a bipartite peptide-binding interface, involving the first helix of each N domain repeat (helix A1 and A5). Hsp104 variants featuring point mutations in either interface are defective in protein disaggregation. While helix A5 mutants are impaired in Hsp70 interaction, mutations in helix A1 are defective in substrate binding and can be rescued by Hsp70/40 chaperones. We propose that the N domain plays an important role in protein disaggregation by facilitating both substrate binding and Hsp70 interaction to accommodate a variety of Hsp104 substrates.
Results
Crystal Structures of Hsp104ND1
To provide a molecular understanding of the functional role of the Hsp104 N domain, we solved the 2.8 Å resolution crystal structure of an N-terminal fragment of yeast Hsp104 (residues 1–360; Hsp104ND1) comprising the N domain and an extended D1-large domain featuring the first α helix of the D1-small domain, termed the C1 helix (residues 343–351)39 (Fig. 1a). Despite a sequence identity of 42% between yeast Hsp104 and bacterial ClpB (see Supplementary Fig. S1), no structure solution could be found by molecular replacement using the previously determined crystal structure of Thermus thermophilus ClpB (PDB ID: 1QVR)39 as a search model, even when performing the analysis with the individual N and D1 domains. To solve the phase problem, we crystallized an engineered Hsp104ND1 construct featuring three methionine substitutions at Leu36, Phe118, and Leu248 (Hsp104MMM), and determined the crystal structure of this variant by seleno-methionine (SeMet) MAD phasing (Table 1). The unbiased experimental map enables tracing of residues 4 to 352 (Fig. 1b). No unaccounted electron density was observed that could be attributed to bound nucleotide, irrespective of the 5 mM nucleotide (ADP or ADPNP) that was added during crystallization, suggesting that Hsp104ND1 was crystallized in the nucleotide-free state and indicating that a complete AAA+ domain featuring both the D1-large and -small domains are needed for nucleotide binding.
Table 1.
Data collection, MAD phasing, and refinement statistics.
| Native | Se-Met (Hsp104MMM) | |||
|---|---|---|---|---|
| Data Collection Statistics | ||||
| Space group | P 6522 | P 6522 | ||
| Unit Cell | a = 179.1 Å, b = 179.1 Å, c = 69.7 Å | a = 179.5 Å, b = 179.5 Å, c = 69.1 Å | ||
| α = 90°, β = 90°, γ = 120° | α = 90°, β = 90°, γ = 120° | |||
| Source | NSLS-X25 | NSLS-X25 | ||
| Wavelength (Å) | λ = 1.10 | λ1 = 0.9789 | λ2 = 0.9792 | λ3 = 1.0024 |
| Resolution (Å) | 41.53–2.82 | 44.8–3.5 | 44.8–3.5 | 44.8–3.8 |
| Completeness (%)a | 84.2 (12.7) | 99.8 (100) | 99.8 (99.5) | 99.9 (99.9) |
| Redundancy | 14.7 (1.7) | 6.9 (6.8) | 6.9 (6.5) | 6.7 (6.9) |
| Rsym a,b | 0.076 (0.386) | 0.086 (0.393) | 0.091 (0.497) | 0.088 (0.320) |
| I/σc | 17.6 | 12.9 | 11.1 | 10.7 |
| MAD Phasing Statistics | ||||
| Riso d | 0.222 | 0.044 | 0.053 | |
| RCullis e | 0.62 | 0.72 | ||
| Phasing Powerf | 2.38 | 2.07 | 1.16 | |
| Figure of Meritg | 0.63 (centric) | 0.60 (acentric) | ||
| Refinement Statistics | ||||
| Resolution (Å) | 41.53–2.82 | |||
| No. reflections | 14036 | |||
| Rcryst/Rfree | 0.210/0.279 | |||
| No. atoms | 2725 | |||
| Protein | 2721 | |||
| Water | 4 | |||
| B-factors | 94.1 | |||
| Protein | 94.1 | |||
| Water | 74.3 | |||
| rmsd bond (Å) | 0.002 | |||
| rmsd angle (°) | 0.399 | |||
| Ramachandran | ||||
| Favored (%) | 97.1 | |||
| Outliers (%) | 0.0 | |||
aValues for the highest resolution shell are given in parentheses. bRsym = Σhkl|I(hkl) − < I(hkl) > |/ΣhklI(hkl), where <I(hkl)> is the mean of the symmetry equivalent reflections of I(hkl). cBased on unmerged data. dRiso = Σ|FPH − FP|/ΣFp, where Fp is the peak (λ1), and FPH are the inflection (λ2), low energy remote (λ3) or native structure factor amplitudes. eRCullis = Σ||FPH ± FP| − FH calc|/Σ|FPH ± FP| for centric reflections only. fIsomorphous phasing power = Σ|FH|/Σ||FPH obs| − |FPH calc||; anomalous phasing power = Σ|F″H|/Σ||ADobs| − |ADcalc||. gFigure of Merit = weighted mean of the cosine of the deviation from αbest.
The 3D structure of our N and D1-large domains is nearly identical to those of the isolated S. cerevisiae Hsp104 N domain9 and Chaetomium thermophilum Hsp104 D1-large domain10 that were recently reported, and superimpose pair-wise with an RMSD of only 0.60 ± 0.09Å2 (N domain) and 1.59 ± 0.12 Å2 (D1-large domain). The N domain can be divided into two repeats related by a pseudo two-fold symmetry. However, unlike bacterial ClpA/B proteins, the N domain of Hsp104 features a four amino acid insertion that extends the length of an intervening loop region (residues 40–49) conferring asymmetry to the yeast N domain structure (Fig. 1b). The structure of the extended D1-large domain shares the canonical α/β-fold of bacterial Hsp100 AAA+ ATPases (Fig. 1b), featuring the conserved Walker A, Walker B, sensor-1, and arginine (Arg)-finger motifs required for ATP binding and hydrolysis40 (see Supplementary Fig. S1).
The crystal structure of the N domain features a peptide-binding site
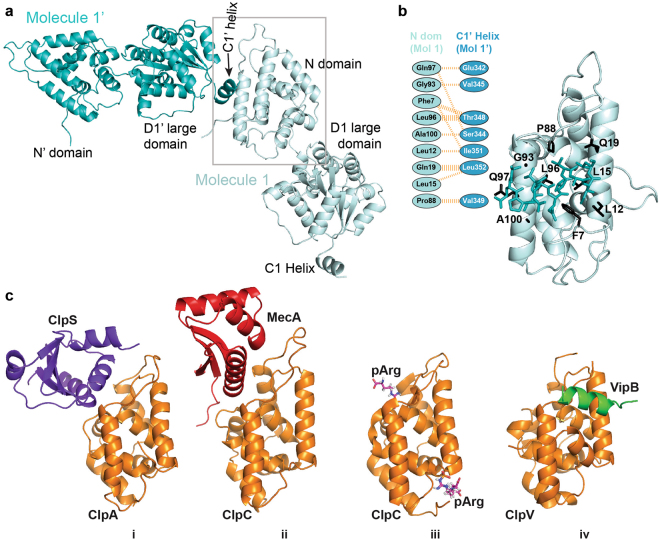
Surprisingly, we find that the N domain of Hsp104ND1 interacts with the C1 helix of the neighboring D1-small domain (Fig. 2a,b and Supplementary Fig. S2). Binding interactions are hydrophobic in nature consistent with the notion that molecular chaperones bind misfolded and aggregation-prone protein conformers that expose hydrophobic residues on their surface41. N domain segments involved in peptide binding include helix A1 (Leu12, Leu15, and Gln19), helix A5 (Gly93, Leu96, Gln97, and Ala100), and additional hydrophobic residues (Phe7 and Pro88) (Fig. 2b). Notably, this peptide-binding site differs from binding sites reported for ClpA42, ClpC43,44, and ClpV45 (Fig. 2c), but overlaps with the proposed peptide-binding site recently reported for ClpB36. However, to our knowledge, this is the first time that an interaction with a helical peptide is observed.
Figure 2.
The substrate-binding interface of the Hsp104 N domain. (a) Molecular content of two asymmetric units showing the interaction between the N domain of one molecule and the neighboring C1 helix (C1′). (b) Specific interactions between the N domain and the C1′ helix. N domain residues in contact with the C1′ helix are shown as stick model. The substrate-binding interface consists of hydrophobic interactions. Molecular interactions between the N domain and the neighboring C1 helix were analyzed using PDBsum54. The thickness of the orange dashed line correlates with the number of interactions made. (c) The substrate-binding site of the Hsp104 N domain is distinct from protein-binding interfaces in other Hsp100 proteins. The orientation of Hsp100 N domains is similar to that shown in Fig. 2b. (i) N domain of ClpA bound to its adaptor ClpS42, (ii) ClpC bound to its adaptor MecA43, and (iii) ClpC bound to a phosphorylated arginine mimicking substrate interaction44, and (iv) ClpV bound to a segment of its VipB substrate45.
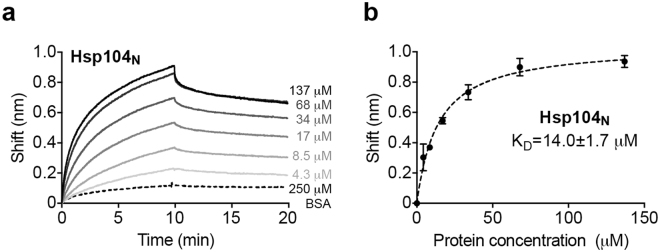
To confirm that this Hsp104 N domain-peptide interaction is also observed in solution, we synthesized a peptide with its sequence derived from the C1 helix (GEVAEPSVRQTVAILRGLQ) and measured the binding interaction using biolayer interferometry (Fig. 3a,b). Our results show that the C1 peptide bound to the isolated N domain with a KD of 14.0 ± 1.7 μM.
Figure 3.
Direct binding of Hsp104N to the C1 peptide determined by biolayer interferometry. (a) Binding of Hsp104N (solid lines) or BSA (dotted line) to immobilized biotinylated C1 peptide was measured by light distance shift (nm). A representative curve set of association (0–10 min) and dissociation (10–20 min) curves for various Hsp104N concentrations (4.3 to 137 µM) or BSA control (250 µM) from two independent measurements are shown. (b) Averaged equilibrium light distance shift at different Hsp104N protein concentrations from two independent measurements.
The N domain contributes towards protein disaggregation when present
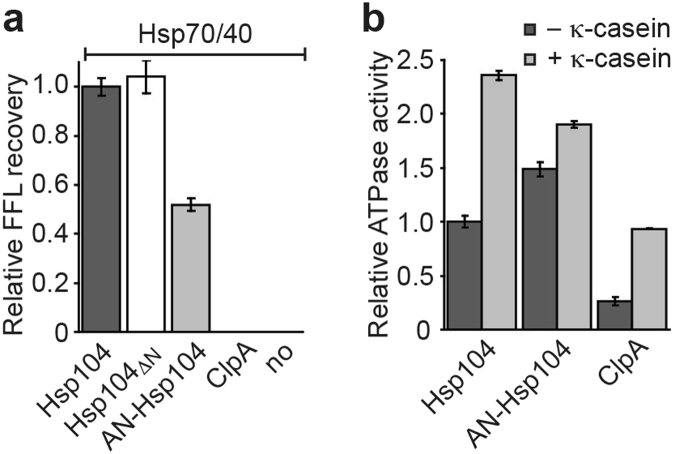
Although the Hsp104 N domain is essential for curing yeast prions by Hsp104 overexpression in vivo 16,37, an Hsp104 variant lacking the N domain (Hsp104ΔN) is functional and cooperates with Hsp70/40 in protein disaggregation in vitro (Fig. 4a). Because the N domain adopts a stably folded conformation that does not make strong contacts with the D1 domain in the Hsp104 hexamer11–13, we asked whether the N domain can be replaced with the structurally homologous N domain of ClpA. ClpA lacks an M domain and does not cooperate with Hsp70/40 in protein disaggregation (Fig. 4a). Instead, ClpA associates with the ClpP protease to degrade ssrA-tagged proteins46. However, because the ClpAP system is converted into a protein disaggregase in the presence of the ClpA adaptor ClpS47, ClpA must feature, at least in principle, all of the structural elements required for protein disaggregation. We find that the AN-Hsp104 chimera featuring the N domain of E. coli ClpA reduces protein disaggregation nearly 50% compared to the native Hsp104 bi-chaperone system (Fig. 4a). The latter is not the result of an impaired ATPase activity, which is elevated compared to both Hsp104 and ClpA, and can be further stimulated by κ-casein (Fig. 4b). Hence, the N domain must feature sequence elements that are important to Hsp104’s protein disaggregase function.
Figure 4.
The Hsp104 N domain affects protein disaggregation when present. (a) Recovery of FFL activity by Hsp104, Hsp104ΔN, AN-Hsp104, and ClpA in the presence of Hsp70/40. (b) Basal (black) and κ-casein stimulated (grey) ATPase activities of AN-Hsp104 and ClpA expressed relative to the basal ATPase activity of Hsp104. Averages of three independent measurements ± SD are shown.
N domain mutants impaired in Hsp70 binding are defective in protein disaggregation
The crystal structure of Hsp104ND1 showed that the N domain contacts the C1 helix of a neighboring molecule (Fig. 2a,b). The observed N domain interactions involve helix A1 (residues 9–24) in the first repeat, and helix A5 (residues 90–105) in the second repeat that overlaps with an Hsp104 segment (residues 91–111) mediating a species-specific interaction with Hsp7021. A functional role for helix A1 has not been established for Hsp104, and could facilitate substrate binding. To test this, we selected conserved hydrophobic residues from each interface, Phe7, Leu15, and Leu96 (Set-1), and Leu92, Leu96, and Phe110 (Set-2), mutated them to alanine either alone or in combination, and determined their effect on Hsp104 ATPase activity (see Supplementary Fig. S3). Both sets included Leu96 that is part of a hydrophobic network and likely contributes binding energy (Fig. 5a). Since the N domain does not form strong contacts with the D1 domain, we expect the impact of these mutations to be small. We find that the ATPase activity is unchanged for Set-1 mutants and elevated for Set-2 mutants compared to Hsp104, but below the ATPase activity observed for Hsp104ΔN that lacks the N domain altogether (see Supplementary Fig. S3).
Figure 5.
The Hsp104 N domain is bipartite and mediates both substrate binding and Hsp70 interaction. (a) Key residues involved in substrate binding are shown in red (Set-1), and those involved in Hsp70 interaction21 in blue (Set-2). (b,c) Set-1 and Set-2 mutants affect protein disaggregation differently. Disaggregase activities of Hsp104 and Hsp104 variants (Set-1 red; and Set-2 blue) in the presence of Hsp70/40. Recoveries of (b) FFL and (c) G6PDH activity expressed relative to the native bi-chaperone system. (d,e) Set-1T4L and Set-2T4L show substantial defects in protein disaggregation in a constitutively active background. (d) Impaired recovery of β-gal activity by Set-1T4L (red) and Set-2T4L (blue). (e) Hsp70/40 restores the protein disaggregation defect of Set-1T4L (red), but not of Set-2T4L (blue). Averages of three independent measurements ± SD are shown.
Although it is intriguing that N domain mutations should have any effect on the ATPase activity, comparing the ATPase activity of Hsp104 variants is not a good measure of chaperone function40. We therefore evaluated our Set-1 and Set-2 mutants using a coupled-chaperone assay (Fig. 5b,c). To avoid substrate-specific effects, we used chemically denatured firefly luciferase (FFL) and heat-aggregated Glucose-6-phosphate dehydrogenase (G6PDH) as model substrates11. We expect that all of our Hsp104 N domain mutants remain functional protein disaggregases because they maintain the critical M domain:Hsp70 interaction required for Hsp104 activation. Although none of the single Set-1 mutants (Hsp104F7A or Hsp104L15A) or their double (Hsp104F7A/L15A) and triple combination (Set-1) are defective in recovering FFL activity in the presence of Hsp70/40, Hsp104 variants carrying one or more Set-2 mutation (Hsp104L92A, Hsp104F110A, Hsp104L92A/F110A, and Set-2) are substantially impaired (Fig. 5b). The recovery of aggregated FFL by Set-2 mutants is on average ~21.3% less than that observed with Hsp104ΔN suggesting that the N domain contributes to Hsp104’s protein disaggregating activity when present. Similarly, Set-2 but not Set-1 mutants were also impaired in protein disaggregation when using heat-aggregated G6PDH as substrate (Fig. 5c), arguing against a substrate-specific effect. Because residues 91–111 mediate an interaction with the cognate Hsp70 chaperone, the observed protein disaggregation defect of Set-2 mutants is likely the direct result of an impaired interaction between the N domain and Hsp70 needed to facilitate substrate binding or recruitment.
Set-1 mutants are defective in substrate interaction
The negligible effect on protein disaggregation observed with Set-1 mutants in the presence of Hsp70/40 was unexpected (Fig. 5b,c). Because Hsp70 binding is essential to unleash the protein disaggregating activity of Hsp104, Hsp70/40 may have masked any N domain defects in substrate binding in the coupled-chaperone assay. To test this, we introduced the Set-1 (F7A/L15A/L96A) and Set-2 (L92A/L96A/F110A) triple mutations into the constitutively active Hsp104T4L variant that recovers functional protein in the absence of Hsp70/407,11. Uncoupling the Hsp70/40 requirement showed a clear defect in protein disaggregation (Fig. 5d). Both Set-1T4L and Set-2T4L are substantially impaired in protein disaggregation compared to Hsp104T4L, and have a 28.9% (Set-1T4L) and 43.5% reduced activity (Set-2T4L), respectively, in recovering heat-aggregated β-gal activity (Fig. 5d). In the presence of Hsp70/40, the ability of Set-1T4L to recover functional β-gal is restored to Hsp104T4L levels (Fig. 5e), whereas β-gal recovery by Set-2T4L remains similar to the level observed with Hsp104T4LΔN lacking the N domain (Fig. 5e). These findings suggest that Set-1 mutants are defective in substrate binding, a defect that is rescued in the presence of the Hsp70 system.
The N domain modulates Hsp104 function
A functional role of the N domain in substrate binding has been reported for ClpB27,29,36,48. However, because of an inability to uncouple the Hsp70 requirement from protein disaggregation in the Hsp104:Hsp70/40 bi-chaperone system, these studies could not differentiate between defects in substrate binding and defects in Hsp70 interaction, not to mention defective Hsp70-mediated substrate binding.
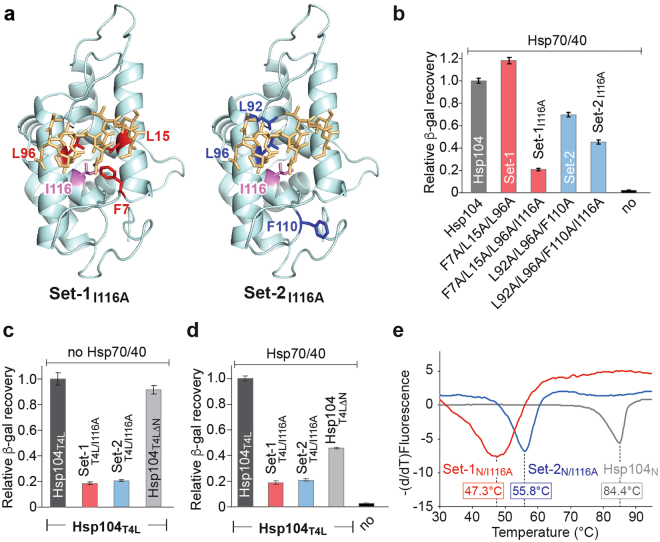
It was recently reported that a T. thermophiles ClpB variant featuring four alanine substitutions in the N domain abrogates protein disaggregation by the bacterial bi-chaperone system36. These four residues correspond to Phe7, Leu15, Leu96, and Ile116 in yeast Hsp104 and are identical to the Set-1 triple mutant with the addition of the Ile116 to alanine mutation. Neither Hsp104I116A (see Supplementary Fig. S4) nor the Set-1 triple mutant shows impairment in protein disaggregation (Fig. 5b,c). We therefore introduced the I116A mutation into the Set-1 and Set-2 variants resulting in Set-1I116A and Set-2I116A, respectively (Fig. 6a). Interestingly, the quadruple Set-1I116A mutant nearly abolished protein disaggregation (Fig. 6b) underscoring the structural and functional conservation between the bacterial and yeast bi-chaperone system. Similarly, the quadruple Set-2I116A mutant further reduced the activity of the Set-2 triple mutant (Fig. 6b). Because none of the Set-1 single, double, or triple mutants are defective, the combined effect of all four mutations is necessary to abrogate protein disaggregation of Set-1I116A (Figs 5b,c and 6b and Supplementary Fig. S4).
Figure 6.
Addition of the I116A mutation to Set-1 and Set-2 triple mutants abrogates protein disaggregation. (a) Ribbon diagram of the Set-1I116A and Set-2I116A mutants with Ile116 shown in pink. (b) Recovery of β-gal activity by Set-1 and Set-1I116A (red bars), and Set-2 and Set-2I116A (blue bars). (c,d) Neither Set-1T4L/I116A (red bars) nor Set-2T4L/I116A (blue bars) promotes protein disaggregation without (c) or with Hsp70/40 (d). Averages of three independent measurements ± SD are shown in Fig. 6b–d. (e) The Set-1N/I116A and Set-2N/I116A quadruple mutants have reduced protein stability. Thermal shift assay with the isolated N domain (Hsp104N, grey curve) and its Set-1 (Set-1N/I116A, red curve) and Set2 variants (Set-2N/I116A, blue curve). Representative curves from three independent measurements together with the calculated T m values are shown.
A defect was also observed with Set-1T4L/I116A that is uncoupled from the Hsp70/40 requirement (Fig. 6c). Unlike the Set-1T4L mutant, addition of Hsp70/40 chaperones could not restore the protein disaggregase activity of the Set-1T4L/I116A mutant (Fig. 6d). Addition of the I116A mutation to Set-2T4L further reduced the chaperone activity of the Set-2T4L variant both in the absence and presence of Hsp70/40 (compare Fig. 5d,e with Fig. 6c,d). These findings indicate that the addition of the I116A mutation to Set-1 or Set-2 affected both mutants similarly.
The Hsp104ND1 crystal structure showed that Ile116 is part of a hydrophobic core that stabilizes the N domain (Fig. 6a). Therefore, mutating Ile116 to alanine could affect N domain stability. To test this, we bacterially expressed and purified the isolated N domain (Hsp104N) and its Set-1N and Set-2N mutants, alone and in combination with I116A, then determined their protein stability by dye binding using differential scanning fluorimetry. The melting temperatures (T m) of the quadruple Set-1N/I116A and Set-2N/I116A mutants are severely left shifted compared to Hsp104N, decreasing the apparent T m by 37.1 °C and 28.6 °C, respectively (Fig. 6e), with the addition of the I116A mutation contributing as much as 19.4 °C (see Supplementary Fig. S5). Thus, addition of the I116A mutation to the Set-1 or Set-2 triple mutants destabilizes the N domain and likely inactivated the Hsp104 motor irreversibly possibly by blocking the axial channel.
Discussion
Hsp100 protein disaggregases are powerful molecular machines that harness the energy derived from ATP binding and hydrolysis to disaggregate a wide variety of aggregated proteins. The crystal structure of Hsp104ND1 supports a role for the N domain in binding of a substrate mimic (Fig. 2b). We observe only hydrophobic interactions that would be compatible with accommodating different protein substrates of diverse size, shape, and sequence. Furthermore, the N domain is bipartite and mediates the direct interaction with both substrate and Hsp70, albeit through distinct binding interfaces (Fig. 5a). Although mutations in either interface interfere with protein disaggregation, the underlying mechanism is different. While helix A1 mutants are defective in substrate binding (Fig. 5d), mutations in helix A5 are also impaired in Hsp70 interaction (Fig. 5e). Notably, helix A1 mutants are indistinguishable from Hsp104 in protein disaggregation in the presence of the Hsp70 system (Fig. 5b,c and e), suggesting that Hsp70/40 chaperones can compensate for N domain mutants defective in substrate binding.
Our observations support the notion that some Hsp104 substrates are recognized and bound directly by the Hsp104 motor, while others may depend on Hsp70/40 to be recruited. Thus, in addition to its role as an Hsp104 activator, Hsp70 may serve as an adaptor to increase the Hsp104 substrate spectrum. Our findings also resolve the outstanding question regarding the functional role of the N domain. Although the N domain is dispensable for protein disaggregation by the bi-chaperone system, the N domain facilitates substrate binding and modulates Hsp104 function when present7,16. Thus, we propose that lack of an N domain or Hsp104 N domain mutants defective in substrate binding are masked by the presence of the Hsp70 system needed for Hsp104 activation and function. Simultaneously, Hsp104-specific substrates that require a direct, physical interaction with the N domain, such as the prion form of yeast Sup3515, may depend on the N domain and cannot be rescued by Hsp70/40 chaperones16,37. Together, these findings provide a molecular explanation for the perplexing N domain requirements in distinct Hsp104 activities.
Methods
Cloning
E. coli ClpA was cloned into pET24b. S. cerevisiae Hsp104MMM and Hsp104 mutants (F7A, L15A, L92A, L96A, F110A, I116A, and their combinations) were generated by QuikChange site-directed mutagenesis. Corresponding Hsp104T4L mutants were generated by cassette mutagenesis. To generate AN-Hsp104, the Hsp104 N domain (M1-L164) was replaced with the equivalent N domain of E. coli ClpA (M1-E166) and cloned into pProEX HTb. Isolated N domain constructs consisting of the first 165 residues (Hsp104N) were PCR amplified and cloned into pProEX HTb.
Protein expression and purification
Protein constructs were overexpressed in E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent Technologies, Santa Clara, CA). Protein expression was induced with 0.5 or 1 mM IPTG for 4 h hours at 37 °C (Hsp104ND1), for 2 h at 37 °C (ClpA), for 4 h at 30 °C (His6-Hsp104N and His6-Ydj1), 6 h at 25 °C (His6-hHsp70, His6-Hsp104ΔN, His6-Hsp104, and His6-Hsp104 mutants), or for 16 h at 16 °C (His6-AN-Hsp104, His6-Hsp104T4LΔN, His6-Hsp104T4L and His6-Hsp104T4L mutants). SeMet-labeled Hsp104MMM was prepared by transforming an E. coli methionine auxotroph strain, B834 (DE3) pLysS. Cells were grown in defined medium supplemented with 50 mg/L seleno-DL-methionine. Preparation of SeMet-labeled Hsp104MMM was otherwise identical to Hsp104ND1 49. Cells were lysed using a microfluidizer in buffer A (25 mM Tris-HCl pH 7.5, 300 mM NaCl, 5% glycerol and 5 mM β-mercaptoethanol) containing 30 mM imidazole followed by ultracentrifugation at 45,000 rpm for 1 h at 4 °C. Proteins were purified from cleared lysates by Ni-NTA agarose affinity chromatography. Proteins were eluted in buffer A containing 300 mM imidazole. The N-terminal His6-tag was cleaved by incubating the protein with His6-TEV protease at 4 °C overnight. The cleaved protein was diluted 1:10 with buffer A and reapplied to a Ni-NTA agarose column to remove the liberated His6-tag and His6-tagged TEV. Protein concentration was measured by a colorimetric assay using the Protein Assay Dye Reagent (Bio-Rad, Hercules, CA). Hsp104ND1/Hsp104MMM was further purified in negative binding mode to a DEAE sepharose column followed by cation exchange chromatography. Hsp104N and its variants were further purified by anion exchange chromatography. AN-Hsp104 and Hsp104T4L variants were purified similarly to Hsp104 in buffer A containing 10% glycerol using Ni-NTA agarose followed by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare Bio-Sciences, Pittsburgh, PA). His6-Ydj1 and His6-Hsp70 were purified by Ni-NTA affinity chromatography as described21. His6-Ydj1 was further purified on a TOYOPEARL Butyl-650S column (Tosoh Bioscience LLC, King of Prussia, PA) followed by anion exchange chromatography. His6-hHsp70 was further purified by anion exchange chromatography.
Protein crystallization and data collection
Native crystals were grown at 4 °C by mixing 2 µl of protein solution (19 mg/ml) with 1 µl of reservoir solution containing 33% PEG 400 (v/v), 50 mM HEPES-HCl pH 7.2, and 150 mM sodium formate49. Crystals were harvested into reservoir solution and flash frozen in liquid nitrogen without additional cryo-protectant. Crystals belonged to space group P6522 with unit cell dimensions of a = b = 179.1 Å, c = 69.7 Å, and one molecule per asymmetric unit. Crystals of SeMet-labeled Hsp104MMM protein were grown at 4 °C by the hanging-drop vapor diffusion method by mixing 2 µl of protein (20 mg/ml) in 50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 10 mM MgCl2 and in the presence or absence of 5 mM nucleotide with 2 µl of reservoir solution consisting of 19% PEG4000 (w/v), 100 mM MES, pH 6.5, and 20 mM sodium formate. Crystals were harvested into reservoir solution containing 25% (v/v) glycerol and flash frozen in liquid nitrogen. A three wavelength MAD experiment was performed by collecting data near the absorption peak and the edge of selenium, 0.9789 and 0.9792 Å, corresponding to the maximum f″ and the minimum f′, respectively, and at a low energy remote of 1.0024 Å. Data sets were obtained from a single crystal and were processed using the HKL2000 software package50 (Table 1). A total of six selenium sites were identified from the anomalous difference map that was sufficient to solve the structure of Hsp104MMM to 3.5 Å resolution. The resulting phases were used to calculate the electron density map to build the atomic model. Multiple rounds of refinement were carried out using CNSsolve v.1.151 combined with manual rebuilding using COOT52 (Table 1, see Supplementary Fig. S2). Atomic coordinates and structure factors for Hsp104ND1 have been deposited with the RCSB under accession code 6AMN.
Biolayer interferometry
Biolayer interferometry was used to examine N domain interaction with peptide, and was performed using an Octet RED96 instrument (Pall FortéBio LLC, Fremont, CA). Biotinylated C1 peptide (Biotin-Ahx-GEVAEPSVRQTVAILRGLQ) was synthesized commercially (AAPPTec LLC, Louisville, KY) and immobilized onto streptavidin-coated biosensors at a concentration of 0.5 µM in binding buffer (50 mM Tris-HCl pH 8.5, 20 mM ammonium citrate, 10% PEG4000 (w/v), and 0.1 mg/ml BSA). Biosensors were immersed in a 200 µl protein solution for 10 min to allow for association and subsequently transferred into 200 µl binding buffer to follow dissociation for 10 min. Binding curves were obtained at 25 °C at different Hsp104N concentrations or BSA control in a 96-well black plate under rocking. Curve fitting and analysis were performed using the GraphPad Prism software (GraphPad Software Inc., La Jolla, CA).
Chaperone activity assay
To prepare aggregates of model substrates, firefly luciferase (FFL; 10 µM) was denatured in 7 M urea in refolding buffer (25 mM HEPES-KOH pH 7.5, 150 mM potassium acetate, 10 mM magnesium acetate, and 10 mM DTT) for 30 min at 22 °C. β-galactosidase (β-gal; 0.4 µM) was thermally aggregated in refolding buffer for 40 min at 59 °C. Glucose-6-phosphate dehydrogenase (G6PDH; 200 µM) was thermally aggregated in denaturation buffer (4 M urea, 8% glycerol, and 20 mM DTT) for 5 min at 47 °C, diluted 10-fold in refolding buffer, and treated for 1 min at 47 °C.
Chemically denatured FFL was diluted 125-fold in refolding buffer containing the bi-chaperone system (1 μM Hsp104, 1 μM hHsp70, 1 μM Ydj1), 5 mM ATP, and an ATP regenerating system consisting of 25 mM phosphoenolpyruvate and 2 μM pyruvate kinase. Heat-aggregated G6PDH (2.5 µM) and β-gal (0.2 µM) were mixed with the bi-chaperone system (1 µM each) together with 4 mM ATP, 20 mM phosphoenolpyruvate, and 2 μM pyruvate kinase. The activities of recovered model substrates were measured as described21,40. G6PDH activity was measured at 30 °C in 50 mM Tris-HCl pH 8.0, 5 mM MgCl2, 0.5 mM NAD, and 2 mM glucose-6-phosphate by monitoring the formation of NADH at 340 nm using a UV-1601 spectrophotometer (Shimadzu Corporation, Kyoto, Japan).
ATPase activity assay
Hsp104 and variants (0.5 µM monomer) or Hsp104T4L and variants (0.2 µM monomer) were incubated with 2 mM ATP at 22 °C for 15 min in the absence or presence of 0.2 mg/ml κ-casein. The amount of released inorganic phosphate was measured using the malachite green assay53.
Thermal shift assay
Thermal shift assays were performed in 396-well thin-wall PCR plates. Purified Hsp104N, Set-1N, Set-2N, Set-1N/I116A, and Set-2N/I116A were diluted in buffer consisting of 25 mM HEPES-KOH pH 7.5, 150 mM potassium acetate, and 10 mM DTT. To measure protein stability, 4 mg/ml protein was used per well together with 1x SYPRO Orange (Invitrogen Corporation, Carlsbad, CA). PCR plates were sealed with optical seal, shaken, and centrifuged at 4000 rpm for 2 min after adding the protein. Thermal scanning (20 °C to 95 °C at 0.06 °C/sec) was performed using a Roche Lightcycler 480 (Roche Diagnostics Corporation, Indianapolis, IN). Fluorescence intensity was measured every 1 sec at an excitation wavelength of 465 nm and an emission wavelength of 580 nm. Curve fitting and melting temperature calculations were performed according to manufacturer’s instruction.
Electronic supplementary material
Acknowledgements
We thank the late S. Lindquist for the gift of the yeast Hsp104ND1 expression construct, and J. Choi and B. Sielaff for generating the Hsp104MMM and AN-Hsp104 chimera construct, respectively. Use of the BNL-NSLS and APS-SBC beamlines was supported by the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-98CH10886 and DE-AC02-06CH11357, respectively. The Drug Discovery Core was supported by a CPRIT Core Facility Support Award (RP160805) and Welch Foundation grant (H-Q-0042). This work was supported by grants from the National Institutes of Health (GM104980 and GM111084) and the Welch Foundation (Q-1530).
Author Contributions
J.L., N.S., S.L. designed and performed experiments, analyzed the data, and wrote the manuscript. F.T.F.T. designed experiments, analyzed the data, and wrote the manuscript. J.M.M. analyzed the data and wrote the manuscript. C.F.H. and C.C. performed experiments and analyzed the data. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11474-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sukyeong Lee, Email: slee@bcm.edu.
Francis T. F. Tsai, Email: ftsai@bcm.edu
References
- 1.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 2.Zolkiewski M, Zhang T, Nagy M. Aggregate reactivation mediated by the Hsp100 chaperones. Arch. Biochem. Biophys. 2012;520:1–6. doi: 10.1016/j.abb.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat. Rev. Mol. Cell Biol. 2013;14:617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 4.Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015;2:22. doi: 10.3389/fmolb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosser DD, Ho S, Glover JR. Saccharomyces cerevisiae Hsp104 enhances the chaperone capacity of human cells and inhibits heat stress-induced proapoptotic signaling. Biochemistry. 2004;43:8107–8115. doi: 10.1021/bi0493766. [DOI] [PubMed] [Google Scholar]
- 6.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 7.Sielaff B, Tsai FTF. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 2010;402:30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miot M, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Li J, Weaver C, Lucius A, Sha B. Crystal structures of Hsp104 N-terminal domains from Saccharomyces cerevisiae and Candida albicans suggest the mechanism for the function of Hsp104 in dissolving prions. Acta Cryst. 2017;D73:365–372. doi: 10.1107/S2059798317002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heuck A, et al. Structural basis for the disaggregase activity and regulation of Hsp104. Elife. 2016;5:e21516. doi: 10.7554/eLife.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Sielaff B, Lee J, Tsai FTF. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokom AL, et al. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat. Struct. Mol. Biol. 2016;23:830–837. doi: 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gates SN, et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeny EA, et al. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell. 2015;57:836–849. doi: 10.1016/j.molcel.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 18.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 19.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyffer F, et al. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 2012;19:1347–1355. doi: 10.1038/nsmb.2442. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, et al. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. USA. 2013;110:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 23.Doyle SM, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schirmer EC, Homann OR, Kowal AS, Lindquist S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol. Biol. Cell. 2004;15:2061–2072. doi: 10.1091/mbc.E02-08-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackrel ME, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnett ME, Zolkiewska A, Zolkiewski M. Structure and activity of ClpB from Escherichia coli. Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 2000;275:37565–37571. doi: 10.1074/jbc.M005211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnett ME, Nagy M, Kedzierska S, Zolkiewski M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005;280:34940–34945. doi: 10.1074/jbc.M505653200. [DOI] [PubMed] [Google Scholar]
- 28.Park S-K, et al. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J. Biol. Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 29.Li J, Sha B. Crystal structure of the E. coli Hsp100 ClpB N-terminal domain. Structure. 2003;11:323–328. doi: 10.1016/S0969-2126(03)00030-3. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka N, Tani Y, Hattori H, Tada T, Kunugi S. Interaction of the N-terminal domain of Escherichia coli heat-shock protein ClpB and protein aggregates during chaperone activity. Protein Sci. 2004;13:3214–3221. doi: 10.1110/ps.04780704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow IT, Barnett ME, Zolkiewski M, Baneyx F. The N-terminal domain of Escherichia coli ClpB enhances chaperone function. FEBS Lett. 2005;579:4242–4248. doi: 10.1016/j.febslet.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 32.Beinker P, Schlee S, Groemping Y, Seidel R, Reinstein J. The N-terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 2002;277:47160–47166. doi: 10.1074/jbc.M207853200. [DOI] [PubMed] [Google Scholar]
- 33.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 34.Clarke AK, Eriksson MJ. The truncated form of the bacterial heat shock protein ClpB/HSP100 contributes to development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 2000;182:7092–7096. doi: 10.1128/JB.182.24.7092-7096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno S, Nakazaki Y, Yoshida M, Watanabe YH. Orientation of the amino-terminal domain of ClpB affects the disaggregation of the protein. FEBS Lett. 2012;279:1474–1484. doi: 10.1111/j.1742-4658.2012.08540.x. [DOI] [PubMed] [Google Scholar]
- 36.Rosenzweig R, et al. ClpB N-terminal domain plays a regulatory role in protein disaggregation. Proc. Natl. Acad. Sci. USA. 2015;112:E6872–E6881. doi: 10.1073/pnas.1512783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104: insights into the mechanism of protein unfolding. J. Biol. Chem. 2008;283:30139–30150. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/S0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 40.Biter AB, Lee J, Sung N, Tsai FTF, Lee S. Functional analysis of conserved cis- and trans-elements in the Hsp104 protein disaggregating machine. J. Struct. Biol. 2012;179:172–180. doi: 10.1016/j.jsb.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeng W, Lee S, Sung N, Lee J, Tsai FTF. Molecular chaperones: guardians of the proteome in normal and disease states. F1000Res. 2015;4:1448. doi: 10.12688/f1000research.7214.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo F, Esser L, Singh SK, Maurizi MR, Xia D. Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J. Biol. Chem. 2002;277:46753–46762. doi: 10.1074/jbc.M208104200. [DOI] [PubMed] [Google Scholar]
- 43.Wang F, et al. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 44.Trentini DB, et al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. doi: 10.1038/nature20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietrosiuk A, et al. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 47.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell. 2002;9:673–683. doi: 10.1016/S1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Tek V, Akoev V, Zolkiewski M. Conserved amino acid residues within the amino-terminal domain of ClpB are essential for the chaperone activity. J. Mol. Biol. 2002;321:111–120. doi: 10.1016/S0022-2836(02)00591-0. [DOI] [PubMed] [Google Scholar]
- 49.Lee S, Tsai FTF. Crystallization and preliminary X-ray crystallographic analysis of a 40 kDa N-terminal fragment of the yeast prion-remodeling factor Hsp104. Acta Crystallogr. 2007;F63:784–786. doi: 10.1107/S1744309107038328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Brünger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Kevin C. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Carter SG, Karl DW. Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods. 1982;7:7–13. doi: 10.1016/0165-022X(82)90031-8. [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, et al. PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/S0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.