Abstract
Uromodulin (UMOD) encodes an uromodulin glycoprotein, and its mutation results in uromodulin glycoprotein dysfunction and the occurrence of gout. The aim of our study was to assess whether UMOD methylation could predict the risk of gout. A total of 89 sporadic gout cases and 103 age and gender-matched healthy controls were recruited in this study. UMOD methylation level was determined by quantitative methylation-specific PCR (qMSP) in peripheral blood, and the percentage of methylated reference (PMR) was described to represent the methylation level. Our results showed that UMOD methylation was significantly higher in gout cases than controls (median: 1.45 versus 0.75, P < 0.001). The area under curve (AUC) of UMOD methylation in gout was 0.764 (P = 2.90E-10) with a sensitivity of 65.2% and a specificity of 88.3%. UMOD methylation level was shown to be significantly correlated with the serum level of uric acid (UA) (r = −0.208, P = 0.035). Besides, the luciferase reporter assay showed that UMOD CpG island region was able to upregulate gene expression (fold change = 2, P = 0.004). In conclusion, UMOD methylation assessment might be used to predict the occurrence of gout.
Introduction
Gout is one of the oldest described rheumatic diseases. It affects 1–2% of the global population1. Gout is a complex disease with much severe comorbidity2. There are many risk factors for the development of gout, including hyperuricaemia, dietary factors, alcohol consumption, metabolic syndrome, hypertension, obesity, diuretic use, chronic renal disease, and genetic factors3, 4. Although effective treatments were available for gout, drug uptake remained poor. Many patients may experience repeated acute attacks and greatly reduces quality of life3. Therefore, the exact pathogenesis of gout is still needed to be explored.
DNA methylation is the most common but crucial way of epigenetic mechanisms5. Genes with aberrant DNA methylation contributed to the risk of diseases or disorders such as coronary heart disease6, cancer7, essential hypertension8, leukemia9 and type 2 diabetes10. However, little research about the association between DNA methylation and the pathogenesis of gout has been reported.
Uromodulin (UMOD) is located at the short arm of chromosome 16 and consists of 11 exons11. Previous study showed that gout was associated with UMOD gene mutations12. UMOD gene variants were associated with susceptibility to the risk of chronic kidney disease in several genome-wide association studies13. Besides, UMOD variants were involved in hypertension14, 15 and end-stage renal disease16. Therefore, we supposed that UMOD methylation might play a potential role in the occurrence of gout. In this study, we measured UMOD methylation level in peripheral blood to explore its association with gout in Chinese Han male population.
Materials and Methods
Sample selection
A total of 89 gout cases and 103 age-matched controls were selected from Ningbo No. 2 Hospital in Zhejiang province of China. All the individuals were Chinese Han males, and the details of their clinical information were shown in Table 1. The mean age of gout patients was 51.52 ± 14.27 years compared with 49.95 ± 12.04 years of the healthy controls.
Table 1.
The characteristics of cases and controls.
| Characteristics | Case (n = 89) | Control (n = 103) | P value* |
|---|---|---|---|
| Age (yrs) | 51.84 ± 14.09 | 50.01 ± 12.09 | 0.393b |
| ALT (U/L) | 26.00 (19.00, 42.50) | 21.00 (17.00, 26.00) | <0.001 a |
| AST (U/L) | 24.00 (18.50, 31.00) | 22.00 (18.00, 27.00) | 0.093a |
| CRE (mmol/L) | 80.52 ± 16.66 | 77.77 ± 9.79 | 0.159b |
| UA (mmol/L) | 423.46 ± 147.48 | 344.20 ± 67.06 | <0.001 b |
| Glu (mmol/L) | 5.30 (4.83, 6.00) | 4.96 (4.72, 5.25) | <0.001 a |
| Cholesterol (mmol/L) | 4.94 ± 1.17 | 4.42 ± 0.68 | <0.001 b |
| HDL (mmol/L) | 1.23 ± 0.31 | 1.50 ± 0.35 | <0.001 b |
| LDL (mmol/L) | 2.51 ± 0.98 | 2.60 ± 0.52 | 0.427b |
| TG (mmol/L) | 2.52 ± 1.52 | 1.17 ± 0.47 | <0.001 b |
| WBC (×109/L) | 8.97 ± 3.48 | 6.42 ± 1.56 | <0.001 b |
*The value in bold indicates statistical significance.
aNot conform to normal distribution, nonparametric rank test was applied, and the results were described as median (interquartile range). bConform to normal distribution, two-sample t-test was applied, and the variables were described as mean ± SD.
ALT: glutamic pyruvic transaminase; AST: glutamic oxalacetic transaminase; CRE: creatinine; UA: uric acid; Glu: blood glucose; HDL: high-density lipoprotein; LDL: low-density lipoprotein; TG: triglyceride; WBC: white blood cell.
The study protocol was approved by the Ethical Committee of Ningbo No. 2 Hospital. All methods were carried out in accordance with relevant guidelines and regulations. Written informed consent forms were obtained from all subjects.
Plasma levels of glutamic pyruvic transaminase (ALT), glutamic oxalacetic transaminase (AST) were determined by the velocity method17, 18. The concentrations of creatinine (CRE), uric acid (UA), blood glucose (Glu) and triglyceride (TG) in plasma were determined using the classic enzymatic methods19–22. Cholesterol level was measured using automated enzymatic methods23. High-density lipoprotein (HDL) cholesterol concentration was measured by enzymatic colorimetric methods with commercially available kits, and low-density lipoprotein (LDL) cholesterol concentration was measured by homogeneous assay24. The number of white blood cell (WBC) was measured by a standard blood test25.
DNA methylation analysis
The details of human genomic DNA extraction and concentration determination were as previously described10. DNA methylation was modified by EZ DNA Methylation-GoldTM kit (Zymo Research Corporation, Irvine, CA, USA). DNA methylation level was measured by quantitative methylation-specific PCR (qMSP) using the LightCycler® 480 machine (Roche Diagnostics, Mannheim, Germany). To avoid errors that may occur from differences in the loading quantity of the samples, ACTB was taken as the internal reference. We used 100% SssI-treated sperm DNA as a positive control26, and nuclease-free water as a negative control for each panel. The qMSP was performed in a total volume of 10 µl and contained 5 µl of 2× SYBR Green Master Mix, 0.25 µl primers, 4 µl of ddH2O and 0.5 µl DNA. The primers were as follows: UMOD, forward 5′-GTTGTTGTTGGCGGAGTA-3′ and reverse 5′-CGACGATAACCTAACCTACG-3′; ACTB, forward 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′. PCR amplification procedure included an initial denaturation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 20 sec, annealing at 59 °C for 30 sec and extension at 72 °C for 30 sec. A melting curve procedure included 95 °C for 15 sec, 58 °C for 60 sec and 0.11 °C per second up to 95 °C. The amount of methylated DNA (PMR, percentage of methylated reference) at a specific locus was calculated by dividing the UMOD:ACTB ratio of a sample by the UMOD:ACTB ratio of SssI-treated human sperm DNA (presumably fully methylated)26.
Luciferase reporter gene assay
The human embryonic kidney 293 T (HEK293T) cell line was cultured as previously described27. The fragment of UMOD (+7151 bp to +7550 bp), GCKR (−173 bp to +227 bp), COMT (−386 bp to +14 bp) and CCL2 (−537 bp to −138 bp) were chemically synthesized and were digested with XhoI and KpnI (New England Biolabs, Ipswich, MA). The target DNA fragment, purified by Cycle Pure Kit (Omega, Norcross, GA, USA), was cloned to pGL3 Basic vector in the presence of DNA Ligation Kit (TaKaRa, Japan). The empty pGL3-Basic vector was used as negative control, and the pGL3-Control vector, (Promega, Madison city, WI, USA) containing an SV40 promoter upstream of the luciferase gene was used as positive control. Cells were prepared in 96-well plates and the details of plasmids transfection were as described previously28. After 18–72 h of HEK293T cells transfection, renilla and firefly luciferase activity was measured by SpectraMax 190 (Molecular Devices, Sunnyvale, USA). Luciferase activity was determined with the dual luciferase reporter assay system (Dual-Luciferase® Reporter Assay Systems, Promega, Madison city, WI, USA).
Statistical analysis
All the statistical analyses were performed by SPSS software version 18.0 (SPSS, Inc., Chicago, IL, USA). Comparisons of the PMR differences between the gout cases and controls were performed by non-parametric test. The correlations between UMOD methylation and clinical features were assessed by Spearman test. Receiver operating characteristic (ROC) curves were generated to confirm the diagnostic accuracy of UMOD. P value less than 0.05 was considered to indicate a statistically significant difference.
Results
In the current study, only the male samples were selected since gout was predominant in males (a male/female ratio of 4:1)29, 30. As shown in Table 1, a total of 11 clinical characteristics were collected from all the individuals. Significantly lower level of HDL was found in the gout cases than controls (mean ± sd: 1.23 ± 0.31 versus 1.50 ± 0.35, P < 0.001). Meanwhile, significantly higher levels of ALT, UA, Glu, cholesterol, TG and WBC were found in the gout cases than controls (all P ≤ 0.001).
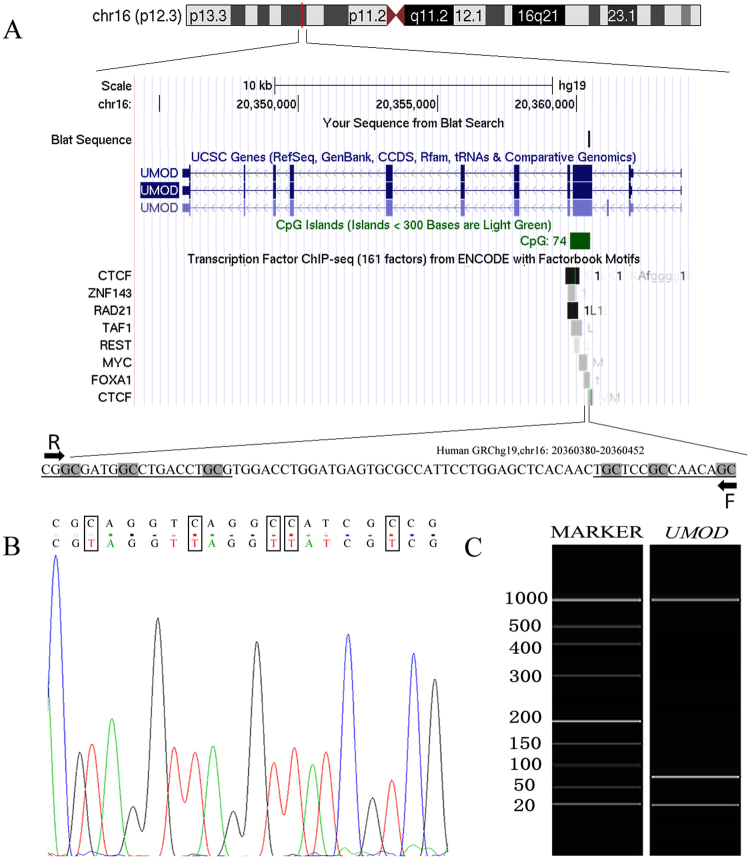
A fragment located in CpG (cytosine-phosphate-guanine) island of UMOD (Chr16: 20,344,373-20,364,037), hg19) was selected for the methylation assay (Fig. 1A). DNA sequence analysis showed that the bisulphite conversion of the template DNA was successful (Fig. 1B). Capillary electrophoresis confirmed that the amplified fragment length was 73 bp (Fig. 1C). As shown in Fig. 2, UMOD hypermethylation was significantly associated with the risk of gout. UMOD methylation was elevated in the gout cases compared with the controls [median (interquartile range): 1.45 (0.87, 3.54) versus 0.75 (0.59, 0.92), P < 0.001]. Subsequently, we analyzed the diagnostic role of UMOD hypermethylation in peripheral blood, obtaining an AUC of 0.763 (P = 2.90E-10, Fig. 3). The ROC curve showed that UMOD methylation was a promising biomarker for gout (sensitivity = 65.2%, specificity of 88.3%).
Figure 1.
The characteristics of target sequences in UMOD gene. Target sequences on UMOD gene CpG island region. (A) The target sequence is located on the CpG island of UMOD gene (location). F stands for forward primer and R stands for reverse primer. (B) Sequencing validation of the MSP product. The top row of the sequences represents the original gene sequence, and the second row shows the converted sequence. (C) The fragment length of MSP product is 73 bp.
Figure 2.
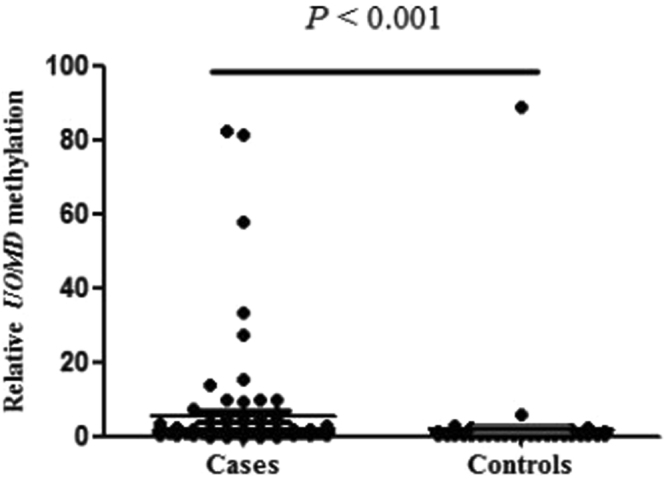
Comparison of relative UMOD methylation levels between gout and controls. The levels of UMOD methylation are represented by percent of methylated reference (PMR). The PMR values of cases and controls are 1.45 (0.87, 3.54) and 0.75 (0.59, 0.92), respectively.
Figure 3.
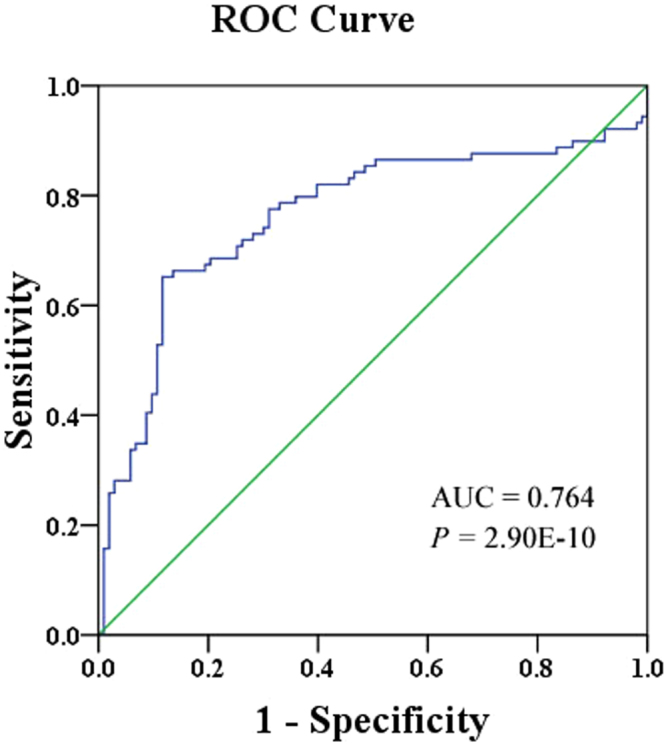
ROC curve for the diagnostic value of UMOD methylation ROC curve analysis of the UMOD gene hypermethylation in gout patients from healthy controls. ROC stands for receiver operating characteristic. AUC stands for the area under the curve. The AUC of UMOD methylation in gout was 0.764 (0.690, 0.836) with a sensitivity of 65.2% and a specificity of 88.3%.
In order to investigate the relationship between UMOD methylation and the pathogenesis of gout, the correlation tests were performed between UMOD methylation levels and clinical features in control samples. Significant inverse correlation was found with UMOD methylation level and UA (r = −0.208; P = 0.035, Table 2). However, there was no significant association between clinical features (age, ALT, AST, CRE, Glu, cholesterol, HDL, LDL, TG, WBC) and UMOD methylation (all P > 0.05, Table 2).
Table 2.
Associations between UMOD methylation levels and clinical indexes features in normal controls.
| Characteristics | r | P value*a |
|---|---|---|
| Age | −0.008 | 0.938 |
| ALT | −0.151 | 0.127 |
| AST | −0.043 | 0.667 |
| CRE | −0.126 | 0.206 |
| UA | −0.208 | 0.035 |
| Glu | 0.074 | 0.455 |
| Cholesterol | 0.001 | 0.988 |
| HDL | 0.088 | 0.374 |
| LDL | 0.041 | 0.681 |
| TG | −0.185 | 0.062 |
| WBC | −0.098 | 0.325 |
*The value in bold indicates statistical significance. aSpearman test was applied. ALT: glutamic pyruvic transaminase; AST: glutamic oxalacetic transaminase; CRE: creatinine; UA: uric acid; Glu: blood glucose; HDL: high-density lipoprotein; LDL: low-density lipoprotein; TG: triglyceride; WBC: white blood cell.
We performed a dual-luciferase reporter assay to check whether the UMOD CpG island region (+7151 bp to +7550 bp) was able to regulate gene expression. Our results showed that the transcriptional activity of recombinant pGL3-UMOD plasmid was higher compared with that of empty vector pGL3-basic (mean ± sd: 36.22 ± 2.15 versus 17.11 ± 0.16, fold change = 2, P = 0.004, Fig. 4).
Figure 4.
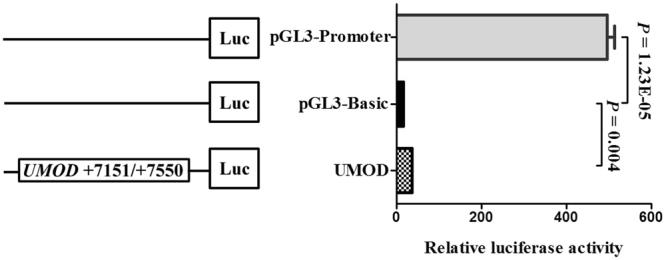
Dual-luciferase reporter assay in HEK-293T cell line. The pGL3 Basic and promoter vectors were used as negative and positive control in this study, respectively. Relative luciferase activity was performed in triplicates.
Discussion
In the present study, we reported for the first time that UMOD hypermethylation was significantly associated with the risk of gout in Chinese male patients. Moreover, the methylation levels of UMOD could be served as a predictive biomarker for the risk of gout.
DNA methylation has been studied in many metabolic diseases. Prdx2 and SCARA3 hypermethylation played an important role in the pathogenesis and progression of diabetes mellitus31. In diabetic ketoacidosis, POMC hypomethylation might make the patients’ condition worse32. Moreover, AR methylation was shown to be associated with hyperuricemia33. However, there were few articles between DNA methylation and gout. Previous studies showed that uromodulin (UMOD) played an important role in gout34. UMOD encoded the uromodulin glycoprotein. The mutations of UMOD led to uromodulin glycoprotein dysfunction and gout35.
As shown in the genecards website, UMOD expression level is able to be detected in the whole blood according to both the microarray and the RNAseq technologies. UMOD expression level is the highest in kidney, and uromodulin is the most abundant urine protein36. Decreased serum uromodulin is often correlated with the increase of serum inflammatory cytokines and the aggravation of diseases including kidney disease, hypertension and diabetes11, 36–38. In addition, the increase of serum uromodulin was a promising prognostic biomarker for recovery from acute kidney injury39. Besides, another kidney-specific gene, Klotho (KL) was reported to be much less expressed in peripheral blood cells compared in kidney40. KL hypermethylation in peripheral blood mononuclear cells was detected to be associated with the aggravation of chronic kidney disease41.
In the current study, elevated UMOD methylation in peripheral blood was shown to be associated with the risk of Gout, which is characterized by urate crystal-induced inflammation42. Since UMOD expression was often inversely associated with the levels of inflammatory cytokines in peripheral blood11, we speculate that elevated UMOD methylation in Gout might reduce the expression of UMOD, which triggers an immune response and leads to the risk of gout. In addition, our study couldn’t exclude the possibility that UMOD hypermethylation (and possibility of other genes) in peripheral blood cells could be secondary to increased circulating levels of uric acid (or of other molecules found to be increased in cases). Future study is warranted to investigate the correlation of UMOD methylation with UMOD expression in peripheral blood, kidney and other tissues.
In our study, a significantly higher serum UA level was found in gout patients than that in normal controls, and this finding might support that an elevated serum UA concentration was the main cause of gout43. But a significant inverse correlation was found between UMOD methylation level and serum UA level in controls. Due to the limited samples, we didn’t measure uromodulin levels in serum or urine in cases and controls in time. Therefore, we couldn’t test the correlation of UMOD expression and UMOD methylation in the samples. The relationship between UMOD methylation and the pathogenesis of gout needs further investigation.
Joint aspiration with synovial fluid analysis for monosodium urate crystals were the reference standard in early diagnosis of gout, however, rarely patients used this method in the early diagnosis of gout due to the risk of infection44. Our ROC curve analysis showed a moderate sensitivity of 65.2% and a high specificity of 88.3%. Moreover, increased levels of uric acid in blood is one of the clinical diagnostic criteria for gout45, 46. However, the blood uric acid index does not seem sensitive enough, patients with early-onset gout do not have a significant increase in uric acid levels47. And the detection rate of gout by using serum uric acid had a relatively low AUC of 0.6148. These findings suggested that UMOD methylation could be a diagnostic biomarker for gout. Dual-luciferase reporter system assay is a common tool to verify whether the cloned DNA fragment can play a regulation role in the expression of the luciferase reporter gene49. HK293T cell line was chosen for its easy culture and transfection. In the current study, pGL3-UMOD recombinant plasmid was constructed, and it was co-transfected into cells along with an internal control vector (pRL-SV40). Our results showed that the specific fragment (+7151 bp to +7550 bp) in UMOD CpG island region could induce a significantly higher expression of reporter gene than the control. Besides, as shown in the Supplementary Figure 1, other 400-bp inserts didn’t show obvious promoter activities, suggesting the UMOD fragment contained DNA elements with gene up-regulation. According to the TCGA dataset (https://genome-cancer.ucsc.edu/), there were five CpGs (cg03140788, cg06294373, cg21996068, cg09792189 and cg00376654) on the 400 bp fragment and three CpGs (cg06294373, cg21996068 and cg09792189) in the 73 bp fragment. Using the TCGA data, we found all the five CpGs were in positive correlation (r > 0.25, P < 0.001), suggesting that the selected CpGs might represent the neighbor CpG sites. In addition, UCSC Genome Browser website showed that the fragment was overlapped with several transcription factors binding sites, such as CTCF and ZNF143. We used P-Match method50 to predict TFBS in the selected fragment, there were Nkx2-5, c-Rel, NF-kappaB(P65), NF-kappaB in this fragment. Further study should be performed to explore the regulatory roles of CpG region in UMOD expression.
In conclusion, our study found that UMOD DNA hypermethylation in peripheral blood might be used to predict the risk of gout.
Electronic supplementary material
Acknowledgements
The research is supported by grants from National Natural Science Foundation of China (81371469) and K.C. Wong Magna Fund in Ningbo University.
Author Contributions
Y.Y., X.C. and S.D. contributed to the conception, design and final approval of the submitted version. Y.J., H.Y., J.D., H.H. and Y.M. contributed to do experiments, interpretation of data and completion of figures and tables. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yong Yang, Xiaoying Chen and Haochang Hu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11627-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang W, et al. EULAR evidence based recommendations for gout. Part I: Diagnosis. Report of a task force of the Standing Committee for International Clinical Studies Including Therapeutics (ESCISIT) Annals of the rheumatic diseases. 2006;65:1301–1311. doi: 10.1136/ard.2006.055251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y, Pandya BJ, Choi HK. Comorbidities of gout and hyperuricemia in the US general population: NHANES 2007-2008. The American journal of medicine. 2012;125:679–687 e671. doi: 10.1016/j.amjmed.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 3.Roddy E, Mallen CD, Doherty M. Gout. Bmj. 2013;347:f5648. doi: 10.1136/bmj.f5648. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama A, et al. GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Annals of the rheumatic diseases. 2017;76:869–877. doi: 10.1136/annrheumdis-2016-209632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng J, et al. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PloS one. 2014;9:e89128. doi: 10.1371/journal.pone.0089128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiaoying C, et al. [The effects of DNA methylation on the homeostasis in vascular diseases] Yi chuan = Hereditas. 2015;37:221–232. doi: 10.16288/j.yczz.14-327. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255–81267. doi: 10.18632/oncotarget.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L, et al. The interactions between alcohol consumption and DNA methylation of the ADD1 gene promoter modulate essential hypertension susceptibility in a population-based, case-control study. Hypertension research: official journal of the Japanese Society of Hypertension. 2015;38:284–290. doi: 10.1038/hr.2014.172. [DOI] [PubMed] [Google Scholar]
- 9.Jiang D, et al. The diagnostic value of DNA methylation in leukemia: a systematic review and meta-analysis. PloS one. 2014;9:e96822. doi: 10.1371/journal.pone.0096822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L, et al. Elevation of peripheral BDNF promoter methylation links to the risk of Alzheimer’s disease. PloS one. 2014;9:e110773. doi: 10.1371/journal.pone.0110773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jian L, Fa X, Zhou Z, Liu S. Functional analysis of UMOD gene and its effect on inflammatory cytokines in serum of essential hypertension patients. International journal of clinical and experimental pathology. 2015;8:11356–11363. [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson TH. gout and the kidney. Current opinion in rheumatology. 2012;24:127–131. doi: 10.1097/BOR.0b013e32834f049f. [DOI] [PubMed] [Google Scholar]
- 13.Kottgen A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nature genetics. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan S, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS genetics. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trudu M, et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nature medicine. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boger CA, et al. Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS genetics. 2011;7:e1002292. doi: 10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nath RL, Ghosh NK. A preliminary report on the determination of the normal values of serum alkaline phosphatase activity by velocity constant method. Bulletin of the Calcutta School of Tropical Medicine. 1962;10:71–72. [PubMed] [Google Scholar]
- 18.Yang X, et al. Kinetic analysis of the lactate-dehydrogenase-coupled reaction process and measurement of alanine transaminase by an integration strategy. Analytical sciences: the international journal of the Japan Society for Analytical Chemistry. 2010;26:1193–1198. doi: 10.2116/analsci.26.1193. [DOI] [PubMed] [Google Scholar]
- 19.Asrow G. Semiautomated enzymic micro methods for blood glucose and lactic acid on a single filtrate. Analytical biochemistry. 1969;28:130–138. doi: 10.1016/0003-2697(69)90164-X. [DOI] [PubMed] [Google Scholar]
- 20.Hunziker P, Keller H. [A mechanized enzymic method for the determination of uric acid (author’s transl)] Zeitschrift fur klinische Chemie und klinische Biochemie. 1975;13:89–96. [PubMed] [Google Scholar]
- 21.Whitlow K, Gochman N. Continuous-flow enzymic method evaluated for measurement of serum triglycerides with use of an improved lipase reagent. Clinical chemistry. 1978;24:2018–2019. [PubMed] [Google Scholar]
- 22.Jaynes PK, Feld RD, Johnson GF. An enzymic, reaction-rate assay for serum creatinine with a centrifugal analyzer. Clinical chemistry. 1982;28:114–117. [PubMed] [Google Scholar]
- 23.Bahijri SM, et al. The relationship of management modality in Saudi patients with type 2 diabetes to components of metabolic syndrome, gamma glutamyl transferase and highly sensitive C-reactive protein. Therapeutic advances in chronic disease. 2016;7:246–254. doi: 10.1177/2040622316658459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie D, et al. Increased high-sensitivity C-reactive protein, erythrocyte sedimentation rate and lactic acid in stroke patients with internal carotid artery occlusion. Archives of medical science: AMS. 2016;12:546–551. doi: 10.5114/aoms.2014.47879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakadate A, et al. Age, Body Mass Index, and White Blood Cell Count Predict the Resumption of Oral Intake in Subacute Stroke Patients. Journal of stroke and cerebrovascular diseases: the official journal of National Stroke Association. 2016;25:2801–2808. doi: 10.1016/j.jstrokecerebrovasdis.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Ogino S, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. The Journal of molecular diagnostics: JMD. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Z, et al. Elevated methylation of CMTM3 promoter in the male laryngeal squamous cell carcinoma patients. Clinical biochemistry. 2016;49:1278–1282. doi: 10.1016/j.clinbiochem.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Ji H, et al. OPRK1 promoter hypermethylation increases the risk of Alzheimer’s disease. Neuroscience letters. 2015;606:24–29. doi: 10.1016/j.neulet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 29.Morris C, Macdonald L, Stubbe M, Dowell A. “It’s complicated” - talking about gout medicines in primary care consultations: a qualitative study. BMC family practice. 2016;17:114. doi: 10.1186/s12875-016-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ankli B. Current epidemiology of gout. Therapeutische Umschau. Revue therapeutique. 2016;73:125–129. doi: 10.1024/0040-5930/a000767. [DOI] [PubMed] [Google Scholar]
- 31.Karachanak-Yankova S, et al. Epigenetic alterations in patients with type 2 diabetes mellitus. Balkan journal of medical genetics: BJMG. 2015;18:15–24. doi: 10.1515/bjmg-2015-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuma I, et al. Cushing Syndrome Due to ACTH-Secreting Pheochromocytoma, Aggravated by Glucocorticoid-Driven Positive-Feedback Loop. The Journal of clinical endocrinology and metabolism. 2016;101:841–846. doi: 10.1210/jc.2015-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inokuchi T, et al. Identification of a new point mutation in hypoxanthine phosphoribosyl transferase responsible for hyperuricemia in a female patient. Metabolism: clinical and experimental. 2004;53:1500–1502. doi: 10.1016/j.metabol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 34.Scolari F, Izzi C, Ghiggeri GM. Uromodulin: from monogenic to multifactorial diseases. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2015;30:1250–1256. doi: 10.1093/ndt/gfu300. [DOI] [PubMed] [Google Scholar]
- 35.Kemter E, et al. Standardized, systemic phenotypic analysis of Umod(C93F) and Umod(A227T) mutant mice. PloS one. 2013;8:e78337. doi: 10.1371/journal.pone.0078337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leiherer A, et al. Serum uromodulin is associated with impaired glucose metabolism. Medicine. 2017;96:e5798. doi: 10.1097/MD.0000000000005798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Risch L, et al. The serum uromodulin level is associated with kidney function. Clinical chemistry and laboratory medicine. 2014;52:1755–1761. doi: 10.1515/cclm-2014-0505. [DOI] [PubMed] [Google Scholar]
- 38.Steubl D, et al. Plasma Uromodulin Correlates With Kidney Function and Identifies Early Stages in Chronic Kidney Disease Patients. Medicine. 2016;95:e3011. doi: 10.1097/MD.0000000000003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Achkar TM, et al. Tamm-Horsfall protein translocates to the basolateral domain of thick ascending limbs, interstitium, and circulation during recovery from acute kidney injury. American journal of physiology. Renal physiology. 2013;304:F1066–1075. doi: 10.1152/ajprenal.00543.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuro-o M, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, et al. Elevated Klotho promoter methylation is associated with severity of chronic kidney disease. PloS one. 2013;8:e79856. doi: 10.1371/journal.pone.0079856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hainer BL, Matheson E, Wilkes RT. Diagnosis, treatment, and prevention of gout. American family physician. 2014;90:831–836. [PubMed] [Google Scholar]
- 43.Hyndman D, Liu S, Miner JN. Urate Handling in the Human Body. Current rheumatology reports. 2016;18:34. doi: 10.1007/s11926-016-0587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qaseem A, McLean RM, Starkey M, Forciea MA. & Clinical Guidelines Committee of the American College of, P. Diagnosis of Acute Gout: A Clinical Practice Guideline From the American College of Physicians. Annals of internal medicine. 2017;166:52–57. doi: 10.7326/M16-0569. [DOI] [PubMed] [Google Scholar]
- 45.Khanna D, et al. American College of Rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia. Arthritis care & research. 2012;64:1431–1446. doi: 10.1002/acr.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruoff G, Edwards NL. Overview of Serum Uric Acid Treatment Targets in Gout: Why Less Than 6 mg/dL? Postgraduate medicine. 2016;128:706–715. doi: 10.1080/00325481.2016.1221732. [DOI] [PubMed] [Google Scholar]
- 47.McQueen FM. Gout in 2013. Imaging, genetics and therapy: gout research continues apace. Nature reviews. Rheumatology. 2014;10:67–69. doi: 10.1038/nrrheum.2013.164. [DOI] [PubMed] [Google Scholar]
- 48.Abhishek A, Valdes AM, Zhang W, Doherty M. Association of Serum Uric Acid and Disease Duration With Frequent Gout Attacks: A Case-Control Study. Arthritis care & research. 2016;68:1573–1577. doi: 10.1002/acr.22855. [DOI] [PubMed] [Google Scholar]
- 49.Xu YZ, Kanagaratham C, Jancik S, Radzioch D. Promoter deletion analysis using a dual-luciferase reporter system. Methods in molecular biology. 2013;977:79–93. doi: 10.1007/978-1-62703-284-1_7. [DOI] [PubMed] [Google Scholar]
- 50.Chekmenev DS, Haid C, Kel AE. P-Match: transcription factor binding site search by combining patterns and weight matrices. Nucleic acids research. 2005;33:W432–437. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.