Abstract
Growth Factor Independence 1 (GFI1) is a transcriptional repressor that plays a critical role during both myeloid and lymphoid haematopoietic lineage commitment. Several studies have demonstrated the involvement of GFI1 in haematological malignancies and have suggested that low expression of GFI1 is a negative indicator of disease progression for both myelodysplastic syndromes (MDS) and acute myeloid leukaemia (AML). In this study, we have stratified AML patients into those defined as having a normal karyotype (CN-AML). Unlike the overall pattern in AML, those patients with CN-AML have a poorer survival rate when GFI1 expression is high. In this group, high GFI1 expression is paralleled by higher FLT3 expression, and, even when the FLT3 gene is not mutated, exhibit a FLT3-ITD signature of gene expression. Knock-down of GFI1 expression in the human AML Fujioka cell line led to a decrease in the level of FLT3 RNA and protein and to the down regulation of FLT3-ITD signature genes, thus linking two major prognostic indicators for AML.
Introduction
The concept of disease stratification promises to provide great improvements in the diagnosis, prognosis, and treatment of cancer, but requires robust and readily measurable biomarkers in order to be feasible. In the case of haematological malignancies, much has been possible through traditional phenotypic categorisation and the identification of associated genetic mutations. More recently, genome wide approaches, both at the level of gene mutations and transcriptome analysis, have enabled refinement of the classification of haematological malignancies, but there is still a great need to identify reliable molecular differences not only as prognostic indicators but also as potential therapeutic targets.
Acute myeloid leukaemia (AML) is a malignant myeloproliferative disease of the bone marrow accounting for ~10% of all haematological disorders1, 2. Currently, risk classification of AML with normal karyotype, representing 50% of the cases, is based on molecular biomarkers including mutations in the genes encoding FLT3, NPM1, NRAS, KRAS, KIT and CEBPA3. FLT3 is a tyrosine kinase receptor that is expressed in early haematopoietic cells and is essential for their proliferation and differentiation4, 5. FLT3 is known to be expressed at elevated levels on the AML blast cells in over 70% of AML patients and it is among the most commonly mutated genes in AML6. The mutations are typically internal tandem duplications within the juxta-membrane domain of the receptor (FLT3-ITD), occurring in 20–30% of AML cases7, 8. FLT3-ITD mutations lead to constitutive signalling and factor-independent cell survival and proliferation9, and are associated with adverse clinical outcome. The leukaemogenic effects of the FLT3-ITD mutations are reflected in a distinct gene regulation signature, which is characterised by up and down regulation of DNase I hypersensitive sites and genes encoding key haematopoietic regulators and functional proteins10.
Growth Factor Independence 1 (GFI1), a zinc-finger transcriptional repressor that plays several critical roles in haematopoietic lineage commitment and development11–13 is being increasingly associated with haematological malignancies14–18. Mutations in GFI1 that lead to a dominant-negative loss of function have been reported in a number of patients with congenital neutropenia16, whilst investigation of single nucleotide polymorphisms (SNP) associated with an increased predisposition towards AML led to the identification of a serine-asparagine substitution in the N-terminal region of GFI1 (GFI136N)17. In line with these findings, Hönes et al. analysed a cohort of 524 de novo AML cases and concluded that low expression of GFI1 is linked with an inferior prognosis18.
In this study we have further analysed publicly available AML data with respect to the link between GFI1 expression and prognosis, but have stratified patients on the basis of the mutational status of their disease. This analysis has revealed that in cytogenetically normal AML (CN-AML) patients, high GFI1 expression predicts a significantly inferior overall survival. This higher GFI1 expression correlated with higher FLT3 levels and a gene expression profile reminiscent of that seen in AML with the FLT3-ITD mutation, thus suggesting an important molecular connection between these factors in CN-AML.
Results
CN-AML patients with high GFI1 expression have a worse clinical outcome
The analysis of AML gene profiling array data presented by Hönes et al.18 focused on the importance of low GFI1 expression as a marker of inferior outcome in AML/MDS patients. The authors of the latter study considered all patients in the cohort reported by Verhaak et al.19, encompassing a broad spectrum of mutations including gene fusions with core transcriptional activators or repressors. We were interested to know how patients with CN-AML might represent a distinct subset in terms of the mechanisms underpinning leukaemogenesis, in particular in relation to the importance of GFI1 and FLT3 expression, and FLT3 mutational status.
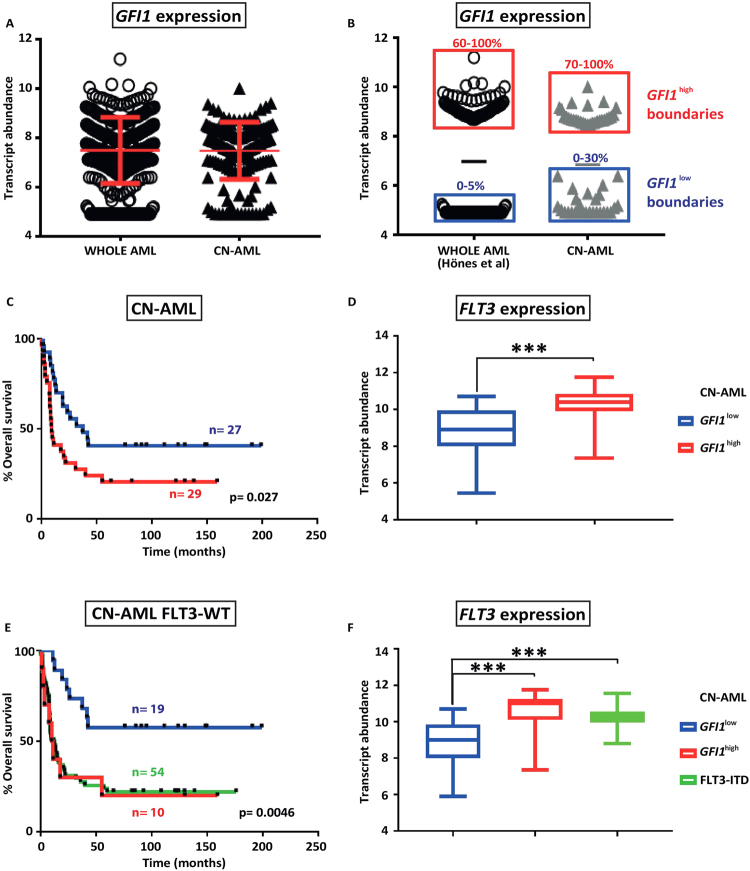
For our analysis, we took the same data set as described by Verhaak et al.19 and compared survival outcomes for the whole cohort with those for patients with CN-AML, distinguishing patients with low or high expression of GFI1. Our CN-AML category (n = 178) excluded patients displaying abnormal or unknown karyotype or classified in the M3 FAB subgroup, as those patients are treated differently. The distribution of GFI1 expression amongst all AML patients is not significantly different to that seen in the CN-AML subgroup (Fig. 1A). From the CN-AML group we selected the bottom 30% of the whole GFI1 expression range as low expressers (GFI1 low, n = 27) and the top 30% as high expressers (GFI1 high, n = 29), which closely encompasses the range of GFI1 expression surveyed by Hönes et al.18 (Fig. 1B). In this cohort, GFI1 low patients demonstrated a significantly inferior outcome compared to the GFI1 high patients (p = 0.027) (Fig. 1C).
Figure 1.
High GFI1 expression associates with raised FLT3 expression and inferior survival in CN-AML patients. (A) Scatter plot representation of GFI1 expression for patient samples from the whole Verhaak et al.19 dataset (WHOLE AML) or selected on the basis of having a normal karyotype (CN-AML). (B) Representation of GFI1 low and GFI1 high expression boundaries as reported by Hönes et al.18 (0–5% low expression, 60–100% high expression) and in the present study (0–30% low expression, 70–100% high expression). (C) Kaplan-Meyer representation of overall survival for CN-AML patients from the Verhaak et al.19 dataset subdivided in low and high GFI1 expressers. (D) Box plot representing FLT3 transcript levels in GFI1 low versus GFI1 high CN-AML patient samples. Statistical significance: ***< 0.001, **< 0.01, *< 0.05. (E) Kaplan-Meyer estimates of overall survival for GFI1 low and GFI1 high FLT3-WT CN-AML samples in comparison with FLT3-ITD CN-AML patients from the Verhaak et al.19 dataset. (F) Boxplot depicting FLT3 transcript levels in GFI1 low and GFI1 high FLT3-WT CN-AML in comparison with FLT3-ITD CN-AML samples.
Similar to the observations of Hönes et al.18 when considering the total AML cohort, analysis of the mutational status of our CN-AML subgroups revealed that FLT3-ITD and NPM1c mutations were more common in GFI1 high samples, these being found in 59% (p = 0.0001) and 52% (p = 0.079) of patients, when compared to the GFI1 low subgroup in which they were found in 11.2% and 29% of the patients, respectively (Table 1). No statistically significant difference was observed for IDH1, IDH2, NRAS, KRAS, CEBPA and EVI1 expression. Since the FLT3-ITD mutation is generally associated with higher FLT3 expression, we next analysed the abundance of FLT3 transcript and observed this to be higher in GFI1 high samples (p = 0.0002, Fig. 1D). No differences between GFI1 low and GFI1 high groups were seen with respect to FAB classification, age, or sex (Table 1). Considering that FLT3-ITD is the poorest prognostic marker in CN-AML, we speculated that this could account for the inferior survival observed in the GFI1 high patient subgroup. We therefore sub-classified the GFI1 high CN-AML patients according to the presence or absence of FLT3-ITD. Surprisingly, GFI1 high FLT3-WT patients (n = 10) still displayed lower overall survival (p = 0.0046) (Fig. 1E) and higher FLT3 expression (p = 0.000033) that is comparable to the levels seen in patients carrying FLT3-ITD mutations (Fig. 1F). Similar to the sub-cohort of CN-AML patients carrying FLT3-ITD mutations, no differences in the FAB classification, age or sex were observed.
Table 1.
Genetic and phenotypic characteristics of the GFI1low and GFI1high leukaemias in the CN-AML category.
| GFI1low (n = 27) | GFI1high (n = 29) | p-value | |
|---|---|---|---|
| Mutations | |||
| IDH1 | 1 | 4 | 0.1990 |
| IDH2 | 3 | 2 | 0.4653 |
| NPM1 | 8 | 15 | 0.0792 |
| FLT3-ITD | 3 | 17 | 0.0002 |
| NRAS | 3 | 1 | 0.2787 |
| KRAS | 2 | 0 | 0.2453 |
| EVI1 | 2 | 0 | 0.2543 |
| CEBPA | 4 | 1 | 0.1544 |
| FAB Classification | |||
| M0 | 1 | 2 | 1 |
| M1 | 7 | 10 | 0.56 |
| M2 | 6 | 3 | 0.2884 |
| M4 | 4 | 4 | 1 |
| M5 | 7 | 10 | 0.5678 |
| M6 | 2 | 0 | 0.2279 |
| Sex | 0.391 | ||
| Male | 12 | 14 | |
| Female | 15 | 15 | |
| Median age | 48.33 | 46.27 | |
The table shows data relating to the 56 patient AMLs included in the CN-AML sub group, including the occurrence of common specific leukaemia-associated mutations, the FAB categorisation, and the sex and age of the patients.
High GFI1 expression corresponds with a FLT3-ITD gene expression signature in FLT3-WT CN-AML
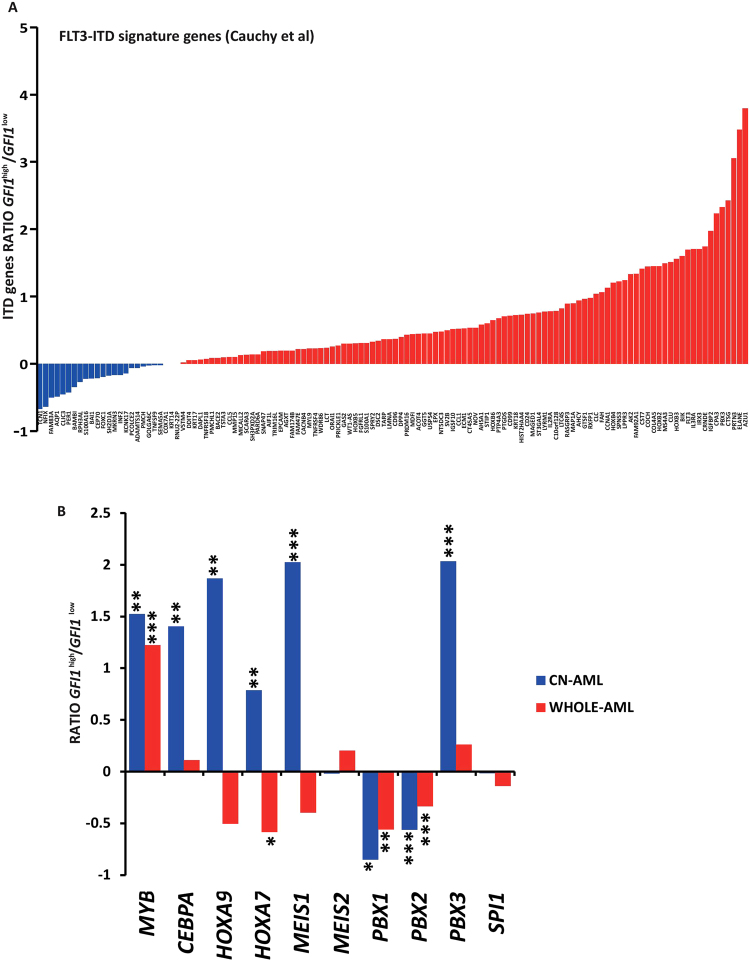
It was recently reported that FLT3-ITD is associated with a distinct gene expression profile, the specific signature being defined as those genes expressed at least one log2-fold higher in FLT3-ITD AML compared to CD34+ peripheral blood stem cells (PBSC), excluding those genes linked to mature myeloid differentiation that are expressed in CD14+ bone marrow cells at a level twice as high in PBSC10. This FLT3-ITD AML gene expression signature incorporates 134 genes, which were further validated against the dataset from Verhaak et al.19.
We therefore investigated whether high GFI1 expression in the CN-AML FLT3-WT leukaemias corresponds to higher abundance of the FLT3-ITD molecular signature genes. Using the AML gene expression dataset of Verhaak et al.19, we determined the expression of the FLT3-ITD signature genes in FLT3-WT CN-AML patients and stratified these according to the level of GFI1 RNA. Strikingly, this analysis revealed that in GFI1 high AML the expression of genes making up the FLT3-ITD signature was largely elevated compared to AML with low GFI1 expression (Fig. 2A). Further to validate these findings, we confirmed the association between high GFI1 levels and FLT3-ITD signature genes in another independent CN-AML cohort from a study performed on 251 CN-AML samples, reported by Kohlmann et al.20 (Fig. 1).
Figure 2.
Analysis of FLT3-ITD signature genes and known FLT3 regulators in CN-AML (A) Histogram representing gene expression ratio of FLT3-ITD molecular signature in GFI high versus GFI1 low FLT3-WT CN-AML patient samples from the Verhaak dataset. (B) Analysis of expression levels of known regulators of FLT3 activity in leukaemia, comparing GFI1 low versus GFI1 high FLT3-WT CN-AML samples (blue bars) or WHOLE AML samples (red bars). Statistical significance: ***< 0.001, **< 0.01, *< 0.05
Known regulators of FLT3 are over expressed in GFI1high CN-AML
Recent reports have demonstrated a mechanism for the Gfi1-dependent acceleration of K-Ras driven myeloproliferative disorders in mice through the over expression of HoxA9 and other AML-related genes15, 21, eventually leading to the development of AML. This study also revealed elevated expression of the HoxA9 partner proteins Meis1 and Pbx1, and of their known target genes Myb and Spi1/PU.1. Considering that these transcription factors, together with CEBPα, are among the main regulators of FLT3 activity in AML22–24, we next sought to determine whether high GFI1 levels are associated with differences in their expression in CN-AML patients. Comparing GFI1 high versus GFI1 low FLT3-WT CN-AMLs, we found higher expression in the former of MYB (p = 0.0029), CEBPA (p = 0.0043), HOXA9 (p = 0.0052), HOXA7 (p = 0.0033), MEIS1 (p = 0.000043) and PBX3 (p = 0.0001), while lower abundance was observed for PBX1 (p = 0.011) and PBX2 (p = 0.00004). No differences were observed in the levels of MEIS2 and PU.1 (Fig. 2B). The latter is unsurprising as we have previously shown that PU.1 is directly involved in regulating FLT3 in haematopoietic progenitor cells but not in leukaemic cells23–25. While the lower levels of PBX1 and PBX2 observed in high GFI1 patients are in agreement with the findings presented by Horman et al.15, higher expression of HOXA9 and its partner MEIS1 are in contrast with their observations. This difference could be explained by the fact that our analysis was performed in CN-AML patients only, therefore excluding patients harbouring chromosomal translocations, such as those involving MLL, in which HOXA9 expression is often deregulated26, 27. To address this, we performed the same analysis using patient samples from the whole AML cohort, classified into GFI1 low and high expressers. In line with previous observations, this analysis revealed lower expression levels of HOXA9, HOXA7 and MEIS1 (Fig. 2B).
Reduction of GFI1 in AML cell lines leads to decreased expression of FLT3-ITD signature genes
In order to assess the significance of our observed correlation between GFI1 expression and the level of FLT3-ITD signature genes in FLT3-WT CN-AML, we next investigated whether manipulation of GFI1 expression in a human AML cell line would lead to corresponding changes in the expression of the FLT3-ITD signature genes. First, we measured the relative expression of GFI1 RNA in human AML cell lines characterised by the expression of wild type FLT3 receptor, including KG1a, Kasumi-1, THP1, and Fujioka. Quantitative RT-PCR revealed that GFI1 is least expressed in KG1a and most highly expressed in Fujioka cells (Fig. 3A). To determine if higher expression of the FLT3-ITD signature genes also correlates with GFI1 level in these cells lines, we then tested the transcript abundance of the FLT3-ITD signature genes NFIX, FAM92A1, BIK, IGFBP2, LYRM1, IRX3, PBX3, PRTN3, AK2, CTSG, FLT3, ELANE and AZU1. Interestingly, this analysis demonstrated higher expression of most of these genes, with the exception of FAM92A1, in those cell lines with the highest expression of GFI1 (Fig. 3B).
Figure 3.
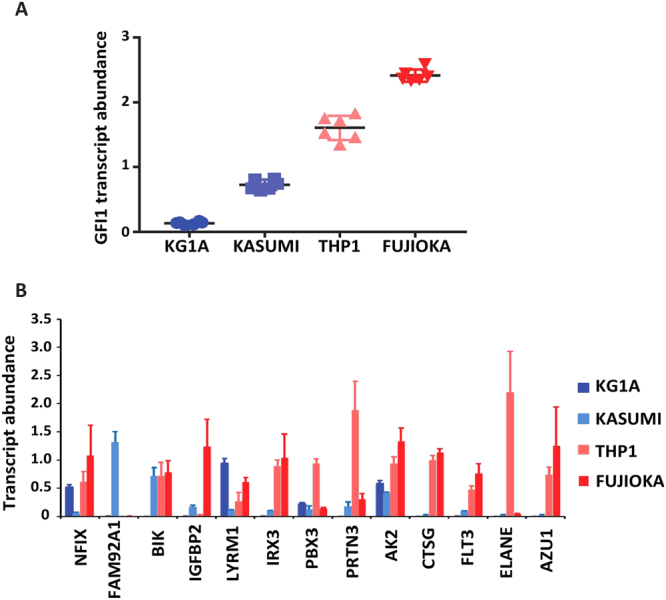
Expression analysis of a set of FLT3-ITD molecular signature genes in human AML cell lines (A) Scatter plot showing the relative abundance of GFI1 transcript in KG1A, Kasumi-1, THP1 and Fujioka human AML cell lines. (B) Histogram depicting the relative transcript abundance of FLT3-ITD signature component in the Fujioka cell line.
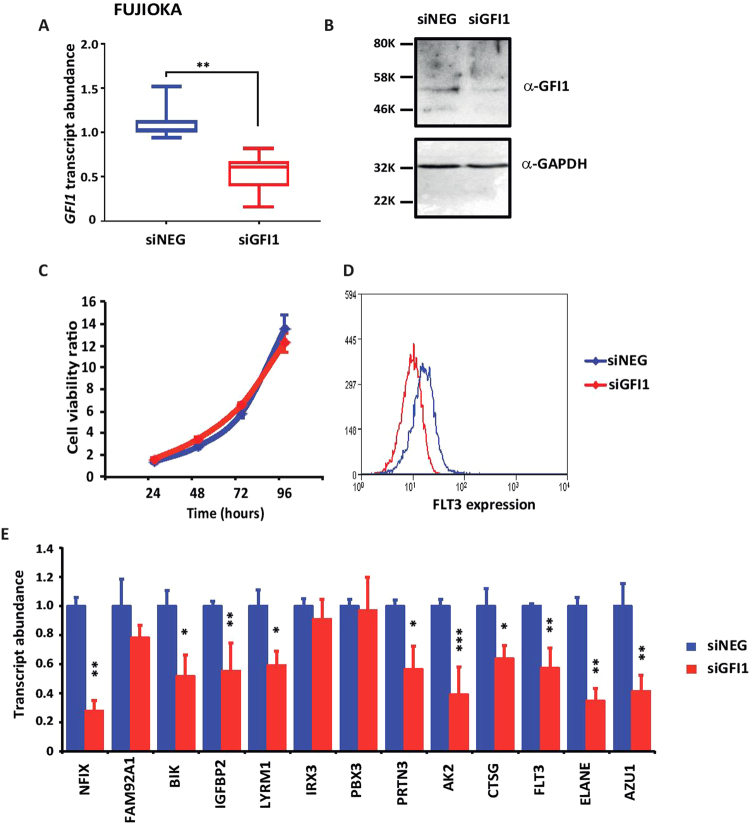
Next, we performed siRNA-mediated knockdown of GFI1 in the Fujioka line to determine if the highlighted FLT3-ITD signature genes were dependent on the high level of GFI1 expression in these cells. Fujioka cells were electroporated with siRNA targeting either GFI1 or a scrambled negative control. Cells were harvested 24 hours post transfection to assess GFI1 expression levels by quantitative RT-PCR, revealing a highly significant down regulation of both GFI1 RNA (p < 0.001) (Fig. 4A). Cells were additionally collected at 48 hours for preparation of protein extracts and assessment of GFI1 protein levels by immunoblotting (Fig. 4B). Cell counts performed every 24 hours post transfection revealed that GFI1 down regulation had no significant effect on cell growth (Fig. 4C). Fujioka cells were maintained in culture for 96 hours post transfection at which point the expression of several surface markers associated with myeloid differentiation was tested by immunofluorescence / flow cytometry. CD34, CD38, CD11b, CD14, and CD56 showed no measurable differences (data not shown), whereas FLT3 surface expression was significantly decreased (Fig. 4D). Importantly, analysis of the expression of FLT3-ITD signature genes demonstrated a reduction in the abundance of several of these, the most notable being NFIX, BIK, IGFBP2, LYRM1 PRTN3, AK2, CTSG, FLT3, ELANE, and AZU1 (Fig. 4E).
Figure 4.
siRNA-mediated downregulation of GFI1 expression in Fujioka cells (A) GFI1 transcript abundance was determined by quantitative-PCR in cells transfected with GFI1 siRNA and negative control siRNA. Results are representative of 5 independent experiments. Statistical significance was calculated using student’s t-test. ***< 0.001, **< 0.01, *< 0.05. (B) Immunoblot analysis of GFI1 protein in extracts from cells transfected with GFI1 siRNA and negative control siRNA. GAPDH was used as internal control. For detection of GFI1 the membrane was cut into two just below the 46 KDa ladder mark to avoid detection of a cross-reacting non-specific band at around 40 KDa. (C) Cell viability was calculated by counting transfected cells every 24 hours for 4 consecutive days. (D) Flow cytometric staining of transfected Fujioka cells with a labelled antibody against FLT3. (E) RNA quantification by q-PCR of FLT3-ITD molecular signature component 24 hours post siRNA transfection in Fujioka cells. Statistical significance was calculated using student’s t-test. ***< 0.001, **< 0.01, *< 0.05.
Discussion
In this study, we show that higher levels of GFI1 expression can be used to predict unfavourable outcome in AML patients with normal cytogenetics, and thereby provide a possible stratification for therapy choices. Additionally, we found that higher GFI1 levels are also associated with higher FLT3 expression and elevated levels of FLT3-ITD signature genes even in patients with non-mutated FLT3.
Compared to earlier findings by other groups, the way in which we have distinguished between CN-AML and AML with abnormal cytogenetics illustrates how stratification based on a single factor such as GFI1 expression can lead to quite different conclusions. Hence, Hönes et al.18 sub-fractioned AML patients purely according to GFI1 expression revealing that overall inferior survival associated with low GFI1 levels. In the present study we first classified AML based on karyotype, dividing them into abnormal karyotype only and CN-AML only and then sub-fractioning these two groups into low and high GFI1 expressers. In the first group we observed that low GFI1 expressers were indeed associated with inferior outcome, in agreement with findings reported previously. Strikingly, when the same criterion was applied to the CN-AML group, we observed that high GFI1 expressers were instead characterized by worse overall survival.
We also considered how the status of FLT3 expression or mutation might correlate with the stratification. In their study of unfractionated AML Hönes et al.18 suggested that FLT3-ITD associates with high GFI1 levels and favourable outcome measures, which is a surprising finding given that FLT3-ITD is the most significant prognostic factor in AML and is associated with unfavourable outcome. However, as we have discussed, patients from this cohort were not subcategorized based on karyotype, raising the possibility that GFI1 impacts on leukaemia in different ways according to the driver mutations. When we looked at the CN-AMLs we also observed an association of FLT3-ITD with the GFI1 high group, suggesting that FLT3-ITD was responsible for the inferior outcome observed in those patients. To test this, we looked instead at only those CN-AML that had a wild type FLT3 status, again classifying into GFI1 low versus GFI1 high. This analysis showed that the overall survival of the GFI1 high AML was much worse, and moreover, these AML exhibited a significantly higher expression of FLT3.
Several studies, mainly in mouse models, have addressed the role of GFI1 in haematological malignancies. In line with an association between high GFI1 expression and poor outcomes it has been reported that Gfi1 cooperates with Pim-1 and Myc in the genesis of T-cell lymphoma28. High levels of GFI1 expression were also found to be important in accelerating T-cell proliferation and preventing induced cell death in Jurkat T-cells29. Furthermore, Khandanpour et al.21 reported a requirement for Gfi1 in the establishment and progression of murine B-cell lymphoma and T-cell acute lymphoblastic leukaemia driven by various genetic lesions, the ablation of Gfi1 leading to significant tumour regression and increased host survival in a p53-dependent manner30. In apparent contrast to these latter findings, the role of Gfi1 in the regulation of HoxA9, Meis1 and Pbx1 in murine myeloid cells appears to prevent predisposition to haematological malignancies, its loss-of-function decreasing the latency of KRas-driven MDS15. These observations were further supported by the identification of a SNP in the GFI1 gene that generates a variant protein (GFI1[36N]), which experimentally accelerated KRas-driven myeloproliferative disorders by inducing epigenetic changes at the HoxA9 locus21. Similarly, studies using a humanized GFI1 knockdown mouse model showed that lower GFI1 levels accelerate the progression of MLL-AF9- and NUP98-HoxD13-driven AML.
These contrasting observations from different model systems clearly imply that the functional consequences of GFI1 activity can be quite different and context dependent, and there is a need to investigate more broadly what mechanisms operate. For example, one could speculate that in AML involving translocations in major chromatin remodellers, such as in MLL-AF4 gene fusions, high levels of GFI1 might antagonise runaway activation while the converse would fail to compensate the latter, resulting in additionally impaired differentiation and thus contributing towards more aggressive leukaemogenesis. In CN-AML however, GFI1 might play a more physiological role with higher expression corresponding to repression of alternative lineages, thus favouring differentiation and thus more manageable AML.
To gain an initial insight into the influence of high GFI1 expression in FLT3-WT CN-AML we sought to determine if there is any correspondence with the signature of gene expression that characterises FLT3-ITD AML. Interestingly, this analysis showed that, despite those samples having wild type FLT3, the molecular signature of FLT3-ITD was prominent in GFI1 high cells. More importantly, in the attempt to validate the relevance of these findings, we observed that a number of the components of the FLT3-ITD molecular signature (IGFBP2, PRTN3, AK2, CTSG, ELANE and AZU1) were down regulated after siRNA-mediated silencing of GFI1 in Fujioka human AML cell line. This suggests that one of the main mechanisms of GFI1 leading to inferior outcome might directly or indirectly act through these genes.
Taken together, our results call for a more elaborate stratification of AML patients to ensure proper diagnosis and effective treatment and demonstrate that high GFI1 expression is a reliable and powerful prognostic indicator for CN-AML. Our findings also suggest that it will be fruitful to investigate in detail how GFI1 is linked to driver mutations, including those involving FLT3.
Materials and Methods
Patient profiling arrays
Log2 transformed, MASS normalised microarray expression data from Verhaak et al.19 and Kohlmann et al.20 were retrieved from GEO accession number GSE6891 and GSE15434, respectively. CN-AML patients reported in those cohorts were ranked according to their GFI1 expression into GFI1 high (top 30% of expression range) and GFI1 low (bottom 30%). Gene expression fold changes were expressed as log2 ratios of GFI high /GFI1 low gene expression levels. Up- and down-regulated genes were identified as those displaying log2 fold change ≥ 1 and p < 0.05. For data from Cauchy et al.10, and gene expression levels and fold changes from Verhaak et al.19 and Kohlmann et al.20 were retrieved for the previously published 134-gene FLT3-ITD signature.
AML cell lines
KG1A, Kasumi-1, THP1, and Fujioka human AML cell lines were grown in RPMI 1640 medium supplemented with 10% foetal bovine serum, 50 u/ml penicillin, 50 µg/ml streptomycin, and 2mM L-Glutamine. Cells were maintained at 0.5 × 106 cells/ml and were washed with phosphate buffered saline solution between passages.
Transfections, cell viability assays, and flow cytometry analysis
In total, 5 × 106 Fujioka cells were electroporated with 300 nM of either GFI1 siRNA (s199938, Ambion – Life Technologies) or scrambled control siRNA (4390843 Silencer Select Negative Control #1) using the BIORAD Gene Pulser XCell (BIORAD, Hercules, California, US). Cell proliferation assays were performed in triplicate with Fujioka cells transfected with GFI1 siRNA and negative control. Cells were counted every 24 hours for 4 consecutive days and growth curves were obtained by quantifying the number of viable cells. Flow cytometry analysis was performed on transfected Fujioka cells stained using antibodies against CD34, CD38, CD11b, CD14, CD56, and FLT3. All the antibodies were purchased from eBioscience.
Quantitative PCR and Western Blot
RNA extraction was performed 24 hours post siRNA transfection using RNeasy Mini kit (Qiagen) and first-strand cDNA synthesis was performed using standard protocols. Quantitative RT-PCR analysis of GFI1, FLT3, GAPDH, and the components of the FLT3-ITD molecular signature was performed using predesigned Taqman gene expression assays (Applied Biosystems). Total proteins obtained from Fujioka cells transfected with either GFI1 siRNA or scrambled control were used for Western Blot analysis. Antibodies were as follows: anti-GFI1 mouse monoclonal (1:500, Santa Cruz Biotechnology) and anti-GAPDH mouse monoclonal (1:10000 dilution, Abcam).
Data availability
All patient data described were obtained from publically available databases.
Electronic supplementary material
Acknowledgements
The authors would like to thank Dr Maarten Hoogenkamp for critical reading of the manuscript and Niall Gilding for providing the Fujioka cell line. The work was supported by a Bloodwise Programme grant held by JF and PG, and through funding provided by the College of Medical and Dental Sciences.
Author Contributions
G.V. conceived and designed the research, analysed the data, performed the in vitro experiments, and wrote the manuscript; D.S.W., D.E.G. and D.B. carried out the genetic manipulation studies; C.W. and P.C. performed bioinformatics analysis; D.J.L.C. and P.N.C. provided reagents/materials and useful discussions; P.G. and J.F. supervised the research and wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Paloma Garcia and Jon Frampton Jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11718-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Giacomo Volpe, Email: g.volpe@bham.ac.uk.
Jon Frampton, Email: j.frampton@bham.ac.uk.
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Dohner H, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 3.Gregory TK, et al. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler HG. Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia. 1996;10:588–599. [PubMed] [Google Scholar]
- 5.Rosnet O, et al. Expression and signal transduction of the FLT3 tyrosine kinase receptor. Acta Haematol. 1996;95:218–223. doi: 10.1159/000203881. [DOI] [PubMed] [Google Scholar]
- 6.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 7.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 8.Kelly LM, et al. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.V99.1.310. [DOI] [PubMed] [Google Scholar]
- 9.Kottaridis PD, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.V98.6.1752. [DOI] [PubMed] [Google Scholar]
- 10.Cauchy P, et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015;12:821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancrin C, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- 12.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 13.Vassen L, Okayama T, Moroy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 2007;109:2356–2364. doi: 10.1182/blood-2006-06-030031. [DOI] [PubMed] [Google Scholar]
- 14.Vassen L, et al. Growth factor independent 1b (Gfi1b) and a new splice variant of Gfi1b are highly expressed in patients with acute and chronic leukemia. Int J Hematol. 2009;89:422–430. doi: 10.1007/s12185-009-0286-5. [DOI] [PubMed] [Google Scholar]
- 15.Horman SR, et al. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Person RE, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandanpour C, et al. A variant allele of Growth Factor Independence 1 (GFI1) is associated with acute myeloid leukemia. Blood. 2010;115:2462–2472. doi: 10.1182/blood-2009-08-239822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hones JM, et al. GFI1 as a novel prognostic and therapeutic factor for AML/MDS. Leukemia. 2016;30:1237–1245. doi: 10.1038/leu.2016.11. [DOI] [PubMed] [Google Scholar]
- 19.Verhaak RG, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohlmann A, et al. Gene expression profiling in AML with normal karyotype can predict mutations for molecular markers and allows novel insights into perturbed biological pathways. Leukemia. 2010;24:1216–1220. doi: 10.1038/leu.2010.73. [DOI] [PubMed] [Google Scholar]
- 21.Khandanpour C, et al. The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood. 2012;120:4006–4017. doi: 10.1182/blood-2011-02-334722. [DOI] [PubMed] [Google Scholar]
- 22.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carotta S, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Volpe G, et al. C/EBPalpha and MYB regulate FLT3 expression in AML. Leukemia. 2013;27:1487–1496. doi: 10.1038/leu.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volpe G, et al. Regulation of the Flt3 Gene in Haematopoietic Stem and Early Progenitor Cells. PLoS One. 2015;10:e0138257. doi: 10.1371/journal.pone.0138257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faber J, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt T, et al. Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene. 1998;17:2661–2667. doi: 10.1038/sj.onc.1202191. [DOI] [PubMed] [Google Scholar]
- 29.Karsunky H, Mende I, Schmidt T, Moroy T. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002;21:1571–1579. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- 30.Khandanpour C, et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23:200–214. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All patient data described were obtained from publically available databases.