Abstract
Grapevine fruit development is a dynamic process that can be divided into three stages: formation (I), lag (II), and ripening (III), in which physiological and biochemical changes occur, leading to cell differentiation and accumulation of different solutes. These stages can be positively or negatively affected by multiple environmental factors. During the last decade, efforts have been made to understand berry development from a global perspective. Special attention has been paid to transcriptional and metabolic networks associated with the control of grape berry development, and how external factors affect the ripening process. In this review, we focus on the integration of global approaches, including proteomics, metabolomics, and especially transcriptomics, to understand grape berry development. Several aspects will be considered, including seed development and the production of seedless fruits; veraison, at which anthocyanin accumulation begins in the berry skin of colored varieties; and hormonal regulation of berry development and signaling throughout ripening, focusing on the transcriptional regulation of hormone receptors, protein kinases, and genes related to secondary messenger sensing. Finally, berry responses to different environmental factors, including abiotic (temperature, water-related stress and UV-B radiation) and biotic (fungi and viruses) stresses, and how they can significantly modify both, development and composition of vine fruit, will be discussed. Until now, advances have been made due to the application of Omics tools at different molecular levels. However, the potential of these technologies should not be limited to the study of single-level questions; instead, data obtained by these platforms should be integrated to unravel the molecular aspects of grapevine development. Therefore, the current challenge is the generation of new tools that integrate large-scale data to assess new questions in this field, and to support agronomical practices.
Keywords: grapevine fruit development, seed development, biotic and abiotic stresses, transcriptomics, metabolomics
Introduction
The grapevine (Vitis vinifera), one of the most important fruit crops worldwide, provides berries that can be used as fresh fruit, raisins, and for wine making and distillation of liquors. The grapevine has fleshy berries derived from the ovary of the flower, whose development is a complex process that can be divided into three stages with distinctive physiological and biochemical characteristics (Coombe and McCarthy, 2000). During the first stage (stage I) there is an exponential increase in berry size due to rapid cell division and growth, leading to the establishment of the final number of cells (Coombe and Hale, 1973). Some of the principal compounds that are present in the berry at stage I are tartaric and malic acids, which accumulate mainly in skin and flesh and confer acidity to fruits and wine (Sweetman et al., 2009, 2012). The second stage (stage II) is a lag phase in which important physiological and biochemical changes occur, such as softening and coloring. Within this stage, veraison takes place, characterized by the beginning of the synthesis of anthocyanins, soluble flavonoids compounds that provide color to red varieties (Figure 1) (Boss et al., 1996). Sucrose, originating from leaves, reaches the fruits through the phloem, and is then hydrolyzed forming glucose and fructose (Robinson and Davies, 2000; Kennedy, 2002; Vignault et al., 2005; Deluc et al., 2007; Hayes et al., 2007; Fontes et al., 2011). Stage II is thus a transition between an unripe fruit and the third stage of development (stage III or ripening). The latter involves important morphological and physiological changes, like color development (Boss et al., 1996), turgor reduction and berry enlargement (Chervin et al., 2008), and decreased acidity (Costenaro-da-Silva et al., 2010), among others. In addition, hormonal changes that occur throughout development positively or negatively regulate ripening (Figure 1; Gerós et al., 2012). Therefore, during ripening, a large number of complex transcriptional and/or post-transcriptional regulatory processes are triggered. In this review, we focus on the integration of global approaches, including proteomics, metabolomics, and especially transcriptomics, to understand grape berry development and the influence of environmental factors on this process. Thus, we will cover initial fruit development, with emphasis on seed formation; the importance of coloration and hormonal changes during development, especially on ripening; and finally, the effect of environmental factors on this process will be discussed.
FIGURE 1.
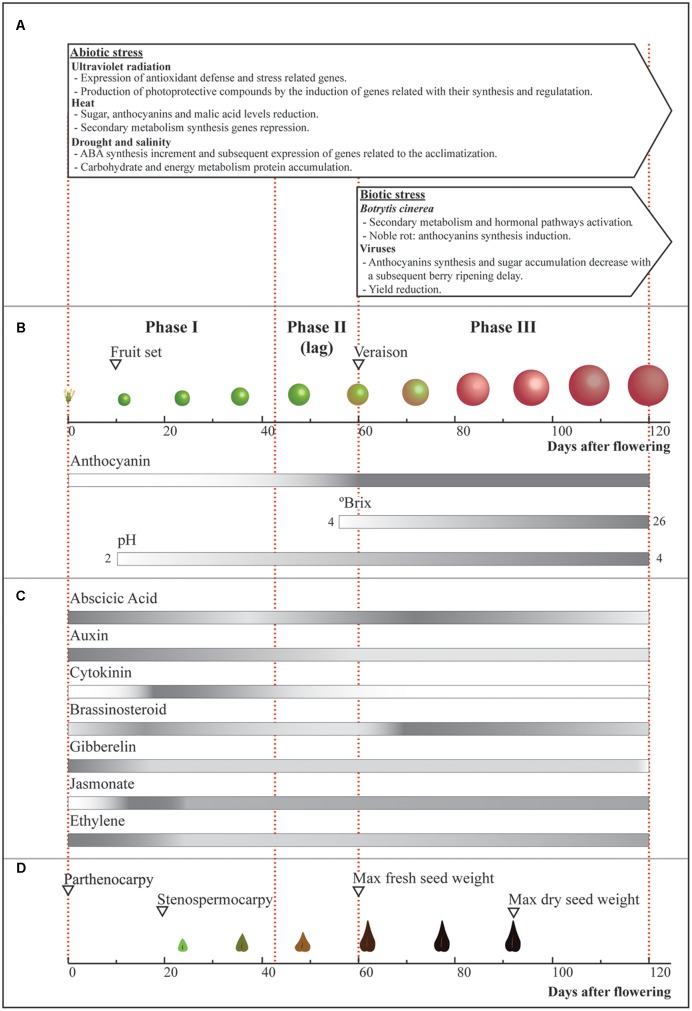
Fruit development and environmental effects. Scheme of the most important changes that berries and seeds undergo during development, and the main environmental factors affecting this process. (A) Boxes indicate the phase where each stress condition (temperature, water-related stress and UV-B radiation, Botrytis cinerea and viruses) affect grape berry and its development. (B) Changes in size, color, brix degree, and pH during berry ripening and (C) variations in hormonal content during grape berry development. (D) Seed development showing the time in which parthenocarpy and stenospermocarpy can take place. The main events are indicated by open triangles. Bars represent the described changes throughout development, in which gray and white represent the higher and lower estimated referential values for each parameter, respectively.
Grape Berry Development from an Omics Perspective
Seed Development and Seedless Fruits
Grape berry development begins after fertilization, when in a process known as fruit set, the ovary changes from a stationary state and experiences an abrupt increase in size that occurs due to cell division and enlargement, leading to rapid pericarp growth. Throughout this period, seed development is an important process, mainly because seeds produce auxins, gibberellins (GAs) and cytokinins, which play multiple roles in grape berry development (Keller, 2010). Seed and berry development are coordinated, and the changes that seeds undergo have an impact on fruit ontogeny. The first stage of berry development is characterized by a rapid increase in seed size, during which embryogenesis and endosperm growth occur. At the second stage, about 10 and 15 days prior to veraison, seeds reach their final size and maximum fresh weight, and at the beginning of the third stage, embryo growth ceases and the endosperm accumulates reserves until the seeds turn dormant (Figure 1; Keller, 2010).
Ripe berries usually contain up to four seeds derived from four ovules (Dokoozlian, 2000; Kennedy, 2002). However, seedless grape varieties have been spontaneously found in nature and have been preserved over the years through asexual propagation. Seedless berries develop naturally via two different mechanisms, parthenocarpy and stenospermocarpy, which generate berries without or with rudimentary seeds, respectively (Varoquaux et al., 2000). In order to understand the main differences between parthenocarpy and stenospermocarpy, we will discuss the few available Omics analyses of early stages of berry development and seed formation.
Parthenocarpy
In parthenocarpic fruits, the stimulus of pollination is sufficient to trigger fruit set (Dokoozlian, 2000). Since the ovary is able to enlarge and form a berry without ovule fertilization, there is no seed in the fruit (Varoquaux et al., 2000). Until now, few parthenocarpic grape cultivars have been described. Of these, cv. Corinto Bianco (CB), a somatic variant of the seeded cv. Pedro Ximenez (PX), constitutes a good model to study seed formation (Vargas et al., 2007). To understand the molecular differences between CB and PX genotypes, flowers at 1 and 10 days pre-anthesis were transcriptionally compared using microarray (Royo et al., 2016). The analyses allowed the identification of 1958 differentially expressed genes between CB and PX. Interestingly, several genes that are specifically expressed in reproductive organs were down-regulated in CB. Processes such as cell wall biosynthesis, cytoskeleton biogenesis, vesicular transport, signaling through G proteins or phosphatidylinositol, among others, were enriched. Also, 14 single-nucleotide polymorphisms (SNP) were identified between both genotypes, which could explain the parthenocarpy phenotype (Royo et al., 2016). Considering that microarrays deliver limited information, a suitable approach to analyze the different stages of development in more detail, would be using RNA-seq technologies, in order to gain further insights into the understanding of seed development and to generate new parthenocarpic varieties.
Stenospermocarpy
During stenospermocarpy, pollination and fertilization occur normally, but a few weeks later, the embryo and/or the endosperm abort and the berries that have been generated possess just traces of seed (Varoquaux et al., 2000). It has been demonstrated that stenospermocarpy occurs in several seedless varieties, and is stable and unaffected by environmental factors (Zhang et al., 2013). However, little is known about the molecular mechanisms that underlie stenospermocarpy in grapes. The most accepted hypothesis proposes the existence of a dominant regulator gene called Seed Development Inhibitor (SDI), which could control three other recessive genes (Bouquet and Danglot, 1996). Different studies based on quantitative trait locus (QTL) analysis have reported a main QTL in linkage group 18 (LG18) (Doligez et al., 2002; Cabezas et al., 2006; Mejía et al., 2007; Costantini et al., 2008), which could explain between 50 and 70% of the seedlessness phenotype in grapes; LG18 could be considered as the SDI locus trait. In this context, VvAGL11 (MADs BOX transcription factor) was in silico mapped to the SDI locus and it has been proposed as the main functional candidate gene for seedlessness (Mejia et al., 2011). In fact, it was demonstrated that the silencing of a VvAGL11 homologous gene in tomato (Solanum lycopersicum L. cv Micro-Tom) generates fruits with few or rudimentary seeds (Ocarez and Mejia, 2016). Based on genome sequencing data, it is known that in the stenospermocarpic variety cv. Thompson Seedless, the VvAGL11 gene has an insertion of 15 bp in the 5′UTR, which could be the cause of the seedless phenotype (Di Genova et al., 2014). In addition, in cv. Sultanine Monococco, which is also a seeded variety of Thompson, the VvAGL11 transcript level is higher in comparison with the seedless variety (Ocarez and Mejia, 2016), supporting the hypothesis that this gene is one of the main regulators of seed formation in grapes.
On the other hand, due to abnormal ovules may be formed during flower development before or after meiosis, through either the abnormal development of the nucellus or ovule integuments, or the degeneration of the egg in the embryo sac (Ebadi et al., 1996). In order to determine the molecular bases of this phenotype, flowers from cv. Thompson Seedless and cv. Thompson Seeded, a spontaneous mutant with seeded berries, were compared through suppression subtractive hybridization (SSH) (Hanania et al., 2007, 2009). The results demonstrated that ch-Cpn21, a gene that encodes for a chloroplastic chaperonin, is repressed in developing flowers of cv. Thompson Seedless. Likewise, the silencing of this gene in tobacco plants (Nicotiana benthamiana) and tomato induces seed abortion (Hanania et al., 2007). The use of somatic variants in combination with current transcriptomic technologies, would be very promising in the study of stenospermocarpy, helping to discover new genes playing important roles in seed abortion.
Based on the studies of Costenaro-da-Silva et al. (2010) and Nwafor et al. (2014), several genes have been associated with early stages of grape berry development. These include VvUBP1, a heterogeneous-nuclear ribonucleoprotein, VvFS41, a putative S1-like ribosomal protein involved in mRNA processing and synthesis of proteins related with cell division during the first days of berry development, VvERF1 and VvERF9, which encode for transcription factors related to several developmental processes, VvDOF1, possibly related to seed development, and VvRIP1 and ABI3, which have been related to hormone signaling, among others. Many of these genes have pleiotropic effects, so it is difficult to estimate their specific molecular contribution to the stenospermocarpy phenotype. Some of them could be involved in this process, but their functional characterization is needed to test this hypothesis.
Non-characterized Mechanisms of Seedlessness
A demonstrated way to produce seedlessness is the exogenous application of GAs before bloom or during anthesis. It is believed that GAs promote seedless grapes by inhibiting pollen germination, allowing unfertilized ovules to enlarge and form fruits, as occurs in parthenocarpy (Kimura et al., 1996; Cheng et al., 2013). However, another study suggests that exogenous GAs interfere with seed development, as described in stenospermocarpy (Cheng et al., 2013). So, the mechanism involved in this response is not clear. A transcriptional analysis by RNA-seq was performed in GA3-treated flowers of the seeded cv. Kyoho and a comparison with non-treated flowers was carried out (Cheng et al., 2015). This study demonstrated that GA3 application modifies the expression profile of genes related to developmental processes, such as cellular metabolism, biosynthesis of different metabolites, stress response, transport, etc. Also, changes in the expression of genes related to flowering, fruit, and embryonic development were found. Within the genes possibly related to seedlessness, the Pelo gene, whose mouse homolog has a role in meiosis and causes embryonic lethality (Adham et al., 2003), was repressed after GA3 treatment, and was correlated with seedlessness in grapes (Cheng et al., 2015). The Pelo gene probably has conserved roles across several species. However, deeper functional studies are needed to corroborate this information in plants and to determine if this gene does indeed fulfill a role in seed development, and more studies are needed to correlate any transcriptional changes with particular phenotypes. Recent studies have demonstrated that reactive oxygen species (ROS) are present throughout the entire seed’s life cycle (Jeevan Kumar et al., 2015). In fact, the oxidative damage induced by an imbalance in plant redox homeostasis can affect normal seed development, leading to abortion (Cheng et al., 2013). Pathways related to ROS scavenging and detoxification are significantly affected after GA3 treatment (Cheng et al., 2015). So, probably, exogenous GA application generates physiological changes that could induce seedless fruits through a ROS-related mechanism, but further research is needed to understand the role of ROS regarding the presence or absence of seeds. Naturally occurring seedlessness could be the result of a series of coordinated transcriptomic switches that cause a global reprogramming of the cell. To date, little is known about the seedless phenotype in grapevines, presenting a great challenge for researchers. The best model for understanding seedlessness is to compare somatic variants (seeded versus seedless) through global approaches, since they have the same genetic background and could be used to discover new genes involved in this phenotype. Even though somatic variants are rare in nature, it is clear that these comparisons are much more informative than the use of two different varieties.
First Stage of Grape Berry Development
The first stage of grape berry development (stage I) is initiated with fruit set. During the first 2 weeks, berry size increases markedly as auxin and GAs directly promote cell division and enlargement (Ojeda et al., 1999; Bottcher et al., 2010; Fortes et al., 2015). Tartaric, malic, and other organic acids, along with different phenolic precursors such as tannins and hydroxycinnamates, are synthesized, modifying the organoleptic properties of the berries (Deluc et al., 2007). Besides, minerals, micronutrients, and aroma-related compounds are present. Transcriptomic analysis of young berries in cv. Shiraz revealed an enrichment of hormone signaling responsive transcripts, suggesting that hormone-controlled metabolic pathways are highly active in early stages of development (Sweetman et al., 2012). During this stage, GAs are the key regulators of fruit set, cell division and cell expansion (Fortes et al., 2015). RNA-seq analysis of cv. Centennial Seedless berries treated with GA3 (12 days after flowering), revealed a repression of an abscisic acid (ABA)-response element binding factor (ABF) and ethylene response factors (ERFs) (Chai et al., 2014). Showing the occurrence of both GA3–ABA and GA3–ethylene crosstalk. The role of jasmonic acid (JA) in grapes remains unclear, but, as has been demonstrated in potato (Solanum tuberosum) leaves, it might stimulate cell division (Ravnikar et al., 1992). In grape, high levels of JA are present during the first stage, which then decrease in mature berries (Fortes et al., 2015). A proteomic study in cv. Muscat Hamburg has reported abundant levels of chloroplast lipoxygenases (LOX), enzymes that provide intermediates for JA biosynthesis during green berry development, followed by a decrease when berries reach a size of 15 mm (Martinez-Esteso et al., 2011). In the case of auxins, it has been proposed that they have a role in fruit growth delaying ripening (Fortes et al., 2015). In berry flesh of cv. Kyoho, a high concentration of auxins has been reported, in particular of indole-3-acetic acid (IAA) during the beginning of stage I, with a rapid decrease at the end of this stage and throughout stage II, consistent with the high rate of cell division observed in the first stage (Zhang et al., 2003). Considering that the final number of cells in the grape berries is defined in the first stage of development (Dokoozlian, 2000), the interaction between hormones regulating cell division is key to cluster progress, and might be an interesting target in studies aimed at improving yield.
Second Stage of Grape Berry Development
Main Changes in Metabolites during Stage II
The second stage of grape berry development (stage II) is a lag phase, where the rate of increase in both fresh and dry weight is very low. At the end of this stage, veraison occurs, which is the transition from the second to the third stage of berry development, and is considered the onset of ripening. Different physiological and biochemical changes take place during veraison, of which anthocyanin synthesis and sugar accumulation are the most characteristic and important processes. In fact, anthocyanins are one of the main pigments present in colored grape berry skins (Souquet et al., 1996), while sugar content is widely considered one of the most important properties that define ripening (Guelfat-Reich and Safran, 1971; Jayasena and Cameron, 2008).
Depending on the cultivar, five types of anthocyanins are frequently found in V. vinifera, which are associated with organoleptic properties such as color (in the case of red wine), bitterness, astringency and also as antioxidant molecules with beneficial effects on human health (Dixon et al., 2005). Anthocyanin biosynthesis occurs through the phenylpropanoid pathway, in which two types of genes are involved: structural genes, encoding for biosynthetic enzymes, and regulatory genes, which are associated with temporal and spatial regulation of the structural genes (Deluc et al., 2007). Both, structural and regulatory genes are present in colorless and colored grapevine cultivars, but in the case of white cultivars, color is not expressed due to multiallelic mutations in MybA1 and MybA2, that prevent the transcription of these two important positive regulators of the phenylpropanoid pathway (Kobayashi et al., 2004). In the case of red cultivars, MYBA1 and MYBA2 transcription factors control anthocyanidin glycosylation through the regulation of flavonoid 3-O-glucosyltransferase (UFGT) expression (Ford et al., 1998). The anthocyanin biosynthesis pathway is not only regulated by MYB transcription factors, as it is also controlled by the critical transcriptional R2R3-MYB/bHLH/WD40 (MBW) complex in grapevine (Wu et al., 2014).
Flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) genes seem to be an important regulatory points in anthocyanin biosynthesis (Castellarin and Di Gaspero, 2007; Matus et al., 2016). Their proteins belong to the cytochrome P450 protein family and compete for a common precursor for the biosynthesis of red and blue anthocyanins, respectively (Bogs et al., 2006). Metabolic and transcriptomic analyses determined that in cv. Cabernet Sauvignon and cv. Shiraz, the F3′5′H gene is up-regulated, whilst that of F3′H is down-regulated (Degu et al., 2014). However, further studies are necessary to understand the fine regulation of the phenylpropanoid pathway, focusing on the anthocyanin branch. In this case, the use of varieties with different berry skin colors would be informative. None of the aforementioned studies consider pink varieties, which have an intermediate color between red and white varieties, and could be used to complete the overview of anthocyanin biosynthesis in a fuller range of colors.
In a recent analysis using Omics approaches to analyze ripe berry skins of five cultivars, Cabernet Sauvignon, Merlot and Pinot Noir (red cultivars), and Chardonnay and Semillon (white cultivars) (Ghan et al., 2015), several transcripts and metabolites were mapped to the phenylpropanoid pathway. A higher transcript abundance for enzymes involved in anthocyanin biosynthesis, such as phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), flavanone 3-dioxygenase (F3H), leucoanthocyanidin dioxygenase (LDOX), and UFGT was observed only in red cultivars. Shikimate was the most abundant metabolite in cv. Cabernet Sauvignon, which acts as a precursor for aromatic amino acid biosynthesis within the shikimate pathway (Maeda and Dudareva, 2012). This intermediary is important because it allows the transfer of the carbon skeleton into anthocyanin structures, and could become a crucial point in the study of anthocyanins in different varieties.
Sugar accumulation (mainly of glucose and fructose) is another important process that begins in veraison and continues throughout ripening. Sugar sensing mechanisms may play important roles during grape berry ripening, as they do in other aspects of plant development (Smeekens et al., 2010; Wind et al., 2010). Thus, the role of sugar is covered in the context of the third stage of berry development (see Third Stage of Grape Berry Development).
Hormonal Control during Stage II
ABA levels are high in young berries and then fall until veraison. A microarray analysis carried out over seven sequential points of berry development in cv. Cabernet Sauvignon, revealed that the transcript abundance of 9-cis-epoxycarotenoid dioxygenase (NCED1), the enzyme that conducts the limiting step in ABA synthesis (Tattersall et al., 2007), increases during the lag phase and peaks at veraison (Deluc et al., 2007). A similar expression pattern was shown for a gene encoding for the ABA signaling transduction protein phosphatase 2C ABI1, while the gene encoding a transcription factor of the same pathway, ABI3/VP1 (Abscisic acid Insensitive 3/Viviparous 1), showed the highest transcript abundance during lag phase (Deluc et al., 2007). Several studies have highlighted the control that ABA exerts over the biosynthesis of anthocyanins; at the transcriptional level by upregulation of biosynthetic genes, and at the metabolic level by increasing anthocyanin content (Wheeler et al., 2009; Giribaldi et al., 2010; Cramer et al., 2014). In this context, A 2-DE proteomic approach in cv. Cabernet Sauvignon showed that ABA treatment before veraison increases three proteins required for flavonoid biosynthesis: chalcone isomerase, dihydroflavonol-4-reductase, and anthocyanidin reductase (Giribaldi et al., 2010).
In non-climacteric fruits, such as grape, the role of ethylene is not fully understood due to the low levels of this hormone during development and the technical difficulties associated with its quantification (Symons et al., 2012). Nevertheless, several reports indicate a possible role of this hormone in grape berry ripening, mainly supported by the consistent presence of a small peak about 2 weeks after veraison (Chervin et al., 2004). These data are consistent with the findings of Pilati et al. (2007), who showed that the expression of the ACC synthase gene, involved in ethylene synthesis, increases just prior to veraison and decreases afterward, together with a peak in expression of ACC oxidase around veraison, which encodes for the enzyme responsible for the last step of ethylene biosynthesis.
Brassinosteroids (BR), on the other hand, are steroid hormones that have been implicated in the ripening of non-climacteric fruits (Symons et al., 2006; Chai et al., 2013). The transcript abundance of the brassinosteroid receptor 1 gene (BRI1) peaks in the entry to lag phase and declines thereafter (Deluc et al., 2007). The expression profile of VvBR6OX1, which encodes for the enzyme that converts 6-deoxocastasterone to castasterone (the bioactive BR in grapes) shows a peak of induction just prior to veraison (Pilati et al., 2007). This evidence is consistent with an increase in BR levels at veraison and the high content observed during ripening in cv. Cabernet Sauvignon berries (Symons et al., 2006). Interestingly, exogenous application of BR increases anthocyanin content leading to premature grape berry coloration, similar to the effect of ABA. The connection between the molecular pathways of BR and ABA that regulate initial events of ripening stages has yet to be clarified. Based on transcriptomic analysis of cv. Merlot berries, it has been hypothesized that BR might be an early signal for ripening, modulating ethylene content (Ziliotto et al., 2012). In this model, the small peak of ethylene could upregulate genes associated with ABA biosynthesis and then initiate all ripening-associated ABA-induced metabolic changes (Ziliotto et al., 2012).
It has been well documented that auxin has a negative role during grape berry ripening. In fact, IAA levels (the active form) remain low from veraison throughout ripening, and auxin treatments during pre-veraison inhibit ripening (Davies et al., 1997; Bottcher et al., 2010, 2011; Ziliotto et al., 2012). Two auxin carriers (an AUX1-like and a PIN1-like) are expressed before veraison, while two auxin response factors (ARFs), ARF5 and ARF18, and an auxin receptor of the ABP family are expressed at pre-veraison and are then repressed during ripening (Pilati et al., 2007; Fortes et al., 2011). It has been suggested that ethylene represses auxin biosynthesis and thus regulates the balance between auxin and ABA to initiate ripening (Ziliotto et al., 2012). Probably, a network coordinated by ABA, BR, ethylene, and auxin levels are regulating the ripening stage, however, the master regulators that connect all these pathways are still unknown.
Third Stage of Grape Berry Development
Main Changes in Metabolites during Stage III
During the third stage of development (Stage III), berries approximately double in size and there is a marked decrease in organic acid concentration and a dramatic accumulation of glucose and fructose (∼1 M each) in the vacuole of flesh cells (Fontes et al., 2011; Dai et al., 2013). The scientific community has gained understanding about the complexity and diversity of sugar-sensing systems, including hexokinase (HXK), protein kinases, as well as, novel molecular regulators, such as trehalose-6-phosphate (T6P) (Li and Sheen, 2016). It has been shown that the HXK enzyme, responsible for the 6-phosphorylation of glucose and fructose, plays a dual-function with both catalytic and regulatory activities and therefore, links gene expression and metabolism in plants (Moore et al., 2003). HXK-dependent signaling represses photosynthetic related-genes in the presence of hexoses, forming a repressive complex that is directly associated with the promoter regions of several genes including those that encode for chlorophyll a/b binding protein (CAB) and carbonic anhydrase (CAA) (Cho et al., 2006).
In grapevine, a genome wide analysis using the completely sequenced V. vinifera genotype PN40024 (cv. Pinot Noir) led to the identification of six members of the HXK family (Çakir, 2014). Four genes that encode for HXKs in cv. Cabernet Sauvignon were analyzed (Gambetta et al., 2010). The authors showed that these genes are highly regulated at the transcriptional level during berry development. Specifically, HXK-1, HXK-2, and HXK-3, which were induced during ripening, while HXK-4 was repressed (Gambetta et al., 2010). Interestingly, under water deficit conditions, HXK-4 was induced during the third stage of berry development compared to the control under well irrigated conditions, indicating that there is both genetic and environmental control of the sugar sensing mechanisms during ripening (Gambetta et al., 2010).
Protein kinases are the major components of intracellular signaling and are responsible for rapid responses to changes in the environment. VviSK1, a protein kinases with sugar signaling function during berry development, whose transcript was shown to be accumulated after sucrose treatments in cv. Cabernet Sauvignon suspension cells (Lecourieux et al., 2010), positively affects sugar accumulation in grape cells and controls glucose transport through the regulation of four genes that encode the hexose transporters VvHT3, VvHT4, VvHT5, and VvHT6. Moreover, during berry development, VviSK1 transcripts decrease after the green stage and increase again after veraison, when sugar is accumulated (Lecourieux et al., 2010). Another protein kinase that may participate in sugar signaling during ripening is SnRK1 (Sucrose-non-fermentative Related kinase 1). In plants, SnRK1 receives inputs from hormones, as well as, sugar phosphates, and has been linked to several developmental processes and the control of primary and secondary metabolism, including photosynthesis and anthocyanin biosynthesis (Baena-Gonzalez et al., 2007; Nunes et al., 2013; Tsai and Gazzarrini, 2014). In grapevine, SnRK1 transcripts accumulate continuously in cv. Cabernet Sauvignon berries from the green stage until ripening (Gambetta et al., 2010). Nonetheless, to our knowledge, the abundance and activity of this protein kinase has not been measured in grapevine berries.
Phosphate sugars are other members of the sugar signaling landscape. Among them, T6P, which is generated by primary metabolism (Lunn et al., 2014) has been recently uncovered as a signal molecule with major implications in plant growth, development, and metabolism (Van Houtte et al., 2013; Wahl et al., 2013). Transcriptomic studies had uncovered that several genes that control T6P abundance are regulated during berry development. In one of the first gene expression profile analyses using AFLP in berry samples from cv. Corvina, the authors reported a T6P-phosphatase as one of the most upregulated genes in postharvest (Zamboni et al., 2008). Moreover, Deluc et al. (2007), using the first commercially available grapevine Affymetrix, identified different profiles for genes encoding T6P-synthase, which was overexpressed in the early days before veraison, and T6P-phosphatase, which was overexpressed at postharvest. Suggesting that the abundance of T6P is highly controlled during grape berry development.
In plants, T6P is linked to sugar signaling and the control of SnRK1, which is linked to developmental processes and control of metabolic pathways, including repression of anthocyanin biosynthesis (Baena-Gonzalez et al., 2007). In grapevine, several transcriptomic studies have shown that orthologs genes of SnRK1 and of the enzymes that control T6P homeostasis are highly regulated during berry development (Deluc et al., 2007; Gambetta et al., 2010). Therefore, the SnRK1/T6P pathway may be an important component of sugar signaling during berry development, and in this context, it remains to be studied whether the activity of SnRK1 protein kinase is actually inhibited by T6P, as found in other plant tissues. The Omics studies shown so far, using mainly transcriptomic and metabolomic approaches, have been useful for the identification of several sugar-signaling components, leading to the proposal that new mechanisms or candidate genes are involved in berry ripening. Nonetheless, specifically in the field of signaling through protein kinases, it is known that transcript accumulation is not the only, or main mechanism that influences their role and activity. In this perspective, it may be necessary to perform more protein-oriented Omics studies such as proteomics or phosphoproteomics. These are powerful technologies and could help to elucidate the importance of protein kinase signaling during berry development.
Hormonal Control during Stage III
Microarray and RNA-seq analyses have uncovered transcriptional reprogramming during ripening (Fasoli et al., 2012). At the onset of ripening in cv. Cabernet Sauvignon, low levels of IAA are required, while the auxin conjugate to aspartate (inactive form) concentration is high (Bottcher et al., 2010). In the case of IAA conjugate formation, the up-regulation of a gene coding for GH3.1 was found at veraison (Bottcher et al., 2010), in contrast to a decrease in GH3.3 expression (Pilati et al., 2007). On the other hand, genes coding for AUX–IAA proteins, transcriptional repressors of auxin-responsive genes, are down-regulated during ripening, while genes coding for IAA19 and IAA16 are up-regulated around veraison. Likewise, a gene homologous to Arabidopsis amidase AtAMI1, that in vitro synthesizes IAA from indole-3-acetamide, decreases its expression during ripening (Pilati et al., 2007) indicating a complex regulation for the maintenance of low levels of active auxin during ripening.
Related to the ethylene metabolism, the role of this hormone during ripening has not been clearly established (Chervin et al., 2004). Nevertheless, it is known that the transcript abundance of genes coding for ACC synthase decrease at veraison, while several genes coding for ACC oxidase are down-regulated and only one is up-regulated during ripening (Terrier et al., 2005). The intricate regulation of the ethylene signaling pathway during ripening seems to be more consistent and clearer during the later stage of this process. Cramer et al. (2014) assessed the transcriptome of Cabernet Sauvignon berries in the late stages of ripening using whole-genome microarrays. They reported that several positive regulators of the ethylene pathway are upregulated, including three different ethylene receptors (VviETR1, VviETR2, and VviEIN4) and several members of the ERF family of transcription factors. Moreover, the negative regulator of ethylene signaling, VviCTR1 is downregulated at the transcript level during late ripening in both pulp and flesh (Cramer et al., 2014). Supporting the idea of an active ethylene signaling role during berry ripening.
Regarding BRs, it has been shown that exogenous application to grape berries significantly promotes ripening, whilst endogenous BR levels dramatically increase at the onset of ripening and then decrease (Symons et al., 2006). These results coincide with the transcript accumulation of the VvBR6OX1 gene observed by Pilati et al. (2007), responsible for the synthesis of the bioactive BR, castasterone. In addition, a gene coding for an enzyme putatively involved in castasterone catabolism (castasterone 26-hydroxylase), leading to the inactivation of this BR, is down-regulated at ripening (Fortes et al., 2011). On the other hand, a gene related to BR biosynthesis that codes for steroid 5-alpha-reductase (DET2), was less expressed around veraison (Fortes et al., 2011). Diminished expression of biosynthetic genes could be associated with negative feedback regulation by increasing levels of BRs.
The ABA concentration increases dramatically during berry ripening (Coombe and Hale, 1973). Several reports suggest that this hormone plays a major role controlling color development (Koyama et al., 2010) and softness (Gambetta et al., 2010). ABA levels are directly related to changes in NCED activity (Wheeler et al., 2009), and indeed, NCED1 transcripts peak around veraison and decrease at advanced ripening (Deluc et al., 2007). A proteomic analysis in berry skins of cv. Barbera at different stages throughout ripening revealed that the most abundant proteins belong to the ABA stress responsive elements (ASR) family, representing nearly 13% of the total protein spot volume in early ripening (Negri et al., 2008). In Arabidopsis, the proposed model of ABA signaling involves the protein kinases SnRK2, which act as positive elements in signaling downstream of ABA. SnRK2s interact with the negative regulators PP2C protein phosphatases that inhibit the activity of SnRK2s. Recently, Liu et al. (2016) identified eight VviSnRK2 genes in the grapevine genome and generated a detailed co-expression network of the ABA signaling components, including transcription factors from the ABF family. They found a high co-expression coefficient of both VviSnRK2.8 and VviSnRK2.11 with VviABF2, which is an important transcriptional regulator of ABA-dependent signaling during grape berry ripening (Nicolas et al., 2014). VviABF2 expression rises from veraison until ripening, and transcriptomic analysis of VviABF2-overexpressing grapevine cells allowed the identification of several co-overexpressed genes regulated by ABA (Wong et al., 2013; Nicolas et al., 2014). The regulation of the ABA signaling pathway is complex; the cellular, physiological and transcriptomic responses to this hormone change dramatically in a tissue-specific manner (Rattanakon et al., 2016), and are also cultivar-dependent (Rossdeutsch et al., 2016). Moreover, as has been mentioned, the gene families that act positively, or negatively downstream of ABA are composed of several genes, and it is plausible that sub-specialization of members in these families exists. The complexity mentioned above is a significant challenge for researchers when attempting to extract mechanisms related to ABA signaling. An important issue to address in the coming years is that of understanding the differential sensitivity to ABA across cultivars, and the crosstalk of ABA with other signals that seem to be important in berry development, such as sugars and other hormones.
The Effect of the Environment on Grape Berry Development
Grape berries are constantly exposed to several biotic and abiotic factors that, to some extent, can affect their normal development and trigger positive or negative changes. In most cases, these factors negatively impact grape cultivation at different stages of plant and berry development during pre- and post-harvest (Armijo et al., 2016a). In this review, some of the most relevant grapevine abiotic and biotic stresses are discussed.
Abiotic Stress
Climate change has caused significant warming in most grape-growing areas, increasing some important abiotic stresses like heat, drought and UV radiation (Teixeira et al., 2013; Keller, 2015). These stresses mainly affect phenolic metabolism and, at the same time, berry composition and development (Figure 1).
Changes in temperatures during vegetative grape development are associated with changes in berry harvest date (Meier et al., 2007). Studies of transcripts, metabolites and proteins also show that sugar accumulation and other parameters related to color and aroma could be affected. Moderate warmer temperatures (∼25°C) lead to higher berry sugar content (Coombe, 1987), while higher temperatures (>30°C) negatively affect photosynthesis, with consequent reductions in sugar, anthocyanin, and malic acid accumulation, followed by a decrease in berry size and weight (Sadras and Moran, 2012; Teixeira et al., 2013; Rienth et al., 2016; de Rosas et al., 2017). Sugar and organic acid metabolism are desynchronized in ripening grapevine fruits at high temperatures, and secondary metabolism is diminished due to the transcriptional repression of their respective genes (Rienth et al., 2016; de Rosas et al., 2017). Thus, high temperatures are a negative regulator of berry development at ripening, but the mechanism behind this is still not clear. Integrated global analyses are required to identify the possible genes associated with the changes in the corresponding physiological traits.
In general, V. vinifera is considered as a salt and drought tolerant species (Tattersall et al., 2007). However, stress caused by water availability is having progressively more impact, due to can generate significant effects on grapevine cultivation. In response to salinity and drought, plants intensify the synthesis of ABA, which is transported to the aerial organs, inducing changes in the expression of genes related to their acclimatization (Tattersall et al., 2007; Cramer et al., 2011). Few studies have addressed salinity and drought stress in berries. For instance, it has been shown that water stress can increase berry flavonol content and affect the expression of genes involved in biosynthesis of stilbene precursors (Teixeira et al., 2013). All these analyses suggest a differential response to water limiting abiotic stresses that is cultivar dependent. Likewise, the optimal growth temperature for grapevines may vary between cultivars, and the activation of ABA and ethylene signaling pathways can differ according to their sensitivity or tolerance to drought. These responses have consequences in grapevine berries, since a common mechanism in response to stress in these organs, is the induction of the anthocyanins accumulation, which act as protective molecules.
The Effect of UV Radiation in Grape Berry Development
Vitis vinifera is often cultivated in Mediterranean climates with varied UV-B radiation dosages (Martinez-Luscher et al., 2013), and it is considered as well adapted to solar radiation due to a variety of physiological responses, mainly based on antioxidant enzyme activities and secondary metabolites. The UV-B spectrum (280–315 nm) can provoke potential damage in macromolecules, including DNA, induce ROS, and disrupt several cellular processes in all living organisms (Frohnmeyer and Staiger, 2003; Jenkins, 2009). In grapevine, several studies have been performed to discover the processes associated with the UV radiation response during berry development. In this context, VviHY5 (ELONGATED HYPOCOTYL 5), VviHYH (VviHY5 HOMOLOGUE), and VviUVR1 (the photomorphogenic factor UV-B RECEPTOR 1) genes were characterized (Loyola et al., 2016). In this work, the authors described that the expression of VviHY5 and VviHYH differs during grape berry and inflorescence development upon exposure to low or high UV-B radiation, while VviUVR1 expression was not regulated by UV-B. Studies performed by Carbonell-Bejerano et al. (2014) indicated that grapevine berries respond to UV-B through the activation of the phenylpropanoid pathway and the production of photoprotective compounds. The accumulation of polyphenolic compounds in the berry involves specific UV-responsive genes that induce the expression of phenylpropanoid pathway related genes and several MYB transcription factors that regulate this pathway (Matus et al., 2009; Berli and Bottini, 2013). Matus et al. (2009) demonstrated that different light conditions increase the accumulation of flavonoid compounds in grape berries, while Loyola et al. (2016) shown that high and low UV-B radiation induce flavonol accumulation in this organ. Carbonell-Bejerano et al. (2014) suggest that UV-B radiation triggers flavonol accumulation in grape berry skin of cv. Tempranillo and induces the expression of VvFLS1 and VvGT5, two flavonol biosynthetic genes. Furthermore, several flavonol biosynthetic genes are regulated by the R2R3-MYB transcription factor VvMYBF1, which triggers flavonol and anthocyanin production in grape berries exposed to solar UV radiation (Czemmel et al., 2009, 2017; Matus et al., 2009; Matus, 2016). Genome-wide microarray studies performed in grape berry skins of cv. Pinot Noir exposed to UV-C light (100–280 nm), showed 238 up-regulated genes (more than fivefold), including several genes encoding for proteins related to stilbene synthesis (Suzuki et al., 2015). These authors also reported that UV-C light increases levels of phenolic compounds like resveratrol and its analogs. Similar results were observed in berries of cv. Tempranillo exposed to solar UV radiation (Carbonell-Bejerano et al., 2014). In general, several volatile compounds accumulate in grape berries during ripening, but the amount of these compounds depends on specific irradiance levels and the type of radiation (Joubert et al., 2016).
Summarizing, there are numerous studies demonstrating that light can affect anthocyanin accumulation in berry skins. Which can be explained by changes in the expression of structural genes related to the phenylpropanoid pathway, as well as regulatory genes such as those of the MYB, bHLH, and WD40 families (Wu et al., 2014).
Biotic Stress
In addition to abiotic stress, grape berry development can be influenced by biotic factors, such as pathogens, of which fungal and viral diseases are the most common and harmful, negatively affecting fruit quality.
Fungal Infections: Botrytis cinerea and Its Dual Effect on Berry Development
The most important fungal disease affecting grape berry development is gray mould, caused by B. cinerea. Grape berries are resistant to the infection until veraison, but are highly susceptible at the onset of ripening and harvest (Kelloniemi et al., 2015). As a necrotrophic pathogen, B. cinerea secretes lytic enzymes and phytotoxins in order to promote cell degradation (Armijo et al., 2016b). Most of the agronomically relevant grapevine cultivars are susceptible to this pathogen, leading to significant losses worldwide.
Different large-scale approaches have been carried out in order to understand the regulatory networks and processes involved in the grape berry–B. cinerea interaction, and to characterize how berry development is affected. Transcriptomic and metabolic analysis of cv. Marselan, comparing B. cinerea berries at veraison with ripe berries, revealed that the former activates an early burst of ROS, together with multiple defense responses, including a salicylate-dependent pathway, resveratrol synthesis and cell-wall strengthening. In contrast, ripe berries activate the JA-dependent pathway against the fungus (Kelloniemi et al., 2015). As a common response, both developmental stages displayed an upregulation of genes encoding WRKY transcription factors, pathogenesis-related proteins, glutathione S-transferase (involved in cellular detoxification), stilbene synthase and PAL (involved in phenylpropanoid biosynthesis), and production of anthocyanins and phytoalexins. Global metabolic changes in cv. Marselan induced by B. cinerea infection correlate the greater resistance of veraison berries with an accumulation of resveratrol and caffeic, ferulic, and chlorogenic acids (Kelloniemi et al., 2015). Also, significantly higher levels of proline, glutamate, arginine, and alanine were detected in B. cinerea-infected ripe berries of cv. Chardonnay, as well as, an accumulation of glycerol, gluconic acid, and succinate, mainly in the berry skin (Hong et al., 2012). A reprogramming of carbohydrate and lipid metabolism toward an increased synthesis of secondary metabolites with antioxidant properties, such as trans-resveratrol and gallic acid, was also observed by Agudelo-Romero et al. (2015) in cv. Trincadeira. During later stages of infection, energy metabolism (photosystem I supercomplex) and secondary metabolism (phenylpropanoid and stilbenoid biosynthesis) also seemed to be downregulated (Agudelo-Romero et al., 2015).
Contrary to the effects caused by gray mould on grape berries, some particular cases of B. cinerea infection can generate favorable effects on wine grapes, in an interaction known as noble rot. Botrytized wines are produced from grapes that have been affected by this fungus under specific environmental conditions, which are typically hot and dry. The infection produces berry dehydration, altering metabolic processes and the saprophytic microbiota (Magyar, 2011). The berry–fungus interaction promotes the accumulation of secondary metabolites that enhance wine grape composition in ripe berries. Transcriptomic and metabolic analyses of noble rot in cv. Semillon determined that anthocyanin biosynthesis is the most consistent hallmark of noble rot. In addition, the biosynthesis of terpenes and fatty acid aroma precursors increase during the infection (Blanco-Ulate et al., 2015). APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2-ERF), and NON APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION FACTOR/CUPSHAPED COTYLEDON (NAC) transcription factors, were up-regulated during noble rot (Blanco-Ulate et al., 2015). Early products of the phenylpropanoid pathway are accumulated in noble-rotted berries, such as rosmarinic acid (a cinnamic acid derivative with antioxidant and aromatic properties). Also, a significant accumulation of several flavonoid glycosides and flavanones was detected, along with build ups of cyanidin-3-rutinoside, delphinidin-3-rutinoside, cyanidin-3-gentiobioside, and delphinidin-3-gentiobioside, anthocyanins that are normally scarce in white-skinned grape berries. Other aromatic compounds such as acetophenones, benzoic acid derivatives, methoxyphenols, and phenolic glycosides showed increased abundance in noble rot, together with gallic acid, a precursor of tannin biosynthesis (Blanco-Ulate et al., 2015).
In conclusion, reprogramming of secondary metabolites and hormonal pathways are common features in B. cinerea-infected grape berries. Additionally, it has been shown that during grape berry development, the fruit undergoes changes that facilitate fungal infection, such as fruit softening, organic acid and sugar level modifications, loss of the preformed defenses and decreased stilbene production, among others. On the other hand, noble rot also alters berry metabolism by inducing stress responses and accelerating ripening to enhance the colonization process. B. cinerea infections can affect the color and sugar concentration, improving wine grape composition. This effect is caused by an imbalance of hormone synthesis and perception, which in turn activates several ripening-associated pathways. However, the mechanism behind this acceleration is still under study.
Viral Diseases and Their Effect on Grape Berry Development
Viral diseases are also common in grapevine plantations. Infections caused by these pathogens are highly complex, due to the large number of viral agents described and the occurrence of multiple infections (Prosser et al., 2007; Martelli, 2014; Jooste et al., 2015; Naidu et al., 2015). Grapevines show no resistance against virus; instead, viruses and host plants establish compatible interactions, where pathogens spread throughout all plant tissues, unimpeded by the resistance responses, generating global cellular stress and developmental defects. However, in compatible interactions, hosts are not passive against the pathogen, and molecular, cellular and physiological responses can be observed (O’Donnell et al., 2003; Ehrenfeld et al., 2005). In general, grapevine viruses infect vegetative organs, but infections also have consequences for berry development, causing a reduction in berry setting, and delayed berry ripening (Martelli, 1993, 2014). Molecular changes during berry ripening in virus-infected grapevine plants have been less characterized than leaf symptomatology. For instance, characterization of the Grapevine leafroll-associated virus 3 (GLRaV-3) infection in the red cv. Cabernet Sauvignon revealed the presence of viral particles in berry tissues together with massive transcriptional changes, which were more pronounced during ripening (Aquea et al., 2011; Vega et al., 2011). Since this virus is restricted to the phloem (Martelli, 2014), GLRaV-3 infection could physically modify sugar accumulation, altering source–sink relationships.
Transcript profiling analyses performed in cv. Cabernet Sauvignon berries at veraison and ripening, using the V. vinifera Affymetrix GeneChip, revealed numerous changes in transcripts related either to viral infection or to berry development (Vega et al., 2011). About 400 genes showed differential expression between veraison and ripening, in uninfected tissues. However, only half of these exhibited such differences when the two stages were compared in infected berries. Thus, viral disease greatly modifies the transcript abundance profile during berry development. The number of differentially expressed genes in infected berries was higher during ripening (146 up- and 86 down-regulated genes) than at veraison (41 up- and 14 downregulated genes), suggesting that the former stage could be more dramatically affected by virus infection. Among the transcripts that change in infected berries, is a group related to sugar transport and metabolism, including ATOCT2, a carbohydrate transmembrane transporter; ATSPS4F, a putative sucrose-phosphate synthase; a short-chain dehydrogenase/reductase (SDR) family protein involved in sugar metabolism; SUS2, sucrose synthase 2; and BFRUCT3, a beta-fructosidase. In agreement with this, glucose and fructose levels also decreased during ripening in infected berries. Several genes from the phenylpropanoid pathway were repressed by viral infection during ripening, such as CHS2 and UFGT, as well as genes that encode for transcription factors MYBPA1 and MYBA (anthocyanin biosynthesis), and FLS1, related to the flavonol biosynthetic pathway. These results were further supported by a decrease in total anthocyanin content and flavonol concentration during ripening in infected berries (Vega et al., 2011).
A characterization of berries of the Italian cv. Nebbiolo harboring a mixed infection of GLRaV-1, Grapevine virus A (GVA) and Rupestris Stem Pitting virus (RSPaV), showed significant differences in bud burst index, berry weight, titratable acidity, and resveratrol content when compared with uninfected berries (Giribaldi et al., 2011). In that study, a proteomic analysis revealed that mixed viral infection affects proteins related to cell structure metabolism in pulp, such as pectin methylesterase, N-acetyl-gamma-glutamyl-phosphate reductase, plastid movement impaired 1, phosphoglycerate kinase, polyphenol oxidase and alpha-tubulin, among others (Giribaldi et al., 2011). A thorough study carried out over three seasons, on the effects of grapevine leafroll disease (GLD) on cv. Merlot, showed that infection impacted greatly on yield, as well as on fruit quality (Alabi et al., 2016). For instance, the authors consistently found a lower fruit yield over the seasons evaluated, supporting previous conclusions that GLD negatively affects vine performance. In virus-infected cv. Merlot plants, developing green berries showed minor compositional changes in comparison to uninfected plants. However, after veraison, dramatic variations were observed as a consequence of viral disease, suggesting that the virus can affect ripening-related processes occurring from veraison onward, as previously shown for cv. Cabernet Sauvignon (Vega et al., 2011).
In general, transcriptomic and metabolic data support the observation that viral diseases delay grape berry ripening, altering several characteristic parameters associated with this stage, such as sugar accumulation and color, among others. However, more studies should be carried out in order to establish how viruses alter grapevine berry ripening, how cultivars and environmental factors interact to produce the complete symptomatology, and how these multiple cues modify berry ripening.
Conclusion
Significant progress has been made toward understanding grape berry development, and how environmental factors can positively or negatively regulate this process. In this field, Omics platforms have been an important tool in the elucidation of the mechanisms underlying these interactions. Due to the lack of transgenic lines and suitable technologies for reverse genetics in grapes, Omics analyses have allowed us to make progress in unraveling the complex mechanisms that take place during berry development. Of the future challenges, the establishment of a robust model to assess biological questions is key. In grapes, the availability of mutant varieties and related cultivars with contrasting phenotypes is an advantage, but differences between cultivars could be more complex than expected. Therefore, global analysis should be carried out. In this context, as Omics provide numerous tools that generate huge data sets from an overall perspective, the integration of this information is the next challenge which needs to be addressed in order to understand the different processes underlying grape berry development. Systems biology deals with the integration of these data sets, advancing the way in which biological processes are studied from gene-by-gene studies toward a global perspective, where the different processes are depicted in regulatory networks. Those networks are useful in the prediction of gene function, while providing new insights into the regulatory mechanisms at a global level. The generation of robust networks to identify new regulators and genome-wide responses to environmental factors requires a vast number of data sets and the integration of multi-omics studies. In grape berries, most of the Omics studies are based on transcriptomic and metabolomic profiles; more-integrated networks are hindered by the lack of proteomic and phosphoproteomic studies. Another challenge in the bioinformatic field is the standardization and centralization of the stored data in order to facilitate the access to, and analysis of, Omics studies. Currently in grape, due to the multiple sources of data and gene annotation, there is a lack of consensus in the integrative tools available. For instance, annotation version 1 (V1) and version 2 (V2) differ in the number of annotated genes, with V2 having around 2000 new genes and 3000 putative long non-coding RNAs (lncRNA). The integration of the different annotations is a task that remains unresolved by the scientific community studying grape. Therefore, to improve our current knowledge, further Omics studies are undoubtedly necessary, yet this new data must be integrated with systems biology tools in order to comprehensively depict the associated regulatory networks.
Author Contributions
The manuscript was written by AS, CE, GA, CI-B, EP, CM-R, CS, FP, AA, and PA-J. AS was involved in revising the manuscript critically for important intellectual content. Manuscript editing was conducted by AS and CE. Figure design was conducted by CM-R. All authors contributed, read and approved the final manuscript. PA-J made the final approval.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ABA
abscisic acid
- ABF
ABA-response element binding factor
- ARF
auxin response factor
- ASR
ABA stress responsive element
- BR
Brassinosteroids
- CAA
carbonic anhydrase
- CAB
chlorophyll a/b binding protein
- CB
Corinto Bianco
- ERF
ethylene response factor
- GAs
gibberellins
- GLD
grapevine leafroll disease
- GLRaV
Grapevine leafroll-associated virus
- GVA
Grapevine virus A
- HXK
hexokinase
- IAA
indole-3-acetic acid
- JA
jasmonic acid
- LOX
chloroplast lipoxygenase
- NCED
9-cis-epoxycarotenoid dioxygenase
- PX
Pedro Ximenez
- QTL
quantitative trait locus
- ROS
reactive oxygen species
- RSPaV
Rupestris Stem Pitting virus
- SDI
seed development inhibitor
- SNP
single-nucleotide polymorphism
- SnRK1
sucrose-non-fermentative related kinase 1
- SSH
suppression subtractive hybridization
- T6P
trehalose-6-phosphate
- UV
ultraviolet
Footnotes
Funding. This work was supported by FONDECYT postdoctoral research 3150608 (AS), Millennium Nucleus of Plant Systems and Synthetic Biology NC130030 and FONDECYT 1150220.
References
- Adham I. M., Sallam M. A., Steding G., Korabiowska M., Brinck U., Hoyer-Fender S., et al. (2003). Disruption of the pelota gene causes early embryonic lethality and defects in cell cycle progression. Mol. Cell. Biol. 23 1470–1476. 10.1128/MCB.23.4.1470-1476.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo-Romero P., Erban A., Rego C., Carbonell-Bejerano P., Nascimento T., Sousa L., et al. (2015). Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 66 1769–1785. 10.1093/jxb/eru517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi O. J., Casassa L. F., Gutha L. R., Larsen R. C., Henick-Kling T., Harbertson J. F., et al. (2016). Impacts of grapevine leafroll disease on fruit yield and grape and wine chemistry in a wine grape (Vitis vinifera L.) cultivar. PLOS ONE 11:e0149666 10.1371/journal.pone.0149666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquea F., Vega A., Timmermann T., Poupin M. J., Arce-Johnson P. (2011). Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep. 30 1087–1097. 10.1007/s00299-011-1015-0 [DOI] [PubMed] [Google Scholar]
- Armijo G., Espinoza C., Loyola R., Restovic F., Santibáñez C., Schlechter R., et al. (2016a). “Grapevine biotechnology: molecular approaches underlying abiotic and biotic stress responses,” in Grape and Wine Biotechnology, eds Morata A., Loira I. (Rijeka: InTech; ). [Google Scholar]
- Armijo G., Schlechter R., Agurto M., Muñoz D., Núñez C., Arce P. (2016b). Grapevine pathogenic microorganisms: understanding infection strategies and host response scenarios. Front. Plant Sci. 7:382 10.3389/fpls.2016.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E., Rolland F., Thevelein J. M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942. 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- Berli F., Bottini R. (2013). UV-B and abscisic acid effects on grape berry maturation and quality. J. Berry Res. 3 1–14. 10.3233/JBR-130047 [DOI] [Google Scholar]
- Blanco-Ulate B., Amrine K. C., Collins T. S., Rivero R. M., Vicente A. R., Morales-Cruz A., et al. (2015). Developmental and metabolic plasticity of white-skinned grape berries in response to Botrytis cinerea during noble rot. Plant Physiol. 169 2422–2443. 10.1104/pp.15.00852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Ebadi A., McDavid D., Robinson S. P. (2006). Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 140 279–291. 10.1104/pp.105.073262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P. K., Davies C., Robinson S. P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111 1059–1066. 10.1104/pp.111.4.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher C., Harvey K., Forde C. G., Boss P. K., Davies C. (2011). Auxin treatment of pre-veraison grape (Vitis vinifera L.) berries both delays ripening and increases the synchronicity of sugar accumulation. Aust. J. Grape Wine Res. 17 1–8. 10.1111/j.1755-0238.2010.00110.x [DOI] [Google Scholar]
- Bottcher C., Keyzers R. A., Boss P. K., Davies C. (2010). Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 61 3615–3625. 10.1093/jxb/erq174 [DOI] [PubMed] [Google Scholar]
- Bouquet A., Danglot Y. (1996). Inheritance of seedlessness in grapevine (Vitis vinifera L.). Vitis 35 35–42. [Google Scholar]
- Cabezas J. A., Cervera M. T., Ruiz-Garcia L., Carreno J., Martinez-Zapater J. M. (2006). A genetic analysis of seed and berry weight in grapevine. Genome 49 1572–1585. 10.1139/g06-122 [DOI] [PubMed] [Google Scholar]
- Çakir B. (2014). Identification and structure of six members of the hexokinase gene family in Vitis vinifera: cloning, expression, and functional analysis of a putative chloroplast stromal-type hexokinase. J. Hortic. Sci. Biotechnol. 89 663–673. 10.1080/14620316.2014.11513135 [DOI] [Google Scholar]
- Carbonell-Bejerano P., Rodriguez V., Royo C., Hernaiz S., Moro-Gonzalez L. C., Torres-Vinals M., et al. (2014). Circadian oscillatory transcriptional programs in grapevine ripening fruits. BMC Plant Biol. 14:78 10.1186/1471-2229-14-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin S. D., Di Gaspero G. (2007). Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol. 7:46 10.1186/1471-2229-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai L., Li Y., Chen S., Perl A., Zhao F., Ma H. (2014). RNA sequencing reveals high resolution expression change of major plant hormone pathway genes after young seedless grape berries treated with gibberellin. Plant Sci. 229 215–224. 10.1016/j.plantsci.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Chai Y.-M., Zhang Q., Tian L., Li C.-L., Xing Y., Qin L., et al. (2013). Brassinosteroid is involved in strawberry fruit ripening. Plant Growth Regul. 69 63–69. 10.1007/s10725-012-9747-6 [DOI] [Google Scholar]
- Cheng C., Jiao C., Singer S. D., Gao M., Xu X., Zhou Y., et al. (2015). Gibberellin-induced changes in the transcriptome of grapevine (Vitis labrusca x V. vinifera) cv. Kyoho flowers. BMC Genomics 16:128 10.1186/s12864-015-1324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Xu X., Singer S. D., Li J., Zhang H., Gao M., et al. (2013). Effect of GA3 treatment on seed development and seed-related gene expression in grape. PLOS ONE 8:e80044 10.1371/journal.pone.0080044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervin C., El-Kereamy A., Roustan J.-P., Latché A., Lamon J., Bouzayen M. (2004). Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci. 167 1301–1305. 10.1016/j.plantsci.2004.06.026 [DOI] [Google Scholar]
- Chervin C., Tira-umphon A., Terrier N., Zouine M., Severac D., Roustan J.-P. (2008). Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiol. Plant. 134 534–546. 10.1111/j.1399-3054.2008.01158.x [DOI] [PubMed] [Google Scholar]
- Cho Y. H., Yoo S. D., Sheen J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589. 10.1016/j.cell.2006.09.028 [DOI] [PubMed] [Google Scholar]
- Coombe B. G. (1987). Influence of temperature on composition and quality of grapes. Acta Hortic. 206 23–36. 10.17660/ActaHortic.1987.206.1 [DOI] [Google Scholar]
- Coombe B. G., Hale C. R. (1973). The hormone content of ripening grape berries and the effects of growth substance treatments. Plant Physiol. 51 629–634. 10.1104/pp.51.4.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe B. G., McCarthy M. G. (2000). Dynamics of grape berry growth and physiology of ripening. Aust. J. Grape Wine Res. 6 131–135. 10.1111/j.1755-0238.2000.tb00171.x [DOI] [Google Scholar]
- Costantini L., Battilana J., Lamaj F., Fanizza G., Grando M. S. (2008). Berry and phenology-related traits in grapevine (Vitis vinifera L.): from quantitative trait loci to underlying genes. BMC Plant Biol. 8:38 10.1186/1471-2229-8-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costenaro-da-Silva D., Passaia G., Henriques J. A., Margis R., Pasquali G., Revers L. F. (2010). Identification and expression analysis of genes associated with the early berry development in the seedless grapevine (Vitis vinifera L.) cultivar Sultanine. Plant Sci. 179 510–519. 10.1016/j.plantsci.2010.07.021 [DOI] [PubMed] [Google Scholar]
- Cramer G. R., Ghan R., Schlauch K. A., Tillett R. L., Heymann H., Ferrarini A., et al. (2014). Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 14:370 10.1186/s12870-014-0370-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer G. R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S., Höll J., Loyola R., Arce-Johnson P., Alcalde J. A., Matus J. T., et al. (2017). Transcriptome-wide identification of novel UV-B- and light modulated flavonol pathway genes controlled by VviMYBF1. Front. Plant Sci. 8:1084 10.3389/fpls.2017.01084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S., Stracke R., Weisshaar B., Cordon N., Harris N. N., Walker A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151 1513–1530. 10.1104/pp.109.142059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z. W., Leon C., Feil R., Lunn J. E., Delrot S., Gomes E. (2013). Metabolic profiling reveals coordinated switches in primary carbohydrate metabolism in grape berry (Vitis vinifera L.), a non-climacteric fleshy fruit. J. Exp. Bot. 64 1345–1355. 10.1093/jxb/ers396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C., Boss P. K., Robinson S. P. (1997). Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 115 1155–1161. 10.1104/pp.115.3.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosas I., Ponce M. T. T., Malovini E., Deis L., Cavagnaro B., Cavagnaro P. (2017). Loss of anthocyanins and modification of the anthocyanin profiles in grape berries of Malbec and Bonarda grown under high temperature conditions. Plant Sci. 258 137–145. 10.1016/j.plantsci.2017.01.015 [DOI] [PubMed] [Google Scholar]
- Degu A., Hochberg U., Sikron N., Venturini L., Buson G., Ghan R., et al. (2014). Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 14:188 10.1186/s12870-014-0188-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L. G., Grimplet J., Wheatley M. D., Tillett R. L., Quilici D. R., Osborne C., et al. (2007). Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8:429 10.1186/1471-2164-8-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Genova A., Almeida A. M., Muñoz-Espinoza C., Vizoso P., Travisany D., Moraga C., et al. (2014). Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol. 14:7 10.1186/1471-2229-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Xie D. Y., Sharma S. B. (2005). Proanthocyanidins–a final frontier in flavonoid research? New Phytol. 165 9–28. 10.1111/j.1469-8137.2004.01217.x [DOI] [PubMed] [Google Scholar]
- Dokoozlian N. (2000). “Grape berry growth and development,” in Raisin Production Manual, ed. Christensen L. P. (Oakland, CA: Agricultural and Natural Resources Publication; ), 30–37. [Google Scholar]
- Doligez A., Bouquet A., Danglot Y., Lahogue F., Riaz S., Meredith P., et al. (2002). Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor. Appl. Genet. 105 780–795. 10.1007/s00122-002-0951-z [DOI] [PubMed] [Google Scholar]
- Ebadi A., Sedgley M., May P., Coombe B. G. (1996). Seed development and abortion in Vitis vinifera L., cv. Chardonnay. Int. J. Plant Sci. 157 703–712. 10.1086/297392 [DOI] [Google Scholar]
- Ehrenfeld N., Canon P., Stange C., Medina C., Arce-Johnson P. (2005). Tobamovirus coat protein CPCg induces an HR-like response in sensitive tobacco plants. Mol. Cells 19 418–427. [PubMed] [Google Scholar]
- Fasoli M., Dal Santo S., Zenoni S., Tornielli G. B., Farina L., Zamboni A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24 3489–3505. 10.1105/tpc.112.100230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes N., Gerós H., Delrot S. (2011). Grape berry vacuole: a complex and heterogeneous membrane system specialized in the accumulation of solutes. Am. J. Enol. Vitic. 62 270–278. 10.5344/ajev.2011.10125 [DOI] [Google Scholar]
- Ford C. M., Boss P. K., Hoj P. B. (1998). Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 273 9224–9233. 10.1074/jbc.273.15.9224 [DOI] [PubMed] [Google Scholar]
- Fortes A. M., Agudelo-Romero P., Silva M. S., Ali K., Sousa L., Maltese F., et al. (2011). Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 11:149 10.1186/1471-2229-11-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes A. M., Teixeira R. T., Agudelo-Romero P. (2015). Complex interplay of hormonal signals during grape berry ripening. Molecules 20 9326–9343. 10.3390/molecules20059326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133 1420–1428. 10.1104/pp.103.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta G. A., Matthews M. A., Shaghasi T. H., McElrone A. J., Castellarin S. D. (2010). Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232 219–234. 10.1007/s00425-010-1165-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerós H., Chaves M., Delrot S. (2012). The Biochemistry of the Grape Berry. Sharjah: Bentham Science Publishers; 10.2174/97816080536051120101 [DOI] [Google Scholar]
- Ghan R., Van Sluyter S. C., Hochberg U., Degu A., Hopper D. W., Tillet R. L., et al. (2015). Five omic technologies are concordant in differentiating the biochemical characteristics of the berries of five grapevine (Vitis vinifera L.) cultivars. BMC Genomics 16:946 10.1186/s12864-015-2115-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribaldi M., Geny L., Delrot S., Schubert A. (2010). Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. J. Exp. Bot. 61 2447–2458. 10.1093/jxb/erq079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribaldi M., Purrotti M., Pacifico D., Santini D., Mannini F., Caciagli P., et al. (2011). A multidisciplinary study on the effects of phloem-limited viruses on the agronomical performance and berry quality of Vitis vinifera cv. Nebbiolo. J. Proteomics 75 306–315. 10.1016/j.jprot.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Guelfat-Reich S., Safran B. (1971). Indices of maturity for table grapes as determined by variety. Am. J. Enol. Vitic. 22 13–18. [Google Scholar]
- Hanania U., Velcheva M., Or E., Flaishman M., Sahar N., Perl A. (2007). Silencing of chaperonin 21, that was differentially expressed in inflorescence of seedless and seeded grapes, promoted seed abortion in tobacco and tomato fruits. Transgenic Res. 16 515–525. 10.1007/s11248-006-9044-0 [DOI] [PubMed] [Google Scholar]
- Hanania U., Velcheva M., Sahar N., Flaishman M., Or E., Degani O., et al. (2009). The ubiquitin extension protein S27a is differentially expressed in developing flower organs of Thompson seedless versus Thompson seeded grape isogenic clones. Plant Cell Rep. 28 1033–1042. 10.1007/s00299-009-0715-1 [DOI] [PubMed] [Google Scholar]
- Hayes M. A., Davies C., Dry I. B. (2007). Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: differential roles in sink and source tissues. J. Exp. Bot. 58 1985–1997. 10.1093/jxb/erm061 [DOI] [PubMed] [Google Scholar]
- Hong Y. S., Martinez A., Liger-Belair G., Jeandet P., Nuzillard J. M., Cilindre C. (2012). Metabolomics reveals simultaneous influences of plant defence system and fungal growth in Botrytis cinerea-infected Vitis vinifera cv. Chardonnay berries. J. Exp. Bot. 63 5773–5785. 10.1093/jxb/ers228 [DOI] [PubMed] [Google Scholar]
- Jayasena V., Cameron I. (2008). °BRIX/acid ratio as a predictor of consumer acceptability of crimson seedless table grapes. J. Food Qual. 31 736–750. 10.1111/j.1745-4557.2008.00231.x [DOI] [Google Scholar]
- Jeevan Kumar S. P., Rajendra Prasad S., Banerjee R., Thammineni C. (2015). Seed birth to death: dual functions of reactive oxygen species in seed physiology. Ann. Bot. 116 663–668. 10.1093/aob/mcv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. I. (2009). Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 60 407–431. 10.1146/annurev.arplant.59.032607.092953 [DOI] [PubMed] [Google Scholar]
- Jooste A. E. C., Molenaar N., Maree H. J., Bester R., Morey L., de Koker W. C., et al. (2015). Identification and distribution of multiple virus infections in Grapevine leafroll diseased vineyards. Eur. J. Plant Pathol. 142 363–375. 10.1007/s10658-015-0620-0 [DOI] [Google Scholar]
- Joubert C., Young P. R., Eyeghe-Bickong H. A., Vivier M. A. (2016). Field-grown grapevine berries use carotenoids and the associated xanthophyll cycles to acclimate to UV exposure differentially in high and low light (shade) conditions. Front. Plant Sci. 7:786 10.3389/fpls.2016.00786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. (ed.). (2010). “Chapter 2 - phenology and growth cycle,” in The Science of Grapevines, (San Diego, CA: Academic Press), 49–83. 10.1016/B978-0-12-374881-2.00002-7 [DOI] [Google Scholar]
- Keller M. (ed.). (2015). “Chapter 7 - environmental constraints and stress physiology,” in The Science of Grapevines, 2nd Edn, (San Diego, CA: Academic Press), 267–341. 10.1016/B978-0-12-419987-3.00007-8 [DOI] [Google Scholar]
- Kelloniemi J., Trouvelot S., Heloir M. C., Simon A., Dalmais B., Frettinger P., et al. (2015). Analysis of the molecular dialogue between gray mold (Botrytis cinerea) and grapevine (Vitis vinifera) reveals a clear shift in defense mechanisms during berry ripening. Mol. Plant Microbe Interact. 28 1167–1180. 10.1094/mpmi-02-15-0039-r [DOI] [PubMed] [Google Scholar]
- Kennedy J. (2002). Understanding grape berry development. Pract. Winery Vineyard 4 1–5. [Google Scholar]
- Kimura P. H., Okamoto G., Hirano K. (1996). Effects of gibberellic acid and streptomycin on pollen germination and ovule and seed development in Muscat bailey A. Am. J. Enol. Vitic. 47 152–156. [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304:982 10.1126/science.1095011 [DOI] [PubMed] [Google Scholar]
- Koyama K., Sadamatsu K., Goto-Yamamoto N. (2010). Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Funct. Integr. Genomics 10 367–381. 10.1007/s10142-009-0145-8 [DOI] [PubMed] [Google Scholar]
- Lecourieux F., Lecourieux D., Vignault C., Delrot S. (2010). A sugar-inducible protein kinase, VvSK1, regulates hexose transport and sugar accumulation in grapevine cells. Plant Physiol. 152 1096–1106. 10.1104/pp.109.149138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Sheen J. (2016). Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 33 116–125. 10.1016/j.pbi.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Y., Chen N. N., Cheng Z. M., Xiong J. S. (2016). Genome-wide identification, annotation and expression profile analysis of SnRK2 gene family in grapevine. Aust. J. Grape Wine Res. 22 478–488. 10.1111/ajgw.12223 [DOI] [Google Scholar]
- Loyola R., Herrera D., Mas A., Wong D. C., Holl J., Cavallini E., et al. (2016). The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67 5429–5445. 10.1093/jxb/erw307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J. E., Delorge I., Figueroa C. M., Van Dijck P., Stitt M. (2014). Trehalose metabolism in plants. Plant J. 79 544–567. 10.1111/tpj.12509 [DOI] [PubMed] [Google Scholar]
- Maeda H., Dudareva N. (2012). The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 63 73–105. 10.1146/annurev-arplant-042811-105439 [DOI] [PubMed] [Google Scholar]
- Magyar I. (2011). Botrytized wines. Adv. Food Nutr. Res. 63 147–206. 10.1016/b978-0-12-384927-4.00006-3 [DOI] [PubMed] [Google Scholar]
- Martelli G. P. (1993). Graft-Transmissible Diseases of Grapevines. Rome: FAO. [Google Scholar]
- Martelli G. P. (2014). Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 96 1–136. 10.4454/JPP.V96I1SUP [DOI] [Google Scholar]
- Martinez-Esteso M. J., Selles-Marchart S., Lijavetzky D., Pedreno M. A., Bru-Martinez R. (2011). A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. J. Exp. Bot. 62 2521–2569. 10.1093/jxb/erq434 [DOI] [PubMed] [Google Scholar]
- Martinez-Luscher J., Morales F., Delrot S., Sanchez-Diaz M., Gomes E., Aguirreolea J., et al. (2013). Short- and long-term physiological responses of grapevine leaves to UV-B radiation. Plant Sci. 213 114–122. 10.1016/j.plantsci.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Matus J. T. (2016). Transcriptomic and metabolomic networks in the grape berry illustrate that it takes more than flavonoids to fight against ultraviolet radiation. Front. Plant Sci. 7:1337 10.3389/fpls.2016.01337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus J. T., Cavallini E., Loyola R., Höll J., Finezzo L., Dal Santo S., et al. (2016). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 91 220–236. 10.1111/tpj.13558 [DOI] [PubMed] [Google Scholar]
- Matus J. T., Loyola R., Vega A., Peña-Neira A., Bordeu E., Arce-Johnson P., et al. (2009). Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60 853–867. 10.1093/jxb/ern336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier N., Pfister C., Wanner H., Luterbacher J. (2007). Grape harvest dates as a proxy for Swiss April to August temperature reconstructions back to AD 1480. Geophys. Res. Lett. 34 1–6. 10.1029/2007GL031381 [DOI] [Google Scholar]
- Mejía N., Gebauer M., Muñoz L., Hewstone N., Muñoz C., Hinrichsen P. (2007). Identification of QTLs for seedlessness, berry size, and ripening date in a seedless x seedless table grape progeny. Am. J. Enol. Vitic. 58 499–507. [Google Scholar]
- Mejia N., Soto B., Guerrero M., Casanueva X., Houel C., Miccono Mde L., et al. (2011). Molecular, genetic and transcriptional evidence for a role of VvAGL11 in stenospermocarpic seedlessness in grapevine. BMC Plant Biol. 11:57 10.1186/1471-2229-11-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336. 10.1126/science.1080585 [DOI] [PubMed] [Google Scholar]
- Naidu R. A., Maree H. J., Burger J. T. (2015). Grapevine leafroll disease and associated viruses: a unique pathosystem. Annu. Rev. Phytopathol. 53 613–634. 10.1146/annurev-phyto-102313-045946 [DOI] [PubMed] [Google Scholar]
- Negri A. S., Prinsi B., Rossoni M., Failla O., Scienza A., Cocucci M., et al. (2008). Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9:378 10.1186/1471-2164-9-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P., Lecourieux D., Kappel C., Cluzet S., Cramer G., Delrot S., et al. (2014). The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol. 164 365–383. 10.1104/pp.113.231977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., Primavesi L. F., Patel M. K., Martinez-Barajas E., Powers S. J., Sagar R., et al. (2013). Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 63 89–98. 10.1016/j.plaphy.2012.11.011 [DOI] [PubMed] [Google Scholar]
- Nwafor C. C., Gribaudo I., Schneider A., Wehrens R., Grando M. S., Costantini L. (2014). Transcriptome analysis during berry development provides insights into co-regulated and altered gene expression between a seeded wine grape variety and its seedless somatic variant. BMC Genomics 15:1030 10.1186/1471-2164-15-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocarez N., Mejia N. (2016). Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits. Plant Cell Rep. 35 239–254. 10.1007/s00299-015-1882-x [DOI] [PubMed] [Google Scholar]
- O’Donnell P. J., Schmelz E. A., Moussatche P., Lund S. T., Jones J. B., Klee H. J. (2003). Susceptible to intolerance - a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33 245–257. 10.1046/j.1365-313X.2003.01619.x [DOI] [PubMed] [Google Scholar]
- Ojeda H., Deloire A., Carbonneau A., Ageorges A., Romieu C. (1999). Berry development of grapevines: relations between the growth of berries and their DNA content indicate cell multiplication and enlargement. Vitis 38 145–150. [Google Scholar]
- Pilati S., Perazzolli M., Malossini A., Cestaro A., Demattè L., Fontana P., et al. (2007). Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at vèraison. BMC Genomics 8:428 10.1186/1471-2164-8-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S. W., Goszczynski D. E., Meng B. (2007). Molecular analysis of double-stranded RNAs reveals complex infection of grapevines with multiple viruses. Virus Res. 124 151–159. 10.1016/j.virusres.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Rattanakon S., Ghan R., Gambetta G. A., Deluc L. G., Schlauch K. A., Cramer G. R. (2016). Abscisic acid transcriptomic signaling varies with grapevine organ. BMC Plant Biol. 16:72 10.1186/s12870-016-0763-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnikar M., Vilhar B., Gogala N. (1992). Stimulatory effects of jasmonic acid on potato stem node and protoplast culture. J. Plant Growth Regul. 11:29 10.1007/bf00193840 [DOI] [Google Scholar]
- Rienth M., Torregrosa L., Sarah G., Ardisson M., Brillouet J.-M., Romieu C. (2016). Temperature desynchronizes sugar and organic acid metabolism in ripening grapevine fruits and remodels their transcriptome. BMC Plant Biol. 16:164 10.1186/s12870-016-0850-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. P., Davies C. (2000). Molecular biology of grape berry ripening. Aust. J. Grape Wine Res. 6 175–188. 10.1111/j.1755-0238.2000.tb00177.x [DOI] [Google Scholar]
- Rossdeutsch L., Edwards E., Cookson S., Barrieu F., Gambetta G., Delrot S., et al. (2016). ABA-mediated responses to water deficit separate grapevine genotypes by their genetic background. BMC Plant Biol. 16:91 10.1186/s12870-016-0778-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo C., Carbonell-Bejerano P., Torres-Pérez R., Nebish A., Martínez Ó., Rey M.,et al. (2016). Developmental, transcriptome, and genetic alterations associated with parthenocarpy in the grapevine seedless somatic variant Corinto bianco. J. Exp. Bot. 67 259–273. 10.1093/jxb/erv452 [DOI] [PubMed] [Google Scholar]
- Sadras V. O., Moran M. A. (2012). Elevated temperature decouples anthocyanins and sugars in berries of Shiraz and Cabernet Franc. Aust. J. Grape Wine Res. 18 115–122. 10.1111/j.1755-0238.2012.00180.x [DOI] [Google Scholar]
- Smeekens S., Ma J., Hanson J., Rolland F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13 274–279. 10.1016/j.pbi.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Souquet J.-M., Cheynier V., Brossaud F., Moutounet M. (1996). Polymeric proanthocyanidins from grape skins. Phytochemistry 43 509–512. 10.1016/0031-9422(96)00301-9 [DOI] [Google Scholar]
- Suzuki M., Nakabayashi R., Ogata Y., Sakurai N., Tokimatsu T., Goto S., et al. (2015). Multiomics in grape berry skin revealed specific induction of the stilbene synthetic pathway by ultraviolet-C irradiation. Plant Physiol. 168 47–59. 10.1104/pp.114.254375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman C., Deluc L. G., Cramer G. R., Ford C. M., Soole K. L. (2009). Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70 1329–1344. 10.1016/j.phytochem.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Sweetman C., Wong D. C., Ford C. M., Drew D. P. (2012). Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genomics 13:691 10.1186/1471-2164-13-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Chua Y. J., Ross J. J., Quittenden L. J., Davies N. W., Reid J. B. (2012). Hormonal changes during non-climacteric ripening in strawberry. J. Exp. Bot. 63 4741–4750. 10.1093/jxb/ers147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Davies C., Shavrukov Y., Dry I. B., Reid J. B., Thomas M. R. (2006). Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol. 140 150–158. 10.1104/pp.105.070706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall E. A., Grimplet J., DeLuc L., Wheatley M. D., Vincent D., Osborne C., et al. (2007). Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct. Integr. Genomics 7 317–333. 10.1007/s10142-007-0051-x [DOI] [PubMed] [Google Scholar]
- Teixeira A., Eiras-Dias J., Castellarin S. D., Geros H. (2013). Berry phenolics of grapevine under challenging environments. Int. J. Mol. Sci. 14 18711–18739. 10.3390/ijms140918711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N., Glissant D., Grimplet J., Barrieu F., Abbal P., Couture C., et al. (2005). Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development. Planta 222 832–847. 10.1007/s00425-005-0017-y [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Gazzarrini S. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5:119 10.3389/fpls.2014.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houtte H., Lopez-Galvis L., Vandesteene L., Beeckman T., Van Dijck P. (2013). Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signal. Behav. 8:e23209 10.4161/psb.23209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas A. M., Vélez M. D., de Andrés M. T., Laucou V., Lacombe T., Boursiquot J.-M., et al. (2007). Corinto bianco: a seedless mutant of Pedro Ximenes. Am. J. Enol. Vitic. 58 540–543. [Google Scholar]
- Varoquaux F., Blanvillain R., Delseny M., Gallois P. (2000). Less is better: new approaches for seedless fruit production. Trends Biotechnol. 18 233–242. 10.1016/S0167-7799(00)01448-7 [DOI] [PubMed] [Google Scholar]
- Vega A., Gutierrez R. A., Pena-Neira A., Cramer G. R., Arce-Johnson P. (2011). Compatible GLRaV-3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera. Plant Mol. Biol. 77 261–274. 10.1007/s11103-011-9807-8 [DOI] [PubMed] [Google Scholar]
- Vignault C., Vachaud M., Cakir B., Glissant D., Dedaldechamp F., Buttner M., et al. (2005). VvHT1 encodes a monosaccharide transporter expressed in the conducting complex of the grape berry phloem. J. Exp. Bot. 56 1409–1418. 10.1093/jxb/eri142 [DOI] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., et al. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339 704–707. 10.1126/science.1230406 [DOI] [PubMed] [Google Scholar]
- Wheeler S., Loveys B., Ford C., Davies C. (2009). The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust. J. Grape Wine Res. 15 195–204. 10.1111/j.1755-0238.2008.00045.x [DOI] [Google Scholar]
- Wind J., Smeekens S., Hanson J. (2010). Sucrose: metabolite and signaling molecule. Phytochemistry 71 1610–1614. 10.1016/j.phytochem.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Wong D. C., Sweetman C., Drew D. P., Ford C. M. (2013). VTCdb: a gene co-expression database for the crop species Vitis vinifera (grapevine). BMC Genomics 14:882 10.1186/1471-2164-14-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. H., Cao Y. G., Guan L., Xin H. P., Li J. H., Li S. H. (2014). Genome-wide transcriptional profiles of the berry skin of two red grape cultivars (Vitis vinifera) in which anthocyanin synthesis is sunlight-dependent or -independent. PLOS ONE 9:e105959 10.1371/journal.pone.0105959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., Minoia L., Ferrarini A., Tornielli G. B., Zago E., Delledonne M., et al. (2008). Molecular analysis of post-harvest withering in grape by AFLP transcriptional profiling. J. Exp. Bot. 59 4145–4159. 10.1093/jxb/ern256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Gong P., Wei R., Li S., Zhang X., Yu Y., et al. (2013). The metacaspase gene family of Vitis vinifera L.: characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes. Gene 528 267–276. 10.1016/j.gene.2013.06.062 [DOI] [PubMed] [Google Scholar]
- Zhang X., Luo G., Wang R., Wang J., Himelrick D. G. (2003). Growth and developmental responses of seeded and seedless grape berries to shoot girdling. J. Am. Soc. Hortic. Sci. 128 316–323. [Google Scholar]
- Ziliotto F., Corso M., Rizzini F. M., Rasori A., Botton A., Bonghi C. (2012). Grape berry ripening delay induced by a pre-veraison NAA treatment is paralleled by a shift in the expression pattern of auxin- and ethylene-related genes. BMC Plant Biol. 12:185 10.1186/1471-2229-12-185 [DOI] [PMC free article] [PubMed] [Google Scholar]