Abstract
Surface properties of bacteria are determined by the molecular composition of the cell wall and they are important for interactions of cells with their environment. Well-known examples of bacterial interactions with surfaces are biofilm formation and the fermentation of solid materials like food and feed. Lactococcus lactis is broadly used for the fermentation of cheese and buttermilk and it is primarily isolated from either plant material or the dairy environment. In this study, we characterized surface hydrophobicity, charge, emulsification properties, and the attachment to milk proteins of 55 L. lactis strains in stationary and exponential growth phases. The attachment to milk protein was assessed through a newly developed flow cytometry-based protocol. Besides finding a high degree of biodiversity, phenotype-genotype matching allowed the identification of candidate genes involved in the modification of the cell surface. Overexpression and gene deletion analysis allowed to verify the predictions for three identified proteins that altered surface hydrophobicity and attachment of milk proteins. The data also showed that lactococci isolated from a dairy environment bind higher amounts of milk proteins when compared to plant isolates. It remains to be determined whether the alteration of surface properties also has potential to alter starter culture functionalities.
Keywords: gene-trait matching, cell surface hydrophobicity, surface charge, attachment to milk proteins, emulsion stability, bacteria-protein interactions, cell wall composition, Lactococcus lactis
Introduction
The bacterial surface is important for interactions of the cell with the environment, especially when it comes to surface adhesion (Bellon-Fontaine et al., 1996; Ly et al., 2006; Boks et al., 2008). Examples for such interactions are the fermentation of solid substrates like fermented foods (Sieuwerts et al., 2008), woody materials and straw (Bayer et al., 2004), bioremediation of soil (Groudev et al., 2010), the formation of biofilms (Decho, 2000; Sutherland, 2001; Prouty et al., 2002; Newman et al., 2013) or during attachment of bacterial cells to the intestinal tract (Kleerebezem and Vaughan, 2009; Bron et al., 2012). Microbial surface properties are especially important for the initial contact with and adhesion to a surface, which can occur via fimbriae, pili, flagella, or EPS (Van Houdt and Michiels, 2010). Once attached, cells can start to produce different polymeric components such as polysaccharides, glycoproteins, proteins, glycolipids, cellulose, and extracellular DNA (Van Houdt and Michiels, 2010), which can lead to biofilm formation and can further accelerate bacterial adhesion (Kumar and Anand, 1998; Decho, 2000).
The cell surface itself is characterized by properties like charge and hydrophobicity (Ly et al., 2008), which are determined by the molecular composition of the cell wall. The cell wall consists of peptidoglycan (Chapot-Chartier and Kulakauskas, 2014), polysaccharides (Ruas-Madiedo et al., 2002), proteins, teichoic, and lipoteichoic acids, lipids (Pelletier et al., 1997) and can be decorated with a sugar pellicle (Chapot-Chartier et al., 2010), pili (Telford et al., 2006; Oxaran et al., 2012; Meyrand et al., 2013; Castelain et al., 2016), and/or an S-layer (van der Mei et al., 2003). The charge of the cell surface is determined by positively and negatively charged groups on teichoic and lipoteichoic acids, polysaccharides, proteins, and pili (Delcour et al., 1999; Boonaert and Rouxhet, 2000; Chapot-Chartier et al., 2010), while its hydrophobicity is related to the presence of polysaccharides, LPS, and (glyco-) proteinaceous material (Pelletier et al., 1997; Firoozmand and Rousseau, 2016), as well as pili (Tarazanova et al., 2016). Although the bacterial surface contains positively and negatively charged molecules, the net surface charge of bacteria is mostly negative (Neu and Marshall, 1990). The surface composition is species and strain specific (Wicken et al., 1983), and can vary between different growth substrates (Wicken et al., 1983) and growth phases (Boonaert and Rouxhet, 2000; Schär-Zammaretti and Ubbink, 2003).
The interactions between cell surface and substrate can be electrostatic. For example, in sand, a strong negative charge of the cell surface causes electrostatic repulsion and thus prevents bacterial adhesion and increases cell transport through the sand matrix while cells with high hydrophobicity are retained by that matrix (Jacobs et al., 2007). Other types of interactions occurring are hydrophobic, van der Waals, and Lewis acid–base forces. An example is the biofilm formation in which Brownian motion, hydrogen bonding, and electrostatic forces play a predominant role during the initial cell attachment, while cell hydrophobic forces as well as dipole-dipole, ionic bonding become more prominent during the phase of “irreversible” attachment of the bacteria to the surface (Neu and Marshall, 1990; Kumar and Anand, 1998). In other words, during bacteria-substrate interactions, a combination of all forces is present: at initial interactions long-range forces are most important but once attachment is achieved, the short-range forces may predominate. Factors like pH, temperature and ionic strength influence the interactions and add complexity to explanations of bacteria-substrate interactions.
Lactococcus lactis is widely used as a starter culture in the production of cheese, sour cream, and buttermilk (Leroy and De Vuyst, 2004), where it is responsible for food preservation, flavor formation, and textural properties (Leroy and De Vuyst, 2004). It is classified into the subspecies (ssp.) lactis including ssp. lactis biovar. diacetylactis, ssp. cremoris, and ssp. hordniae. The molecular composition of the L. lactis cell wall and its interactions with food components were reviewed by Burgain et al. (2014). It was shown that within L. lactis the diversity in cell surface charge, hydrophobicity and the ability to stabilize emulsion is very high (Ly et al., 2006). Most L. lactis strains originate either from a dairy environment or from plant material, and literature suggests that strains of dairy origin have evolved from plant isolates (van Hylckama Vlieg et al., 2006). The transition from the plant to the dairy environment was analyzed by comparative genomics (Siezen and van Hylckama Vlieg, 2011) or experimental evolution (Bachmann et al., 2012) and the results consistently describe similar metabolic adaptations. The main alterations during the plant-dairy transition are the loss of genes for the utilization of carbohydrate those only occur in plant material and the improved utilization of milk proteins. However, nothing is known about possible effects of the environmental transition on surface properties.
In this study, we investigated the surface properties of 55 L. lactis strains of which 25 were isolated from plant material and 30 from a dairy environment. We measured the cell surface hydrophobicity (CSH) and charge as well as emulsion stabilizing properties and the attachment of the bacterial cells to milk proteins. Genotype-phenotype matching (GTM) (Siezen et al., 2008; Bayjanov et al., 2012, 2013) allowed identifying key molecules involved in L. lactis surface properties. An analysis based on phylogeny and strain origin revealed that dairy isolates have a much higher capacity to bind milk proteins.
Materials and methods
Bacterial strains and culturing conditions
L. lactis used in this study (Table 1) were grown as standing cultures at 30°C in M17 (Oxoid, Thermo Scientific, Hampshire, UK) broth supplemented with 1% glucose (GM17) or 1% lactose (LM17). E. coli E10 containing pUC19 with an erythromycin resistance gene, pUC19E, was grown in tryptone yeast extract broth (TYB) at 37°C under vigorous shaking and access of oxygen. When required, antibiotics were added to the media: erythromycin (Em) was used at 10 μg/ml; chloramphenicol (Cm)—10 μg/ml; nisin—10 ng/ml. Optical density at 600 nm (OD600) was measured using a single cell spectrophotometer (Ultrospec 2000, Pharmacia Biotech, Centerville, USA). Exponentially growing cells were prepared by diluting an overnight culture to an OD600 of 0.01 and subsequent incubation until an OD600 of 0.45 ± 0.04 was reached. Stationary cells were prepared similarly by growing a culture for 16–18 h.
Table 1.
The 55 L. lactis strains and plasmid used in this study.
| Strain | Genotype | Origin | References | |
|---|---|---|---|---|
| 1 | ATCC19435 | L. lactis ssp. lactis | Milk (dairy starter) | Siezen et al., 2011; Backus et al., 2017 |
| 2 | HP | L. lactis ssp. cremoris | Dairy starter | Siezen et al., 2011; Wels et al., 2017 |
| 3 | P7266 | L. lactis ssp. lactis | Litter on pastures | Siezen et al., 2011; Backus et al., 2017 |
| 4 | NCDO895 | L. lactis ssp. lactis | Dairy starter | Siezen et al., 2011; Backus et al., 2017 |
| 5 | LMG8520 | L. lactis ssp. Hordniae | Leaf hopper | Siezen et al., 2011; Backus et al., 2017 |
| 6 | N41 | L. lactis ssp. cremoris | Soil and grass | Siezen et al., 2011; Wels et al., 2017 |
| 7 | M20 | L. lactis ssp. lactis biovar. diacetylactis | Soil | Siezen et al., 2011; Backus et al., 2017 |
| 8 | ML8 | L. lactis ssp. lactis | Dairy starter | Siezen et al., 2011; Backus et al., 2017 |
| 9 | V4 | L. lactis ssp. cremoris | Raw sheep milk | Siezen et al., 2011; Wels et al., 2017 |
| 10 | Li-1 | L. lactis ssp. lactis | Grass | Siezen et al., 2011; Backus et al., 2017 |
| 11 | UC317 | L. lactis ssp. lactis | Dairy starter | Siezen et al., 2011; Backus et al., 2017 |
| 12 | E34 | L. lactis ssp. lactis | Silage | Siezen et al., 2011; Backus et al., 2017 |
| 13 | N42 | L. lactis ssp. lactis | Soil and grass | Siezen et al., 2011; Backus et al., 2017 |
| 14 | DRA4 | L. lactis ssp. lactis biovar. diacetylactis | Dairy starter | Siezen et al., 2011; Backus et al., 2017 |
| 15 | AM2 | L. lactis ssp. cremoris | Dairy starter | Siezen et al., 2011; Wels et al., 2017 |
| 16 | P7304 | L. lactis ssp. lactis | Litter on pastures | Siezen et al., 2011; Backus et al., 2017 |
| 17 | LMG8526 | L. lactis ssp. lactis | Chinese radish seeds | Siezen et al., 2011; Backus et al., 2017 |
| 18 | LMG9446 | L. lactis ssp. lactis | Frozen peas | Siezen et al., 2011; Backus et al., 2017 |
| 19 | LMG9447 | L. lactis ssp. lactis | Frozen peas | Siezen et al., 2011; Backus et al., 2017 |
| 20 | LMG14418 | L. lactis ssp. lactis | Bovine milk | Siezen et al., 2011; Backus et al., 2017 |
| 21 | NIZO2244B | L. lactis ssp. lactis | Mustard and cress | Siezen et al., 2011; Backus et al., 2017 |
| 22 | FG2 | L. lactis ssp. cremoris | Dairy starter | Siezen et al., 2011; Wels et al., 2017 |
| 23 | K231 | L. lactis ssp. lactis | White kimchi | Siezen et al., 2011; Backus et al., 2017 |
| 24 | KF7 | L. lactis ssp. lactis | Alfalfa sprouts | Siezen et al., 2011; Backus et al., 2017 |
| 25 | KF24 | L. lactis ssp. lactis | Alfalfa sprouts | Siezen et al., 2011; Backus et al., 2017 |
| 26 | KF146 | L. lactis ssp. lactis | Alfalfa and radish sprouts | Siezen et al., 2011; Backus et al., 2017 |
| 27 | KW10 | L. lactis ssp. cremoris | Kaanga way | Siezen et al., 2011; Wels et al., 2017 |
| 28 | K337 | L. lactis ssp. lactis | White kimchi | Siezen et al., 2011; Backus et al., 2017 |
| 29 | KF67 | L. lactis ssp. lactis | Grapefruit juice | Siezen et al., 2011; Backus et al., 2017 |
| 30 | KF134 | L. lactis ssp. lactis | Alfalfa and radish sprouts | Siezen et al., 2011; Backus et al., 2017 |
| 31 | KF196 | L. lactis ssp. lactis | Japanese kaiwere shoots | Siezen et al., 2011; Backus et al., 2017 |
| 32 | KF201 | L. lactis ssp. lactis | Sliced mixed vegetables | Siezen et al., 2011; Backus et al., 2017 |
| 33 | KF282 | L. lactis ssp. lactis | Mustard and cress | Siezen et al., 2011; Backus et al., 2017 |
| 34 | LMG6897 | L. lactis ssp. cremoris | Cheese starter | Siezen et al., 2011; Wels et al., 2017 |
| 35 | NCDO763 | L. lactis ssp. cremoris | Dairy starter | Siezen et al., 2011; Wels et al., 2017 |
| 36 | SK11 | L. lactis ssp. cremoris | Dairy starter | Siezen and Renckens, 2005; Wels et al., 2017 |
| 37 | MG1363 | L. lactis ssp. cremoris | Cheese starter | Wegmann et al., 2007 |
| 38 | KF147 | L. lactis ssp. lactis | Mung bean sprouts | Siezen et al., 2010; Backus et al., 2017 |
| 39 | IL1403 | L. lactis ssp. lactis | Dairy starter | Bolotin et al., 2001 |
| 40 | MG1299 | L. lactis ssp. cremoris | Dairy starter | Wegmann et al., 2012 |
| 41 | B40 | L. lactis ssp. cremoris | Dairy starter | van Kranenburg et al., 1997; Wels et al., 2017 |
| 42 | NCDO712 | L. lactis ssp. cremoris | Dairy starter | Tarazanova et al., 2016 |
| 43 | SK110 | L. lactis ssp. cremoris | Dairy starter | Sijtsma et al., 1988; Wels et al., 2017 |
| 44 | MG1362 | L. lactis ssp. cremoris | Dairy starter | Gasson, 1983 |
| 45 | MG1063 | L. lactis ssp. cremoris | Dairy starter | Gasson, 1983 |
| 46 | MG1261 | L. lactis ssp. cremoris | Dairy starter | Gasson, 1983 |
| 47 | MG1365 | L. lactis ssp. cremoris | Dairy starter | Gasson, 1983 |
| 48 | TIFN1 | L. lactis ssp. cremoris | Dairy starter | Erkus et al., 2013 |
| 49 | TIFN2 | L. lactis ssp. lactis biovar. diacetylactis | Dairy starter | Erkus et al., 2013 |
| 50 | TIFN3 | L. lactis ssp. cremoris | Dairy starter | Erkus et al., 2013 |
| 51 | TIFN4 | L. lactis ssp. lactis biovar. diacetylactis | Dairy starter | Erkus et al., 2013 |
| 52 | TIFN5 | L. lactis ssp. cremoris | Dairy starter | Erkus et al., 2013 |
| 53 | TIFN6 | L. lactis ssp. cremoris | Dairy starter | Erkus et al., 2013 |
| 54 | TIFN7 | L. lactis ssp. cremoris | Dairy starter | Erkus et al., 2013 |
| 55 | NZ9000 | L. lactis ssp. cremoris | Dairy starter | Linares et al., 2010 |
| PLASMID USED FOR GENE OVER-EXPRESSION | ||||
| Plasmid | Host organism | Reference | ||
| pNZ8150 | L. lactis | Mierau and Kleerebezem, 2005 | ||
Cell surface charge (mV)
Cells were harvested by centrifugation at 2,676 g for 3 min at room temperature and the cell pellet was washed 2x with 1 volume of 10 mM phosphate buffer (PB; pH = 6.7) and re-suspended in the same buffer to an optical density OD600 of 1. Approximately 2 ml of this cell suspension was filled into the ZetaSizer DST1070 cuvette, which was inserted into ZetaSizer (Nano-ZS, Malvern, Malvern, UK). The electrophoretic mobility of cells was measured at 20°C and automatically re-calculated into the values of zeta potential (mV).
Cell surface hydrophobicity (CSH, %)
Cell surface hydrophobicity (CSH, %) was measured as described previously (Rosenberg et al., 1980) with the following modifications: 5 ml of cell suspension (OD600 = 1) in PB was mixed with 2 ml of either petroleum or hexane (both from Sigma-Aldrich Chemie Gmbh, Munich, Germany) in surfactant-free glass tubes with a surfactant-free stopper. Tubes were vortexed for 2 min and kept still for 15 min at room temperature to allow phase separation to occur. Subsequently, 1 mL of the aqueous phase was transferred to a spectrophotometer cuvette and optical density (OD600) of the cell suspension was measured at 600 nm. The surface hydrophobicity was calculated according to the following formula:
Cell Surface Hydrophobicity (CSH, %) = (A0−A1)/A0·100, in which A0 represents the initial OD600 of cell suspension before mixing and A1 is the OD600 of the water phase after mixing with petroleum or hexane and subsequent phase separation.
Emulsion stability (E24, %)
Emulsion Stability (E24, %) was determined as described earlier (Khopade et al., 2012; Padmapriya, 2012). Initially samples of cells in exponential and stationary growth phases were prepared in the same way as described for CSH with slight modifications. Briefly, 5 ml of cell suspension (OD600 = 1) in PB was mixed with 2 ml hydrocarbon, vortexed for 2 min and left standing for 24 h. The E24 index is given as a percentage according to: E24(%) = h1/h2*100%, in which h1 is the height of the emulsified layer and h2 is the total height of the emulsified layer and liquid column, both in mm.
Cell binding capacity to milk proteins
To 1 ml of cell suspension in either stationary or exponential growth phase prepared as described above (OD600 = 1) 1 μL Syto 9 was added (Green Fluorescent Nucleic Acid Stain, Life Technologies, Bleiswijk, The Netherlands) after which the suspension was incubated in the dark for 30 min at room temperature. The cells were washed twice with PB to remove free dye.
Preparation of proteins
Sodium caseinate and sodium para-caseinate suspensions were prepared by dissolving 10 g of protein powder in 100 ml sterile demineralized water, followed by incubation for 20 min at 30–40°C to bring the proteins into solution and adjustment of the pH to 6.7 with 0.1 M NaOH or with 0.1 M HCl. One half of the prepared sodium caseinate solution was heated to 90°C for 10 min which allows denaturation of the residual whey proteins and their interaction with caseins. For protein staining 400 μl Nile blue A (Sigma-Aldrich Chemie Gmbh) was added to 100 ml of each protein solution, mixed, and incubated for 15 min at 21°C in the dark. To remove surplus dye the protein solution was transferred to a cellulose membrane tube with a molecular weight cut-off of 14 kDa (Sigma-Aldrich D9777-100FT, 25 mm width, 60 cm in length). Membrane tubes were pre-soaked in the sterile demineralized water for 1 h at room temperature. The protein solution in the membrane tube was dialyzed against sterile PB for 24 h at 4°C in the dark. After dialysis, the protein concentration was quantified with Pierce BCA Protein Assay Kit (ThermoFisher, Bleiswijk, The Netherlands) according to the manufacturer's instructions. The Nile blue A-stained protein solution was divided over sterile Eppendorf tubes and stored at −40°C. The protein solutions were diluted to a final concentration of 1% prior to using them in the experiments.
For attachment measurements, 0.1 ml of Syto9-stained cells and 0.1 ml of Nile blue A-stained proteins were mixed with a vortex for 10–15 s and incubated for 1 min. Subsequently, this solution was analyzed in a Flow Cytometer (BD FACSaria II Cell Counter, BD BioSciences, Sparks, MD, USA). Excitation/emission wavelengths were 635/660 ± 20 nm for Nile blue A and 485/530 ± 30 nm for Syto 9, respectively. The results were analyzed using Flowing Software version 2.5.0 (http://www.flowingsoftware.com/).
Sorting procedure
Automatic (CST) and “Accudrop Drop Delay” calibration (BD BioSciences, USA) of the flow cytometer was performed with 70 μm nozzle and threshold for FSC and SSC of 1,500. A total of 10,000 events from the area of interest were sorted in 1 ml of sterile PB. Serial dilutions (10−1, 10−2, 10−3) of the sorted events were prepared in the sterile PB and 100 μl of each dilution was plated on LM17 or GM17 agar plates. The agar plates were incubated overnight at 30°C and colony counts were determined.
Fluorescence microscopy
Cells and proteins were stained as described above; 100 μl of stained cells (OD600 = 1) were mixed with the same volume of 1% protein solution. Subsequently, 1–2 μl of this mixture was placed on a microscope slide, covered with a cover slip and examined at 100-fold magnification using an Olympus BX41 microscope (Olympus Corporation, Tokyo, Japan) with excitation wavelengths of 485 ± 10 and 635 ± 10 nm and emission wavelengths of 530 ± 30 and 660 ± 20 nm, respectively. Images were acquired with a charge-coupled-device camera with identical acquisition settings for all images; exposure to excitation light was for 200 ms for the Syto 9-stained cells and for 2,000 ms for the Nile Blue A-stained proteins. Image overlays were generated using ImageJ version 1.45s (https://imagej.nih.gov/ij).
Data analysis
Gene-trait matching (GTM) was performed using PhenoLink (Bayjanov et al., 2013). Data visualization was done using R (https://cran.r-project.org/bin/windows/base/). The heatmap.plus function using Euclidian distance matrices and average hierarchical clustering and data scaling was used for the generation of heat maps.
Gene overexpression or deletion
Genes targeted for overexpression were PCR amplified using the hot-start KOD polymerase (Novagen, Madison, USA) according to the protocol of the manufacturer with primers listed in Table S1. Amplicons were purified using MSB® Spin PCRapase (Invitek, Gmbh, Berlin, Germany), digested with the restriction enzymes ScaI and XbaI (Fermentas GmbH, St. Leonn-Rot, Germany) and ligated into plasmid pNZ8150 (Table 1) digested with the same enzymes using T4 DNA ligase (Invitrogen, Breda, the Netherlands). DNA purification was carried out according to the protocols supplied by the manufacturers with Wizard®SV gel and PCR Clean-Up system (Promega, Leiden, The Netherlands). Ligations were carried out at 16°C for 16–18 h and the ligated product, which was precipitated with 3 M sodium acetate (pH 5.5) and 70% ethanol, was used to transform electrocompetent cells of L. lactis NZ9000 (Holo and Nes, 1989). After electroporation, cells were plated on GM17 agar plates containing 10 μg/ml chloramphenicol and incubated at 30°C for 3 days. Single colonies were isolated and insert DNA in the plasmid was confirmed using colony PCR with the appropriate primers (Table S1A).
Double crossover knock-outs of 4 genes were made in L. lactis MG1363 using pUC19 harboring an erythromycin (Em) resistance gene, pUC19E. Upstream and downstream flanking regions (left flank, LF; right flank, RF) of the target genes were amplified with the primer pairs described in Table S1B. Typically the left amplified flanking regions contained a sequence overlapping with the right flanking region (termed +vlag in the primer name in Table S1B) which allowed to perform a splicing by overlap extension (SOE) PCR (Horton et al., 2013) to generate amplicons which were digested and subsequently ligated into similarly digested pUC19E. The ligation mixture was used to transform L. lactis MG1363 as described previously (Holo and Nes, 1989). Strains that were the result of a single crossover event were selected after plating and incubating the transformation mixture for 2 days at 30°C on GM17 agar plates supplemented with erythromycin. These single cross-over strains were grown for at least 100 generations in GM17 broth without Em to obtain strains that were the results of a double crossover event and that became Em-sensitive. The presence of the correct, clean gene deletions was confirmed by PCR using specific primers (Table S1B).
Protein overexpression was verified with Sodium Dodecyl Sulfate—(10%) Polyacrylamide Gel Electrophoresis (SDS-PAGE) according to NuPAGE technical guide (Invitrogen, Carlsbad, CA) in cell extracts and supernatant fractions of exponentially growing cells (OD600 = 0.42–0.45) grown for 5h after addition of 10 ng/ml nisin.
Cell extracts were obtained by collecting the cell pellet from 10 ml culture (2,927 g for 5 min) and protein in supernatants was precipitated using trichloroacetic acid (TCA). The cell pellet was re-suspended in sterile demineralized water to a final OD600 of 5 and transferred to microfuge tubes containing 1 g of zirconium beads and cooled on ice for 5 min. This was followed by 3 × 30 s of bead beating with a FastPrep FP100 bead-beater (Qbiogene, Cedex, France) and 1 min off cooling on ice in between the three cycles. After this treatment the tubes were left on ice for 5 min to allow the beads to sink and the supernatant was transferred to sterile Eppendorf tubes and kept at −20°C.
TCA precipitation was conducted by adding 2.5 ml 100% trichloracetic acid to 10 ml of culture supernatant, followed by vigorous mixing and incubation for 30 min on ice. Subsequently the tubes were centrifuged at 2,927 g at 4°C for 30 min, the supernatant was discarded carefully and the pellet was dissolved in 0.5 ml cold acetone. This was followed by centrifugation at the 2,081 g at 4°C for 15 min, acetone was discarded; pellet was dried on air and r-suspended in sterile reverse osmosis water.
Results
Bacteria-protein interactions
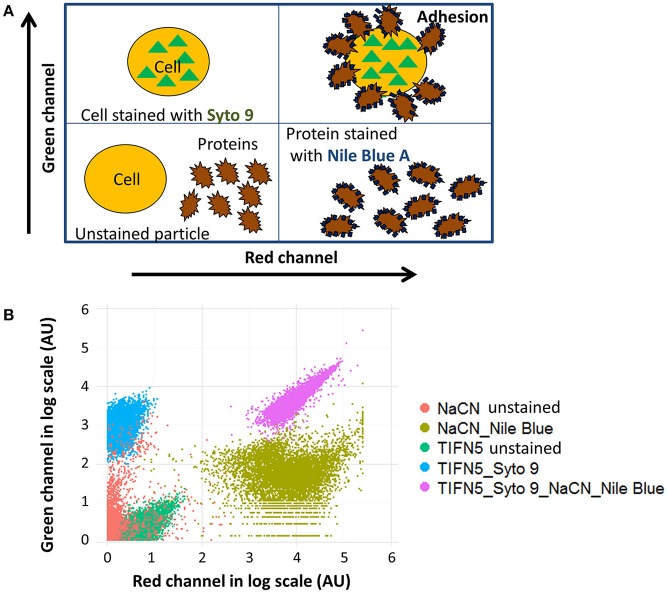
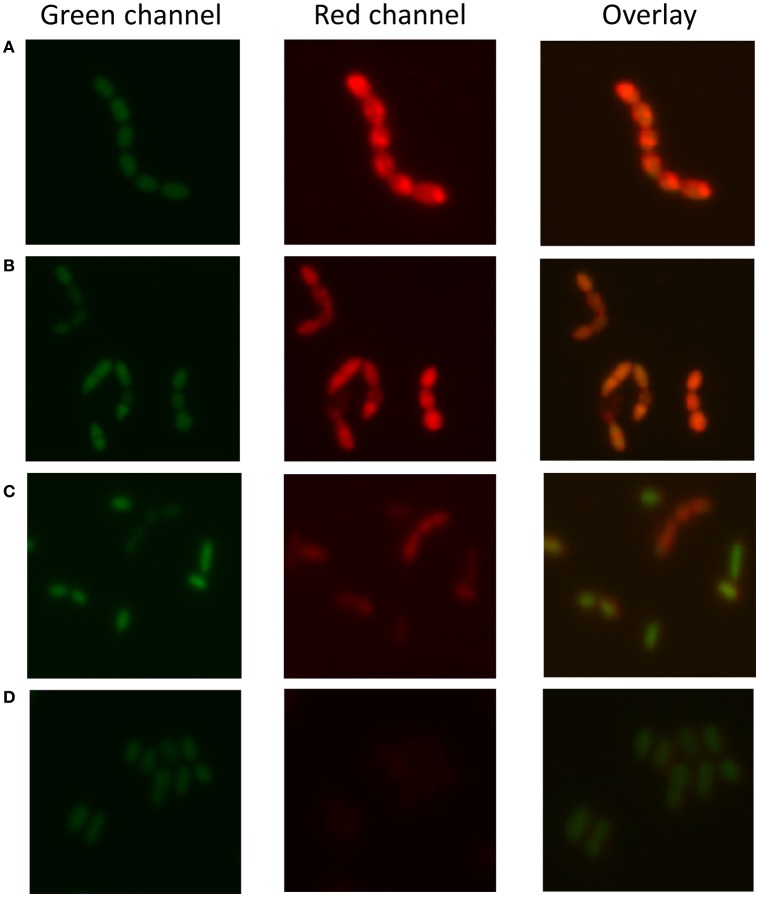
As 30 out of 55 strains used in this study were isolated from a dairy environment, we decided to examine the affinity of L. lactis to different milk proteins. For this purpose, we developed a protocol that allowed quantifying the attachment of proteins to microbial cells. Bacterial cells were stained with the fluorescent DNA stain Syto 9, while proteins were colored with the fluorescent dye Nile Blue A. The emission spectra of these two dyes show little overlap and the individually stained particles could thus be distinguished by flow cytometry (Figure 1A). Cells give a high fluorescent emission signal at 530 nm while the proteins emit the highest signal at 680 nm. When Nile Blue A-stained protein binds to a Syto 9-labeled bacterial cell, the resulting particle should show high fluorescence at both wavelengths while this should not be the case when the two do not interact. This basic concept was tested using L. lactis TIFN5 (Figure 1B). We performed two additional experiments to validate the methodology with independent techniques. In the first experiment, Syto-9 labeled cells were mixed with stained proteins. The strain used was predicted to give limited to no binding to the protein based on flow cytometry results. Using the flow cytometer we sorted 10,000 events from the population identified as protein based on the described method. The plating of these proteins showed that ~5% of the sorted events led to the formation of a colony. On the other hand, sorting of events from the population identified as cells resulted in ~95% of the cases leading to growth of a colony. These results were consistent with what was expected with an acceptable error margin of ~5%. In the second validation experiment, cells that were identified by flow cytometry as either binding or not binding to milk proteins were incubated with the protein of interest and subsequently visualized by light microscopy (Figure 2). Clear differences in Nile Blue A fluorescence intensity indicate different levels of protein binding to the cells. The level of protein binding to a cell, as detected by fluorescence microscopy at 680 nm (Figure 2), is in agreement with the protein binding levels observed with flow cytometry, which decreases from strain HP to SK11, KW10 and P7266 respectively (Figure S2, Table S2).
Figure 1.
Measuring protein attachment to bacteria using flow cytometry. (A) Schematic view of bacteria-protein interaction and their expected appearance in a flow cytometer measurement. Unstained proteins and cells are expected in the lower left quadrant. The lower right quadrant shows proteins stained with a red fluorescent dye while the upper the left quadrant shows cells stained with a green fluorescent dye. Bacteria covered with surface-bound protein should appear in the upper right quadrant while two separate clouds should be seen if the proteins do not attach to the cell surface. (B) Example of attachment of sodium caseinate (NaCN) to L. lactis TIFN5. Values on the both axes are log-transformed. Unstained proteins (red) and cells (dark green) are located in the bottom left quadrant; stained cells are shown in blue, stained protein are colored light green, and events representing proteins-attached-to-cells are located in the upper right corner (purple).
Figure 2.
Fluorescence microscopy to determine protein (sodium caseinate) attachment to L. lactis. Strains are sorted from (A–D) by decreasing attachment strength as determined by the flow cytometry-based method. (A) L. lactis HP, (B) L. lactis SK11, (C) L. lactis KW10, (D) L. lactis P7266. The green channel represents the DNA stain the red channel the stained protein bound to the cell surface and the overlay combines both signals. The results indicate decreased attachment of sodium caseinate to the strains from (A–D), respectively, which is consistent with the findings of the flow cytometric approach.
Sodium caseinate and sodium para-caseinate were selected as the proteins to be studied, as they represent the major milk proteins, i.e., the caseins. In sodium para-caseinate, the C-terminal caseinomacropeptide (CMP) of κ-casein is removed by enzymatic hydrolysis, as in cheese-making. Sodium caseinate was used in two forms: either or not pre-treated for 10 min at 90°C. The heat treatment was performed to account for possible heat-induced changes in the proteins, which is a common processing step when manufacturing fermented milk products (Hashizume and Sato, 1988; Lucey and Singh, 1997).
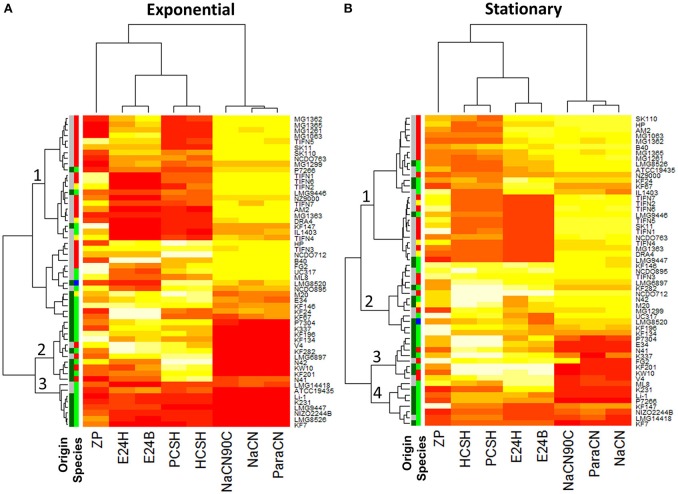
Using this approach, 55 strains of L. lactis were screened for their ability to bind the three milk proteins. The strains bound all proteins to some extent, but clear differences were observed (Table S2, Figure 3, Figures S1–S3). Significant differences in growth phase-dependent protein binding were seen for 12 of the 55 strains (Figure S4). While the ability of L. lactis to bind proteins is strain-specific the results also show that the capacity of binding milk proteins appeared to be larger than 61% with the average between 82 and 97% for strains of dairy origin. These mostly belong to the L. lactis ssp. cremoris and ssp. lactis biovar. diacetylactis. The 10 out of 23 L. lactis strains of plant origin, which belong to the ssp. lactis (indicated as dark and light green in the “origin” and “species” column in Figure 3, respectively), showed either poor (<50%) or no protein binding for both growth phases (Figure 3, Figures S1–S3, Table S2).
Figure 3.
Heat map of surface properties of 55 Lactococcus lactis strains: cell charge (ZP), hydrophobicity measured with petroleum (PCSH) or hexane (HSCH), emulsion stability measured after 24 h (E24B—measured with Petroleum or E24H—measured with Hexane), attachment to milk proteins: para-caseinate (ParaCN), sodium caseinate (NaCN), and sodium caseinate heated for 10 min at 90°C (NaCN90C). (A) Comprises results using the cells from the exponential growth phase, while for (B) cells from the stationary growth phase were taken. Low values are represented by a darker/red color while higher values are represented by a lighter/yellow color of the heat map segment (n = 3). For the charge (ZP) a darker color represents more negatively charged cells. The Origin/Species columns, respectively, indicate plant (green) or dairy (gray) origin and the species lactis (green), cremoris (red), hordniae (blue), or lactis biovar diacetylactis (yellow).
Cell surface hydrophobicity, emulsification properties, and surface charge
Cell surface charge, hydrophobicity, and emulsion stabilizing ability of the individual strains were determined on exponentially growing and stationary cells (Table S2). The results show a considerable phenotypic diversity (Figure 3, Figures S1–S3, S5–S7). The cell surface charge—measured as zeta potential—varied from −3.4 to −42.3 mV between the different strains. CSH is the measure of the extent to which cells suspended in a water phase are attracted to a hydrocarbon phase when both phases are mixed vigorously and left for phase separation to occur. We found that 25 out of the 55 strains have a CSH ranging from 0 to 20% for both growth phases. On the contrary, 9 strains showed >95% hydrophobicity for stationary growth phase, while another 5 strains showed such a high hydrophobicity in the exponential growth phase. In stationary growth phase strains NCDO712 and MG1299 displayed 60–99% hydrophobicity while the plasmid cured derivatives MG1063, MG1261, MG1362. MG1363, MG1365, and NZ9000 showed a hydrophobicity of 5–35% (Figures S5, S8, Table S2). This indicates that hydrophobicity in NCDO712 might be a plasmid-encoded trait.
Hierarchical cluster analysis revealed no correlations between the CSH of the strains as determined with different hydrocarbons and their binding of proteins (Figure 3). Strains in clade 1 show low surface hydrophobicity in the exponential growth phase, but high binding affinity to milk protein is seen. This clade consists mainly of L. lactis ssp cremoris strains of dairy origin. In contrast, strains in clade 2 poorly bind to milk proteins while they have a high CSH. Clade 3-strains show poor protein binding capacity and a low surface hydrophobicity. Clade 2 and 3 consist mainly of L. lactis ssp. lactis strains originating from the plant environment. For cells from stationary growth phase some differences are seen but the overall trends are the same in the clades 1, 3, and 4 (Figure 3B). Together, the results demonstrate that surface hydrophobicity of L. lactis cells and their protein binding capacity are independent parameters. Interestingly, the analysis of the origin of the strain (plant or dairy) and species of L. lactis revealed that the majority of L. lactis ssp. cremoris and L. lactis ssp. lactis biovar. diacetylactis, both of dairy origin, have a high capacity to bind to milk proteins. In contrast, the majority of strains originating from plan material only poorly bind milk proteins (Figure 3).
The stability of the emulsions obtained by mixing the water and hydrocarbon phases varied between the strains from no observed emulsification to total hydrocarbon phase emulsification and stability for at least 24 h. No correlation was found between emulsion stabilizing capacity of the strains and their cell hydrophobicity, between their charge and hydrophobicity, and between their charge/hydrophobicity and capacity to bind proteins.
A comparison of stationary-phase and exponential-phase cells revealed that 12 of the 55 strains show significant growth phase-related differences in their capacity to bind the milk protein samples tested, 3 strains show significant changes in hydrophobicity and 9 of the 55 strains show differences in the E24 measured (Figures S4, S8). The charge of stationary and exponentially growing cells differed significantly for 22 out of 55 strains tested (Figure S7). Taken together, the results indicate that the measured cell surface properties have some growth phase dependency, but strain specific properties are much more determinant. The binding of different milk proteins (regression coefficient r = 0.88–0.93) and cell hydrophobicity measured with different hydrocarbons (r = 0.98) are in relatively good agreement. However, little to no correlation is observed between hydrophobicity, cell surface charge and protein binding. Interestingly, there is a clear overrepresentation of strains of dairy origin in the clusters that show high binding ability of milk proteins which suggests that this trait might be beneficial during evolution in a dairy environment.
Genotype-phenotype matching
A random forest-based genotype-phenotype matching algorithm (Bayjanov et al., 2013) was employed to identify genes potentially involved in cell surface properties. The analysis was run separately for each individual phenotype measured, resulting in candidate lists for genes involved in the individual traits. This resulted initially in 201 candidate genes which were selected based on the highest importance score and gene description. From these 201 candidate genes, 18 were selected for further characterization on the basis of three parameters: (i) the importance score in the individual GTM analyses; (ii) multiple appearances in the GTM analysis for the individual phenotypes; and (iii) the predicted gene function being related to cell surface (Table S3). The following choices were made for the further characterization of these genes. If the presence of a selected candidate gene was found to be associated with a phenotype and this gene is absent in L. lactis MG1363, it was overexpressed in MG1363. Conversely, when gene absence was found to be associated with a phenotype and this gene is present in MG1363, it was knocked-out in MG1363. We selected the longest gene within the orthologous group of the 55 strains for overexpression purposes, to eliminate the risk of working with a truncated protein. Eight of the 18 genes could not be deleted and/or overexpressed, possibly because they are essential or deleterious. Ultimately, 6 of the selected candidate genes were successfully cloned downstream of the nisin-inducible promoter PnisA in pNZ8150 (Table 1) in L. lactis NZ9000, an L. lactis MG1363 derivative, while 4 of the genes were deleted from the chromosome of L. lactis MG1363 (Table 2). While we cannot exclude that the addition of nisin itself has an effect on surface properties we would like to point out that the over-expression results reported here are in relation to the nisin induced control strain which carried an empty plasmid vector. In addition no significant differences in surface properties are seen between the uninduced L. lactis MG1363 and its nisin induced derivative NZ9000 harboring pNZ8150 (Tables S2, S4, Table 1). Over-expression of proteins of the predicted sizes, and their cellular localization were examined with SDS-PAGE after induction with 10 ng/ml nisin of exponentially growing cells carrying the expression plasmids and incubation for another 5 h (Figure S9). The results showed that all 6 proteins could be successfully overexpressed, as they were detected either intracellularly or in the medium.
Table 2.
Genes that were over-expressed or deleted from the chromosome.
| Gene name | Present in strain | Locus tag protein ID | Protein size (aa/kDa) | Growth phase¥ | Gene presence (Pr)¥ Gene absence (Ab) | Predicted phenotype¥,$ | Detected phenotype*,$ | Detected in growth phase* |
|---|---|---|---|---|---|---|---|---|
| OVEREXPRESSION IN L. lactis NZ9000 | ||||||||
| Cell surface protein precursor | B40 | B40_0084 LITC01000011 KZK48299.1 | 930/102.17 | ST | Pr | ZP▾ | CSH▴ | ST |
| Ribose 5-phosphate isomerase A (rpiA) | KF147 | KF147_0667 ABX75739.2 | 243/26.95 | EX | Ab | ParaCN▴ | CSH▴ | ST |
| Internalin, putative (LPXTG motif) | KF282 | KF282_0409 - | 559/61.28 | ST | Pr | ZP▴ | CSH▴ | EX, ST |
| Hypothetical protein (yreB) | IL1403 | L128699 AAK05785.1 | 314/35.64 | EX | Pr | CSH▴ | None | – |
| Cell wall surface anchor family protein | KF147 | LLKF_0311 ADA64081 | 809/87.46 | EX EX | Pr Ab | CSH▴ E24▴ | CSH▴ ParaCN▾ NaCN▾ NaCN90C▾ | EX, ST ST ST ST |
| Endo-beta-N-acetylglucosaminidase (ypcCD) | KF147 | LLKF_1605 ADA65249 | 923/102.53 | ST ST | Ab Ab | ParaCN▴ NaCN▴ | CSH▴ | ST |
| KNOCKOUT IN L. lactis MG1363 | ||||||||
| Conjugal transfer protein (traG) | MG1363 | llmg_1383 CAL97970 | 612 | EX | Ab | ParaCN▾ | ParaCN▾ NaCN▾ NaCN90C▾ | EX EX EX |
| Cell surface protein precursor | MG1363 | llmg_1096 CAL97690 | 387 | ST | Ab | ZP▾ | No | – |
| Hypothetical protein | MG1363 | llmg_1093 CAL97687.1 | 334 | ST | Pr | NaCN▴ | NaCN▾ | EX, ST |
| Sortase SrtA (srtA) | MG1363 | llmg_1449 | 250 | – | – | – | No | – |
These columns indicate if either the presence or absence of a particular gene of cells from exponential (EX) or stationary (ST) growth phase resulted in an altered phenotype.
These columns indicate phenotypic changes that were detected in a particular growth phase in engineered strains where the indicted genes were either overexpressed or deleted.
ZP, charge (mV); ParaCN, attachment to paracaseinate (%); NaCN, attachment to sodium caseinate (%); NaCN90C, attachment to sodium caseinate heated at 90°C for 10 min (%); CSH, cell surface hydrophobicity (%); E24, emulsion stability for 24 h (%); ▴, cell surface property increases; ▾, cell surface property decreases. All phenotype changes indicated are significant with p < 0.01.
Cell surface properties of the recombinant L. lactis strains
The deletion of the gene llmg_1383 decreased the binding of exponentially growing cells of MG1363 to the 3 milk proteins tested by 53 ± 3%. The protein Llmg_1383 of L. lactis MG1363 is annotated as a conjugal transfer protein TraG, which is described to aid in the transfer of DNA during bacterial conjugation. As it is predicted to be involved in membrane pore formation (type IV secretion; Schroder et al., 2002), a role in surface alteration seems plausible.
Similar to llmg_1383, the deletion of the hypothetical gene llmg_1093 also decreased the binding of L. lactis MG1363 to the three milk proteins examined by 60 ± 27% for cells in the exponential growth phase and by 42 ± 37% for cells in the stationary growth phase. Protein Llmg_1093 is a putative secreted protein but its actual function is not known.
As several of the genes studied here harbor sortase dependent LPXTG signals we also deleted the sortase A gene (llmg_1449) from the chromosome of strain L. lactis MG1363. This deletion did not change the cell surface properties of L. lactis MG1363 (Table S4), indicating that SrtA might not be involved directly in cell surface properties. We also tried to obtain a knock-out mutant of the srtC gene but were not able to obtain it after two attempts, indicating a possibly essential role or SrtC for growth.
The cell surface protein precursor B40_0084 is a putative mucus binding protein as it has 4 mucus binding domains (MucBP) (pfam06458) and a LPXTG-motif cell wall anchor domain (TIGR01167). GTM predicted that the presence of this protein leads to a more negative cell surface charge. This prediction was not confirmed, but changes were observed for hydrophobicity. For example, for stationary phase cells where the control strain shows a CSH of 8.8% (±5%), the overexpression of B40_0084 resulted in a CSH of 89% (±7%) (Table S4). Interestingly, while SDS PAGE analysis (Figure S9) verified the overexpression of B40_0084 in exponentially growing cells, this overexpression did not affect the CSH of the cells in this growth phase. Mucus binding proteins are described to be involved in the binding of carbohydrates such as mannose (Pretzer et al., 2005) and they are speculated to be important for probiotic function (Kleerebezem et al., 2010). The effect of the overexpression of B40_0084 only leads to hydrophobicity changes in stationary phase cells, indicating that other surface decoration(s) dominates surface properties in a growth phase-dependent manner.
Another gene identified by gene-trait matching is a putative internalin containing four MucBP domains (pfam06458) and a surface-anchoring domain (COG4932). Internalins were originally described in Listeria monocytogenes as surface proteins that are involved in adhesion to mammalian epithelial cells (Lecuit et al., 1997). For example, protein internalin A (InlA) mediates bacterial adhesion and invasion of epithelial cells in the human intestine through specific interaction with its host cell receptor E-cadherin (Schubert et al., 2002). Here, we overexpressed the ortholog from strain KF282 (KF282_0409). While gene-trait matching associated the presence this gene with lower surface charge, its overexpression in MG1363 increased the CSH, from 8.8% (±5) in the control strain to 49.3% (±21.8). Furthermore, overexpression of a cell wall surface anchor family protein (LLKF_0311) led to the decreased attachment of cells to milk proteins from 98% (±0.7) to about 31% (±47%) in the stationary growth phase and to the increase in CSH from 9 ± 5 to 78 ± 9% in the stationary and exponential growth phases (Table S4) while initially the presence of LLKF_0311 was predicted to effect cells surface hydrophobicity and emulsion stability in exponential phase of growth. (Table S4).
The endo-beta-N-acetylglucosaminidase ypcCD (LLKF_1605) was found to be associated to the binding of cells to milk proteins. Overexpression of this protein did not lead to an alteration in attachment of cells in stationary growth phase to milk proteins, but led to an increase in surface hydrophobicity from about 8.8 ± 5 to 23.7 ± 16.1%.
Overall, the phenotype predicted by gene-trait matching could be confirmed experimentally for 3 out of 10 engineered strains, and for 2 out of 3 strains additional altered surface properties were detected. Overexpression of the selected candidate genes did not influence the protein binding properties. A total of 4 strains showed altered surface properties but not the predicted ones whereas for 3 strains no changes were observed. While not all predictions were correct, the approach did allow identifying targets that are of importance for lactococcal surface properties.
Discussion
This study describes bacterial surface properties such as cell surface charge, hydrophobicity and the attachment to milk proteins for 55 L. lactis strains isolated from either plant material or the dairy environment. A flow cytometry based method for the characterization of protein binding allowed to demonstrate the existence of a large biodiversity in cell attachment to milk proteins. We show that the capacity of cells to bind the milk proteins is growth phase-dependent for some of the strains tested. Importantly, this methodology is not restricted to the use of milk proteins and we expect it to be applicable for the characterization of cell attachment to other proteins, and for other bacterial species. In combination with cell sorting the method may prove useful in enabling the selection of cells with desired surface characteristics.
In contrast to a previous executed GTM study with L. lactis strains, which was done based on comparative genome hybridization data (Bayjanov et al., 2013), we were able to use either draft or complete genome sequences, which should increase the predictive power of the approach. The characterization of selected target genes, by their overexpression and/or deletion from the chromosome, resulted in the identification of 7 genes that are involved in cell surface properties.
The observed biodiversity of cells obtained from different growth phases might be explained by differences in the molecular composition of the cell wall. For example, peptidoglycan modification during exponential growth in L. lactis results only in partial (75%) amidation of the alpha-carboxyl group of the D-Asp cross-bridge to the PG precursor (Courtin et al., 2006; Veiga et al., 2009). In contrast, the amidation of amino acids during peptidoglycan modification for L. casei is almost complete (near 100%) during both growth phases (Chapot-Chartier and Kulakauskas, 2014). However, peptidoglycan is not the major component exposed at the bacterial surface, but it is rather dominated by polysaccharides, teichoic acids, and proteins. The charge of the cell surface is mainly determined by net charge of molecular composition of cell wall. For example, a negative cell surface charge can be partially determined by carboxyl and phosphate groups of LTA and TA, while positive charges partially derive from D-alanine molecules that are esterified to TA and LTA (Delcour et al., 1999).
Overall we see poor or no correlations between properties such as emulsion stabilizing ability and cell hydrophobicity, between charge and hydrophobicity, or between charge/hydrophobicity and the attachment to proteins. This might be caused by the amphiphilic surface properties of bacteria (Van Oss, 2003).
While there is ample literature describing the cell surface and cell wall composition of bacteria in general, available information on key molecules determining cell surface properties of lactic acid bacteria are limited. For instance, the overexpression of the surface anchored protein CwaA from Lactobacillus plantarum NL42 in L. lactis NZ9000 led to cell auto-aggregation, increased hydrophobicity and attachment of the CwaA-producing L. lactis cells to human epithelial HT-29 cells (Zhang et al., 2015). Other studies describe the autolysins AcmA and AcmD, which are involved in cell chaining (Visweswaran et al., 2013), or the expression of pili on the surface of L. lactis. Pili can be plasmid- as well as genome-encoded and they have been shown to cause auto-aggregation (Oxaran et al., 2012; Tarazanova et al., 2016) and to increase attachment to epithelial cells (Meyrand et al., 2013; Zhang et al., 2015). The L. lactis cell wall proteinase PrtP was also shown to be involved in cell surface properties and adhesion to solid surfaces (Habimana et al., 2007).
We successfully verified the influence of 3 proteins on predictions based on the performed gene-trait matching. However, the alteration of protein expression in some of our engineered strains did result in an effect on the cell surface other than the predicted one. The underlying molecular details of these discrepancies are not clear, but we speculate that affected molecules are in competition for space on the cell surface. Alteration of the expression level of one molecule would indirectly affect the overall surface composition, which could result in unexpected phenotypic outcomes. Such a speculation is in line with recent theory on trade-offs that can be determined by physical-chemical constraints such as membrane space (Bachmann et al., 2016, 2017). While most of the genes identified here could be linked to the cell surface we could not find such a link for some identified proteins based on sequence analysis. The over-expression of a ribose-5-phosphate isomerase, an enzyme involved in the pentose phosphate pathway, altered CSH. A direct role of this enzyme in cell surface properties seems unlikely, but a study in L. plantarum suggests that ribose acts as a precursor for alternative cell wall teichoic acids (Bron et al., 2012) and it is therefore conceivable that a change in ribose availability could lead to altered cell wall properties. The identification of such proteins, that cannot be linked to cell surface properties with e.g. sequence based motif analysis, points out the added predictive value of genotype/phenotype matching.
An interesting observation is the fact that strains isolated from a dairy environment show much stronger binding of milk proteins as compared to plant isolates. Literature holds many examples for the role of surface alterations to improve the fitness of an organism in a particular environment. In pathogenic bacteria, for instance, peptidoglycan modifications allow escaping the host's immune system (Foster, 2005) and in soils the capacity to form biofilms is a key factor for microbial fitness (Nazir et al., 2010). The proposed evolutionary transition of L. lactis from the plant to the dairy environment is described to be accompanied by the loss of the ability to synthesize several amino acids or to catabolize typical plant-derived sugars. The occurrence of amino acid auxotrophies in dairy isolates is compensated by improved utilization of milk proteins through e.g. extracellular proteases, dedicated (oligo)peptide transport systems and intracellular peptidases (van Hylckama Vlieg et al., 2006; Bachmann et al., 2012). The fact that binding to milk proteins was selected for in a dairy environment suggests a selective advantage, which seems plausible seen the growth dependency of dairy strains of L. lactis on extracellular amino acids. It will be interesting to see if the alteration of surface properties of L. lactis also impacts on the functionality of starter cultures in pure and mixed-culture fermentations.
Author contributions
MT, TH, JK, HB conceived the research. MT, MB, SvS, PJ carried out the experiments. MT, MW, HB analyzed the data. MT, JK, HB wrote the manuscript. All authors commented on the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Roland Siezen for discussions of the gene-trait matching results. We are grateful to Symeon Karakatsanis and Turki Hazzazi for assistance with measurements of bacterial surface properties.
Footnotes
Funding. The project is funded by TI Food and Nutrition, a public-private partnership on precompetitive research in food and nutrition. The public partners are responsible for the study design, data collection and analysis, decision to publish, and preparation of the manuscript. The private partners have contributed to the project through regular discussion.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01691/full#supplementary-material
References
- Bachmann H., Bruggeman F. J., Molenaar D., Branco dos Santos F., Teusink B. (2016). Public goods and metabolic strategies. Curr. Opin. Microbiol. 31, 109–115. 10.1016/j.mib.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Bachmann H., Molenaar D., Branco dos Santos F., Teusink B. (2017). Experimental evolution and the adjustment of metabolic strategies in lactic acid bacteria. FEMS Microbiol. Rev. 41(Suppl. 1), S201–S219. 10.1093/femsre/fux024 [DOI] [PubMed] [Google Scholar]
- Bachmann H., Starrenburg M. J. C., Molenaar D., Kleerebezem M., van Hylckama Vlieg J. E. (2012). Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22, 115–124. 10.1101/gr.121285.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus L., Wels M., Boekhorst J., Dijkstra A. R., Beerthuyzen M., Kelly W. J., et al. (2017). Draft genome sequences of 24 Lactococcus lactis strains. Genome Announc. 5:e01737-16. 10.1128/genomeA.01737-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E. A., Belaich J.-P., Shoham Y., Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554. 10.1146/annurev.micro.57.030502.091022 [DOI] [PubMed] [Google Scholar]
- Bayjanov J. R., Molenaar D., Tzeneva V., Siezen R. J., van Hijum S. A. (2012). PhenoLink - a web-tool for linking phenotype to ~omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics 13:170. 10.1186/1471-2164-13-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayjanov J. R., Starrenburg M. J. C., van der Sijde M. R., Siezen R. J., van Hijum S. A. F. T. (2013). Genotype-phenotype matching analysis of 38 Lactococcus lactis strains using random forest methods. BMC Microbiol. 13:68. 10.1186/1471-2180-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon-Fontaine M.-N., Rault J., van Oss C. J. (1996). Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B Biointerfaces 7, 47–53. 10.1016/0927-7765(96)01272-6 [DOI] [Google Scholar]
- Boks N. P., Norde W., van der Mei H. C., Busscher H. J. (2008). Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology 154, 3122–3133. 10.1099/mic.0.2008/018622-0 [DOI] [PubMed] [Google Scholar]
- Bolotin A., Wincker P., Mauger S. (2001). The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11, 731–753. 10.1101/gr.GR-1697R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonaert C. J. P., Rouxhet P. G. (2000). Surface of lactic acid bacteria: relationships between chemical composition and physicochemical properties. Appl. Environ. Microbiol. 66:2548. 10.1128/AEM.66.6.2548-2554.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron P., Baarlen P., van Kleerebezem M. (2012). Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 11, 66–78. 10.1038/nrmicro2690 [DOI] [PubMed] [Google Scholar]
- Burgain J., Scher J., Francius G., Borges F., Corgneau M., Revol-Junelles A. M., et al. (2014). Lactic acid bacteria in dairy food: surface characterization and interactions with food matrix components. Adv. Colloid Interface Sci. 213, 21–35. 10.1016/j.cis.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Castelain M., Duviau M.-P., Canette A., Schmitz P., Loubière P., Cocaign-Bousquet M., et al. (2016). The nanomechanical properties of Lactococcus lactis pili are conditioned by the polymerized backbone pilin. PLoS ONE 11:e0152053. 10.1371/journal.pone.0152053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier M.-P., Kulakauskas S. (2014). Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact. 13:S9. 10.1186/1475-2859-13-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier M.-P., Vinogradov E., Sadovskaya I., Andre G., Mistou M.-Y., Trieu-Cuot P., et al. (2010). Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285, 10464–10471. 10.1074/jbc.M109.082958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtin P., Miranda G., Guillot A., Wessner F., Mezange C., Domakova E., et al. (2006). Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188, 5293–5298. 10.1128/JB.00285-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decho A. (2000). Microbial biofilms in intertidal systems: an overview. Cont. Shelf Res. 20, 1257–1273. 10.1016/S0278-4343(00)00022-4 [DOI] [Google Scholar]
- Delcour J., Ferain T., Deghorain M., Palumbo E., Hols P. (1999). The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Van Leeuwenhoek 76, 159–184. 10.1023/A:1002089722581 [DOI] [PubMed] [Google Scholar]
- Erkus O., de Jager V. C. L., Spus M., van Alen-Boerrigter I. J., van Rijswijck I. M. H., Hazelwood L., et al. (2013). Multifactorial diversity sustains microbial community stability. ISME J. 7, 2126–2136. 10.1038/ismej.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozmand H., Rousseau D. (2016). Microbial cells as colloidal particles: pickering oil-in-water emulsions stabilized by bacteria and yeast. Food Res. Int. 81, 66–73. 10.1016/j.foodres.2015.10.018 [DOI] [Google Scholar]
- Foster T. J. (2005). Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958. 10.1038/nrmicro1289 [DOI] [PubMed] [Google Scholar]
- Gasson M. J. (1983). Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudev S., Spasova I., Nicolova M., Georgiev P. (2010). In situ bioremediation of contaminated soils in uranium deposits. Hydrometallurgy 104, 518–523. 10.1016/j.hydromet.2010.02.027 [DOI] [Google Scholar]
- Habimana O., Le Goff C., Juillard V., Bellon-Fontaine N., Buist G., Kulakauskas S., et al. (2007). Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 7:36. 10.1186/1471-2180-7-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume K., Sato T. (1988). Gel-forming characteristics of milk proteins. 1. Effect of heat treatment. J. Dairy Sci. 71, 1439–1446. 10.3168/jds.S0022-0302(88)79706-4 [DOI] [Google Scholar]
- Holo H., Nes I. F. (1989). High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Cai Z., Ho S. N., Pease L. R. (2013). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 54, 528–535. 10.2144/000114017 [DOI] [PubMed] [Google Scholar]
- Jacobs A., Lafolie F., Herry J. M., Debroux M. (2007). Kinetic adhesion of bacterial cells to sand: cell surface properties and adhesion rate. Colloids Surf. B Biointerfaces 59, 35–45. 10.1016/j.colsurfb.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Khopade A., Biao R., Liu X., Mahadik K., Zhang L., Kokare C. (2012). Production and stability studies of the biosurfactant isolated from marine Nocardiopsis sp. B4. Desalination 285, 198–204. 10.1016/j.desal.2011.10.002 [DOI] [Google Scholar]
- Kleerebezem M., Vaughan E. E. (2009). Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 63, 269–290. 10.1146/annurev.micro.091208.073341 [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Hols P., Bernard E., Rolain T., Zhou M., Siezen R. J., et al. (2010). The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 34, 199–230. 10.1111/j.1574-6976.2009.00208.x [DOI] [PubMed] [Google Scholar]
- Kumar C. G., Anand S. (1998). Significance of microbial biofilms in food industry: a review. Int. J. Food Microbiol. 42, 9–27. 10.1016/S0168-1605(98)00060-9 [DOI] [PubMed] [Google Scholar]
- Lecuit M., Ohayon H., Braun L., Mengaud J., Cossart P. (1997). Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65, 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy F., De Vuyst L. (2004). Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15, 67–78. 10.1016/j.tifs.2003.09.004 [DOI] [Google Scholar]
- Linares D. M., Kok J., Poolman B. (2010). Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192, 5806–5812. 10.1128/JB.00533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey J. A., Singh H. (1997). Formation and physical properties of acid milk gels: a review. Food Res. Int. 30, 529–542. 10.1016/S0963-9969(98)00015-5 [DOI] [Google Scholar]
- Ly M. H., Aguedo M., Goudot S., Le M. L., Cayot P., Teixeira J. A., et al. (2008). Interactions between bacterial surfaces and milk proteins, impact on food emulsions stability. Food Hydrocoll. 22, 742–751. 10.1016/j.foodhyd.2007.03.001 [DOI] [Google Scholar]
- Ly M. H., Vo N. H., Le T. M., Belin J.-M., Waché Y. (2006). Diversity of the surface properties of Lactococci and consequences on adhesion to food components. Colloids Surf. B Biointerfaces 52, 149–153. 10.1016/j.colsurfb.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Meyrand M., Guillot A., Goin M., Furlan S., Armalyte J., Kulakauskas S., et al. (2013). Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol. Cell. Proteomics 12, 3935–3947. 10.1074/mcp.M113.029066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau I., Kleerebezem M. (2005). 10 Years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717. 10.1007/s00253-005-0107-6 [DOI] [PubMed] [Google Scholar]
- Nazir R., Warmink J. A., Boersma H., van Elsas J. D. (2010). Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 71, 169–185. 10.1111/j.1574-6941.2009.00807.x [DOI] [PubMed] [Google Scholar]
- Neu T. R., Marshall K. C. (1990). Bacterial polymers: physicochemical aspects of their interactions at interfaces. J. Biomater. Appl. 5, 107–133. 10.1177/088532829000500203 [DOI] [PubMed] [Google Scholar]
- Newman J. A., Rodrigues C., Lewis R. J. (2013). Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis. J. Biol. Chem. 288, 10766–10778. 10.1074/jbc.M113.455592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxaran V., Ledue-Clier F., Dieye Y., Herry J.-M., Péchoux C., Meylheuc T., et al. (2012). Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLoS ONE 7:e50989. 10.1371/journal.pone.0050989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmapriya B. (2012). Isolation and screening of biosurfactants produced by Pseudomonas aeruginosa from oil spilled soils. Int. J. Pharm. Biol. 3, 321–325. [Google Scholar]
- Pelletier C., Bouley C., Cayuela C., Bouttier S., Bourlioux P., Bellon-Fontaine M. N. (1997). Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl. Environ. Microbiol. 63, 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretzer G., Snel J., Molenaar D., Wiersma A., Bron P. A., Lambert J., et al. (2005). Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187, 6128–6136. 10.1128/JB.187.17.6128-6136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty A. M., Schwesinger W. H., Gunn J. S. (2002). Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70, 2640–2649. 10.1128/IAI.70.5.2640-2649.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Gutnick D., Rosenberg E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9, 29–33. 10.1111/j.1574-6968.1980.tb05599.x [DOI] [Google Scholar]
- Ruas-Madiedo P., Hugenholtz J., Zoon P. (2002). An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. Int. Dairy J. 12, 163–171. 10.1016/S0958-6946(01)00160-1 [DOI] [Google Scholar]
- Schär-Zammaretti P., Ubbink J. (2003). The cell wall of lactic acid bacteria: surface constituents and macromolecular conformations. Biophys. J. 85, 4076–4092. 10.1016/S0006-3495(03)74820-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder G., Krause S., Zechner E. L., Traxler B., Yeo H.-J., Lurz R., et al. (2002). TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184, 2767–2779. 10.1128/JB.184.10.2767-2779.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert W. D., Urbanke C., Ziehm T., Beier V., Machner M. P., Domann E., et al. (2002). Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111, 825–836. 10.1016/S0092-8674(02)01136-4 [DOI] [PubMed] [Google Scholar]
- Sieuwerts S., de Bok F. A., Hugenholtz J., van Hylckama Vlieg J. E. T. (2008). Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74, 4997–5007. 10.1128/AEM.00113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., van Hylckama Vlieg J. E. (2011). Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 10:S3. 10.1186/1475-2859-10-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., Bayjanov J. R., Felis G. E., van der Sijde M. R., Starrenburg M., Molenaar D., et al. (2011). Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb. Biotechnol. 4, 383–402. 10.1111/j.1751-7915.2011.00247.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., Bayjanov J., Renckens B., Wels M., Van Hijum S. A., Molenaar D., et al. (2010). Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J. Bacteriol. 192, 2649–2650. 10.1128/JB.00276-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., Starrenburg M. J. C., Boekhorst J., Renckens B., Molenaar D., Van Hylckama Vlieg J. E. T. (2008). Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74, 424–436. 10.1128/AEM.01850-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R., Renckens B. (2005). Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl. Environ. Microbiol. 71, 8370–8383. 10.1128/AEM.71.12.8371-8382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijtsma L., Sterkenburg A., Wouters J. T. (1988). Properties of the cell walls of Lactococcus lactis subsp. cremoris SK110 and SK112 and their relation to bacteriophage resistance. Appl. Environ. Microbiol. 54, 2808–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. 10.1099/00221287-147-1-3 [DOI] [PubMed] [Google Scholar]
- Tarazanova M., Beerthuyzen M., Siezen R., Fernandez-Gutierrez M. M., de Jong A., van der Meulen S., et al. (2016). Plasmid complement of Lactococcus lactis NCDO712 reveals a novel pilus gene cluster. PLoS ONE 11:e0167970. 10.1371/journal.pone.0167970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006). Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519. 10.1038/nrmicro1443 [DOI] [PubMed] [Google Scholar]
- van der Mei H. C., van de Belt-Gritter B., Pouwels P. H., Martinez B., Busscher H. J. (2003). Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf. B Biointerfaces 28, 127–134. 10.1016/S0927-7765(02)00144-3 [DOI] [Google Scholar]
- Van Houdt R., Michiels C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109, 1117–1131. 10.1111/j.1365-2672.2010.04756.x [DOI] [PubMed] [Google Scholar]
- van Hylckama Vlieg J. E., Rademaker J. L., Bachmann H., Molenaar D., Kelly W. J., Siezen R. J. (2006). Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17, 183–190. 10.1016/j.copbio.2006.02.007 [DOI] [PubMed] [Google Scholar]
- van Kranenburg R., Marugg J. D., van Swam I. I., Willem N. J., de Vos W. M. (1997). Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24, 387–397. 10.1046/j.1365-2958.1997.3521720.x [DOI] [PubMed] [Google Scholar]
- Van Oss C. J. (2003). Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J. Mol. Recogn. 16, 177–190. 10.1002/jmr.618 [DOI] [PubMed] [Google Scholar]
- Veiga P., Erkelenz M., Bernard E., Courtin P., Kulakauskas S., Chapot-Chartier M.-P. (2009). Identification of the asparagine synthase responsible for D-Asp amidation in the Lactococcus lactis peptidoglycan interpeptide crossbridge. J. Bacteriol. 191, 3752–3757. 10.1128/JB.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran G. R. R., Steen A., Leenhouts K., Szeliga M., Ruban B., Hesseling-Meinders A., et al. (2013). AcmD, a homolog of the major autolysin AcmA of Lactococcus lactis, binds to the cell wall and contributes to cell separation and autolysis. PLoS ONE 8:e72167. 10.1371/journal.pone.0072167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U., O'Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C., et al. (2007). Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189, 3256–3270. 10.1128/JB.01768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U., Overweg K., Jeanson S., Gasson M., Shearman C. (2012). Molecular characterization and structural instability of the industrially important composite metabolic plasmid pLP712. Microbiology 158, 2936–2945. 10.1099/mic.0.062554-0 [DOI] [PubMed] [Google Scholar]
- Wels M., Backus L., Boekhorst J., Dijkstra A., Beerthuyzen M., Siezen R. J., et al. (2017). Draft genome Sequences of 11 Lactococcus lactis subsp. cremoris strains. Genome Announc. 5, e01739-16. 10.1128/genomeA.01739-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicken A. J., Ayres A., Campbell L. K., Knox K. W. (1983). Effect of growth conditions on production of rhamnose-containing cell wall and capsular polysaccharides by strains of Lactobacillus casei subsp. rhamnosus. J. Bacteriol. 153, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zuo F., Yu R., Zeng Z., Ma H., Chen S. (2015). Comparative genome-based identification of a cell wall- anchored protein from Lactobacillus plantarum increases adhesion of Lactococcus lactis to human epithelial cells. Sci Rep. 5:14109. 10.1038/srep14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.