Abstract
Sinorhizobium fredii is a fast-growing rhizobial species that can establish a nitrogen-fixing symbiosis with a wide range of legume species including soybeans (Glycine max). In soybeans, this interaction shows a high level of specificity such that particular S. fredii strains nodulate only a limited set of plant genotypes. Here we report the identification of a dominant gene in soybeans that restricts nodulation with S. fredii USDA193. Genetic mapping in an F2 population revealed co-segregation of the underlying locus with the previously cloned Rfg1 gene. The Rfg1 allele encodes a member of the Toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat class of plant resistance proteins that restricts nodulation by S. fredii strains USDA257 and USDA205, and an allelic variant of this gene also restricts nodulation by Bradyrhizobium japonicum USDA122. By means of complementation tests and CRISPR/Cas9-mediated gene knockouts, we demonstrate that the Rfg1 allele also is responsible for resistance to nodulation by S. fredii USDA193. Therefore, the Rfg1 allele likely provides broad-spectrum resistance to nodulation by many S. fredii and B. japonicum strains in soybeans.
Keywords: soybean, nodulation, nitrogen fixation, rhizobial symbiosis, symbiosis specificity
Introduction
The leguminous plants are able to establish a symbiotic relationship with nitrogen-fixing soil bacteria called rhizobia. The symbiosis is featured by the formation of root nodules where the bacteria in nodule cells can convert atmospheric nitrogen into ammonia and make it available to the plant. This symbiotic partnership has important implications in sustainable agriculture because it reduces the need for nitrogen-based fertilizers.
The legume-rhizobial symbiosis starts with host perception of bacterially derived lipo-chitooligosaccharides known as nodulation (Nod) factors (Oldroyd et al., 2011). Recognition of Nod factors secreted by compatible bacteria induces cell divisions in the root cortex leading to the formation of nodule primordia, and at the same time initiates the infection process that delivers the bacteria into these primordia. Infection of most legumes such as soybeans (Glycine max) and alfalfa (Medicago sativa) is through root hairs. Bacteria are first entrapped by curled root hairs and multiply to form micro-colonies referred to as infection foci. From these foci, plant-made tubular-like structures, called infection threads, start to develop through local cell wall hydrolysis and invagination of the plant plasma membrane. The infection threads that are colonized by dividing bacteria proceed through the epidermal cell layer into the inner cortex where the nodule primordium has formed. Bacteria are then internalized in an endocytosis-like process and surrounded by a host membrane, where they differentiate into nitrogen-fixing bacteroides and are confined in organelle-like structures called symbiosomes (Jones et al., 2007; Oldroyd et al., 2011).
While some bacteria can nodulate a wide range of hosts, most bacteria have strict host selectivity. As such, particular rhizobial species or strains nodulate only a narrow group of legume species or genotypes (Broughton et al., 2000; Perret et al., 2000; Wang et al., 2012). Understanding the genetic and molecular basis of this specificity is important for developing strategies to improve the agronomic potential of the root nodule symbiosis in agriculture. It has been reported that plant domestication and breeding processes have led to the reduced ability of the modern cultivars to interact with indigenous soil strains as compared to their wild progenitors (Mutch and Young, 2004; Kiers et al., 2007; Kim et al., 2014). In this case, development of cultivars that are promiscuous with indigenous strains would be beneficial. On the other hand, certain indigenous soil bacteria are highly competitive for nodulation with host legumes but with low nitrogen fixation efficiency. Under this latter scenario, it is desirable to grow plants that can restrict nodulation with low-efficient indigenous strains but nodulate preferentially with the effective inoculant strains (Keyser and Li, 1992; Devine and Kuykendall, 1996).
Establishment of a root nodule symbiosis requires mutual recognition of multiple molecular signals between the symbiotic partners (Jones et al., 2007; Deakin and Broughton, 2009; Oldroyd et al., 2011). Therefore, symbiotic specificity can be regulated by multiple mechanisms at different stages of the nodule development (Perret et al., 2000; Wang et al., 2012, 2017; Liu et al., 2014; Yang et al., 2017). In most legumes, bacterial infection and nodule formation is initiated by host recognition of rhizobial Nod factors (Lerouge et al., 1990; Geurts et al., 1997; Limpens et al., 2003; Radutoiu et al., 2003). Nod factors produced by different bacteria carry specific chemical decorations on the chitin backbone, and this structural diversity has been thought to be a major determinant of nodulation specificity in the legume–rhizobal interaction, particularly at the species level (Bras et al., 2000; Radutoiu et al., 2007). Similar to pathogenic bacteria, symbiotic rhizobia also use conserved microbe-associated molecular patterns (MAMPs) or secreted effectors to facilitate their interaction with the host (D’Haeze and Holsters, 2004; Fauvart and Michiels, 2008; Deakin and Broughton, 2009; Soto et al., 2009; Downie, 2010; Wang et al., 2012; Kawaharada et al., 2015). Accordingly, effector- or MAMP-triggered plant immunity mediated by host receptors also plays an important role in regulating host range of rhizobia (Yang et al., 2010; Wang et al., 2012; Faruque et al., 2015; Kawaharada et al., 2015; Tang et al., 2016).
We have cloned several dominant genes in soybeans (e.g., Rj2, Rfg1, and Rj4) that restrict nodulation with specific rhizobial strains (Yang et al., 2010; Tang et al., 2016). In these cases, symbiosis incompatibility is controlled in a similar manner as ‘gene-for-gene’ resistance against plant pathogens (Sadowsky et al., 1990; Devine and Kuykendall, 1996). Rj2 and Rfg1 are allelic genes, each encoding a typical Toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) resistance protein that confers resistance to nodulation by specific strains of Bradyrhizobium japonicum and Sinorhizobium fredii, respectively (Yang et al., 2010), while Rj4 encodes a thaumatin-like pathogenesis-related protein that restricts nodulation by specific strains of B. elkanii (Tang et al., 2016). Moreover, the function of these nodulation-restrictive genes is dependent on the bacterial type III secretion system (Krishnan et al., 2003; Okazaki et al., 2009; Yang et al., 2010; Tsukui et al., 2013; Tsurumaru et al., 2015; Tang et al., 2016; Yasuda et al., 2016). These studies revealed an important role of effector-triggered plant immunity in the regulation of nodulation specificity in soybeans (Wang et al., 2012).
Here we describe the study of symbiotic incompatibility of soybean with S. fredii USDA193. A previous report suggested that restriction of nodulation by this strain is associated with the Rj2 and/or Rj3 loci (Nakano et al., 1997). Our study revealed a single dominant gene responsible for this incompatibility. Genetic mapping in an F2 population showed co-segregation of the underlying locus with the previously cloned Rfg1 gene that confers resistance to nodulation by S. fredii strains USDA257 and USDA205 (Yang et al., 2010). Through complementation tests and CRISPR/Cas9-mediated gene disruption, we demonstrate that the Rfg1 allele also is responsible for restricting nodulation by S. fredii USDA193. Our study suggests that the Rfg1 locus is involved in the determination of nodulation specificity with multiple S. fredii and B. japonicum strains in soybeans.
Materials and Methods
Plant Material, Nodulation Assay and Genotyping
The F2 mapping population was derived by crossing the North American cultivar ‘Williams 82’ with the cultivar ‘Peking,’ an introduction from China. Peking formed nitrogen-fixing nodules when inoculated with S. fredii USDA193 (Nod+) while Williams 82 restricted nodulation by this strain (Nod-) (Keyser et al., 1982). S. fredii USDA193, originally isolated from China (Keyser et al., 1982), was obtained from the National Rhizobium Germplasm Collection (USDA-ARS, Beltsville, MD, United States). The strain was cultured on YEM agar plates (yeast extract, 1.0 g/L; mannitol, 10.0 g/L; dipotassium phosphate, 0.5 g/L; magnesium sulfate, 0.2 g/L; sodium chloride, 0.1 g/L; calcium carbonate, 1.0 g/L; agar, 15.0 g/L) in the dark at 28°C for 4–5 days, and the bacterial paste was then collected and diluted in sterile water to OD600 of 0.1. For nodulation assay, each 1-week-old seedling was flood-inoculated with 10 mL of the bacterial suspension. Plants were grown in sterilized 50/50 Perlite-Turface mix in a growth chamber programmed for 16 h light at 26°C and 8 h dark at 23°C. Nodulation phenotype was assayed 4 weeks post inoculation. Genotyping was conducted by using a CAPS (cleaved amplified polymorphic sequences) marker developed based on a SNP (single nucleotide polymorphism) between the Rfg1 (Williams 82) and rfg1 (Peking) alleles. The primer pair used was 5′TGAGAGTACTGGAATGGTGGAG3′ and 5′TTGCTGATCGAACCACTCTG3′, and the restriction enzyme used was HpaI.
Complementation Tests
The Williams 82 allele of Rfg1 was used for complementation tests. Genomic DNA of the Rfg1 allele was derived from the BAC clone Gm_WBa0019D20 of Williams 82 (Marek and Shoemaker, 1996) by digestion with PstI and BmgBI. The released 10.9-kb fragment included the 4.9-kb coding region, the 4.0 kb upstream of the start codon containing the promoter and 5′ untranslated region, and the 2.0 kb downstream of the stop codon encompassing the 3′ untranslated region. The DNA fragment was cloned into the binary vector pCAMBIA1305.1 through blunt end cloning. The binary vector was transferred into the Peking genetic background through hairy root transformation as described below. The transgenic roots were identified by GUS-staining because of the presence of a GUS expression cassette in the pCAMBIA1305.1 vector.
CRISPR/Cas9-Mediated Gene Knockout
The CRISPR/Cas9 gene knockout constructs were developed based on the pHSE401 vector described by Xing et al. (2014). Two pairs of oligos were designed to specifically target two different sites within the fourth exon of Rfg1. For the first targeted site, we used the oligo pair 5′ATTGATGAGGACTTAAAAAGCTC3′ and 5′AAACGAGCTTTTTAAGTCCTCAT3′. For the second targeted site, we used the oligo pair 5′ATTGACAGTAAGC CTTACTACCT3′ and 5′AAACAGGTAGTAAGGCTTACTGT3′. The underlined sequences represent the targeted positions. The oligo pairs were first annealed to produce a double-stranded fragment with 4-nt 5′ overhangs at both ends, and then ligated into the BsaI-digested pHSE401 vector. The constructs were individually transformed to the Williams 82 (Nod-) background by means of hairy root transformation. To validate the CRISPR/Cas9-mediated gene disruption, the roots that formed nodules were subjected to DNA isolation, PCR amplification, and DNA sequencing. If the initial sequencing suggested the presence of multiple heterogeneous mutant alleles, the PCR product was ligated into pGEM T-Easy Vector System (Promega) and at least 10 colonies were selected for sequencing.
Construction of a Chimeric Gene of Rfg1 and Rj2
The Rfg1 allele that restricts nodulation by S. fredii USDA257 is allelic to Rj2, an allele that restricts nodulation by B. japonicum USDA122 (Yang et al., 2010). At this locus, there exist three type of alleles in natural populations of soybeans, including the Rj2 (rfg1) allele that restricts nodulation with USDA122 but allows nodulation with USDA257, the rj2 (Rfg1) allele that permits nodulation with USDA122 but restricts nodulation with USDA257, and the rj2 (rfg1) allele that nodulates with both strains. These allelic specificities are defined by seven amino-acid substitutions (Yang et al., 2010). However, we did not identify an Rj2 (Rfg1) allele type that prohibits nodulation with both USDA122 and USDA257 in the surveyed soybean lines (Yang et al., 2010). We thus modified the Rfg1 allele by replacing part of polymorphic sequence of Rfg1 with the corresponding sequence of the Rj2 allele, with the expectation to generate a chimeric gene that could lead to restriction of nodulation by both strains. For this purpose, we amplified two overlapping DNA fragments, one that contained the substitutions responsible for the Rj2 allelic function and another that possessed the substitutions required for the Rfg1 allelic function. We then used an overlapping PCR strategy to assemble the two DNA templates into a single one. The amplified fragment was then cloned into the EcoRI digested genomic construct of Rfg1 mentioned above using the In-Fusion Advantage PCR Cloning Kits (Clontech). We tested the function of this chimeric allele by transferring it into Peking, a genotype that formed nodules with both USDA122 and USDA257. To avoid redundancy, we will provide a further description of this experiment in the “Results” section.
Hairy Root Transformation
Agrobacterium rhizogenes-mediated hairy root transformation was carried out based on the protocol described by Kereszt et al. (2007). Briefly, bacterial paste of the A. rhizogenes strain K599 that contains individual binary vectors was injected into the cotyledonary node of 1-week-old seedlings using a latex free syringe with a thin needle (0.4 mm × 13 mm) (1 ml 27G1/2, Becton, Dickinson & Co.). The infected seedlings were grown in sterile vermiculite and covered with plastic bags in a growth chamber to maintain high humidity. Two to three weeks after inoculation, when hairy roots were well developed at the infection sites, the main roots were removed, and the composite plants were inoculated with rhizobia. Nodulation assays were performed 4 weeks post inoculation.
Microscopic Analysis
Assay for root hair curling followed the method described in Yang et al. (2010). For anatomical analysis of nodules, nitrogen-fixing nodules of Peking and rudimentary nodules of Williams 82 were harvested 4 weeks post inoculation and immediately fixed in 4% paraformaldehyde (w/v) overnight at 4°C. The tissues were then dehydrated in a graded ethanol series followed by a graded series of xylene. After infiltrated in 50/50 Epon-Araldite resin and propylene oxide overnight and then in 75/25 Epon-Araldite resin and propylene oxide for 8 h, the samples were embedded in resin. Embedded tissues were sectioned (10 μm thick) with a microtome, stained with Toluidine Blue, and examined with bright-field optics.
Results
Characterization of the Soybean–S. fredii USDA193 Interactions
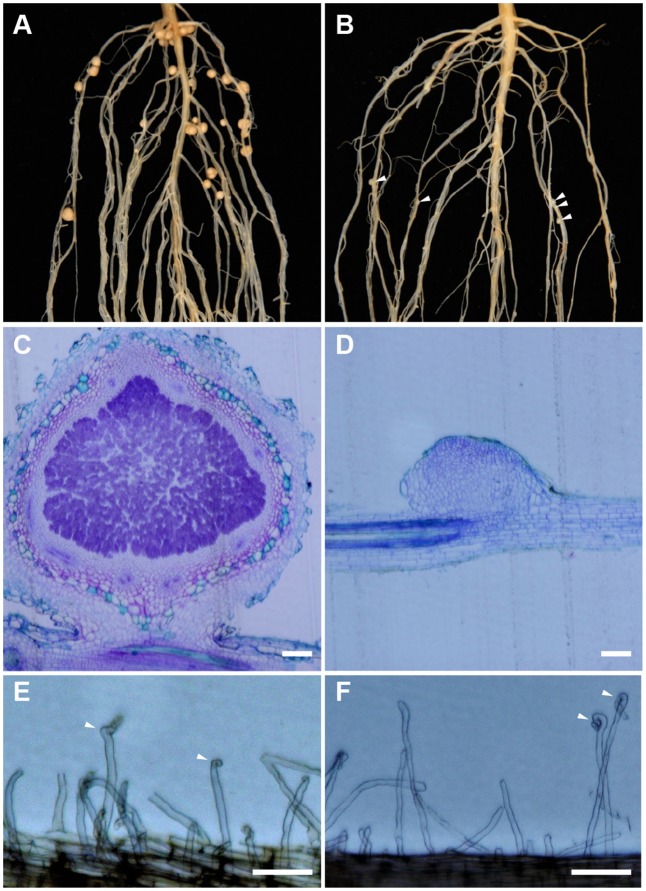
Sinorhizobium fredii USDA193 formed mature nitrogen-fixing nodules on the roots of Peking (Figure 1A) but not on the roots of Williams 82 (Figure 1B). Despite being unable to form functional nodules in the Williams 82 background, the bacterial strain could frequently induce the formation of rudimentary nodules, bump-like small cortical proliferations on the roots (Figure 1B). In contrast to the infected nodules formed in the compatible interaction (Figure 1C), the rudimentary nodules on the roots of Williams 82 were completely devoid of infected bacteria; as such, the cortical cell division ceased at very early stage of the nodule development (Figure 1D). The ability to produce rudimentary nodules with USDA193 on the Williams 82 roots suggested that the early responses of Nod factor perception are not affected. Consistent with this inference, root hair curling, a hallmark of Nod factor responses, occurred in both compatible and incompatible interactions (Figures 1E,F). Thus, we concluded that the restriction of nodulation by S. fredii USDA193 in Williams 82 was not due to a failure in Nod factor perception but caused by the block of bacterial infection.
FIGURE 1.
Nodulation specificity associated with Sinorhizobium fredii USDA193 in soybean. The bacterial strain formed nitrogen-fixing nodules on Peking (A) but only small nodule primordia on Williams 82, as indicated by the arrowheads (B). In the compatible Peking/USDA193 interaction, nodules developed normally and contained bacteria (C), whereas in the incompatible interaction between Williams 82 and USDA193, the nodule primordia did not contain bacteria (D). Bars = 100 μm. However, USDA193 induced root hair curling (indicated by arrowheads) on both Peking (E) and Williams 82 (F). Photographs were taken 5 days post inoculation. Bars = 100 μm.
Restriction of Nodulation with USDA193 Is Controlled by a Single Dominant Gene Mapped to the Rfg1 Locus
We carried out genetic analysis of the symbiosis incompatibility involving S. fredii USDA193 in an F2 population derived from the cross between Williams 82 (Nod-) and Peking (Nod+). From a small population of 122 plants inoculated by USDA193, 95 were Nod- and 27 were Nod+. The segregation statistically fits the 3:1 (Nod- to Nod+) ratio (χ2 = 0.54, df = 1, P = 0.46), suggesting that the restriction of nodulation by USDA193 in Williams 82 is controlled by a single dominant gene. The dominant nature of the nodulation-restrictive allele supports our hypothesis that the incompatible interaction between Williams 82 and USDA193 was not due to a failure in Nod factor signaling but resembles ‘gene-for-gene’ resistance in the plant–pathogen interactions.
Williams 82 and Peking showed the same phenotype when inoculated with S. fredii strains USDA257 and USDA193, and we previously reported that the resistance to nodulation with USDA257 in Williams 82 is controlled by the dominant Rfg1 allele (Glyma16g33780) that encodes a TIR-NBS-LRR protein (Yang et al., 2010). We therefore suspected that Rfg1 possibly also confers resistance to S. fredii USDA193. To test this possibility, we started the mapping experiment by using a polymorphic DNA marker developed from Rfg1. Consistent with our hypothesis, linkage analysis in the aforementioned F2 population revealed co-segregation between the nodulation phenotypes and the marker genotypes. Thus, we considered Rfg1 as a candidate gene that restricts nodulation by S. fredii USDA193.
Rfg1 Is Responsible for Resistance to Nodulation by S. fredii USDA193
We first tested the Rfg1 gene by complementation tests using A. rhizogenes-mediated hairy root transformation. Because the transformation experiments were performed without selection, the hairy roots induced by A. rhizogenes included both transgenic and wild type, which can be readily distinguished by the GUS-staining assay. As shown in Figure 2, introduction of the Rfg1 allele of Williams 82 into the Peking background resulted in complete block of nodule formation on the transgenic roots. From >20 composite transgenic plants that possessed both transgenic and wild-type roots, nodules were formed on the wild-type roots but not on the transgenic roots.
FIGURE 2.
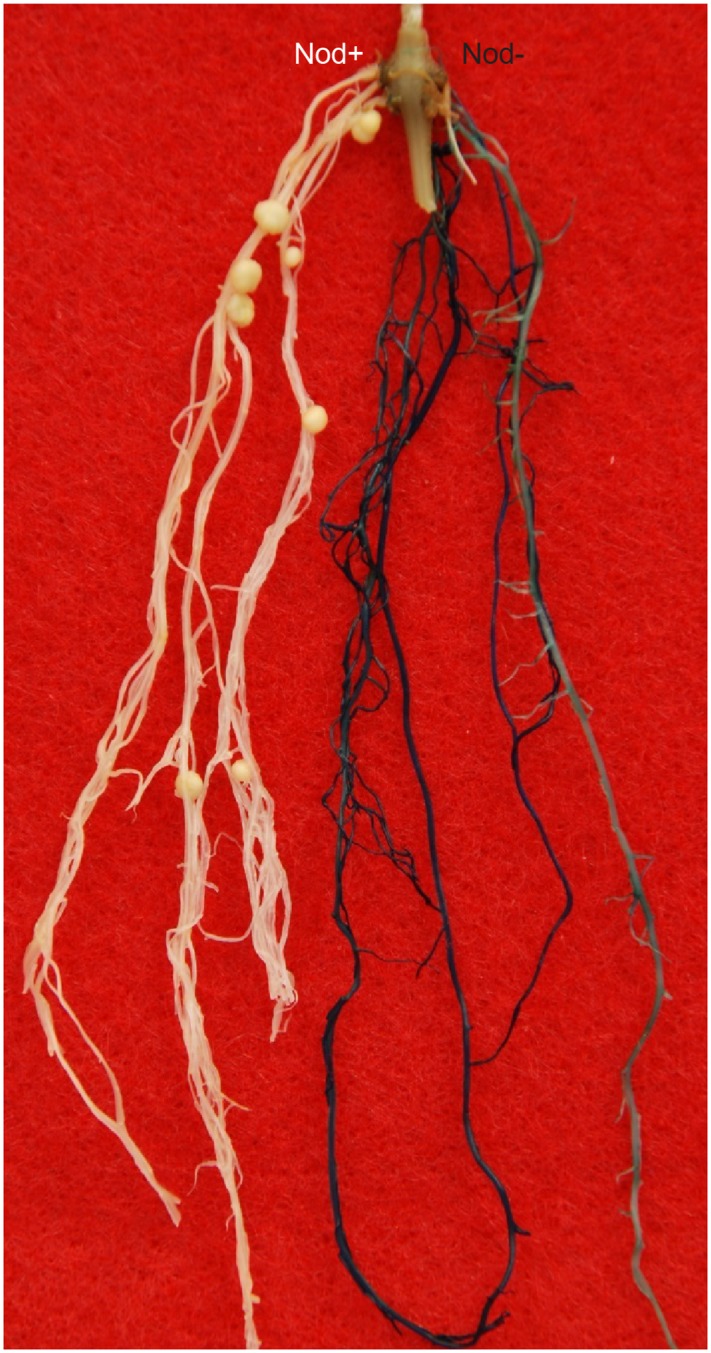
Introduction of the Rfg1 allele of Williams 82 into Peking led to restriction of nodulation on the transgenic roots by USDA193 (blue), but nodulation was normal on the wild-type roots (white).
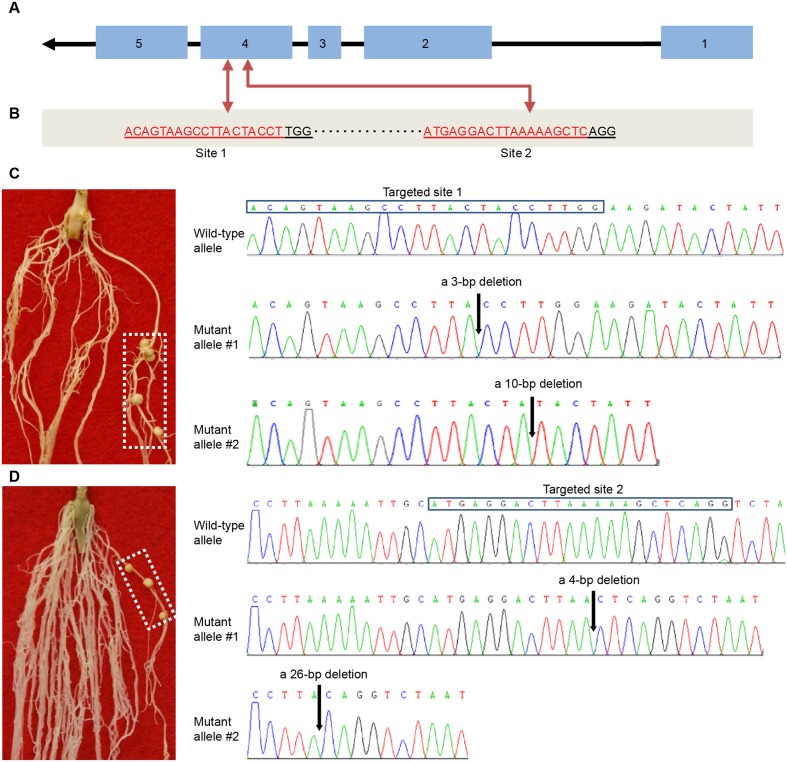
We further used the CRISPR/Cas9-based reverse genetics tool (Doudna and Charpentier, 2014) to knock out the Rfg1 gene in the Williams 82 background (Nod-). For this purpose, we designed two gRNA vectors that individually target two different sites of the fourth exon (Figures 3A,B). The vectors were introduced to A. rhizogenes K599 for hairy root transformation, followed by assaying the nodulation capacity of the hairy roots by inoculation with S. fredii USDA193. For both vectors, we obtained >30 putative transgenic roots from more than 50 independent plants that formed mature nitrogen-fixing nodules. DNA sequencing validated the targeted gene disruption in these roots and these roots did not contain the wild-type allele. Two examples are illustrated in Figures 3C,D. Figure 3C shows that a transgenic root forming nodules resulted from the knockout of Rfg1 at the site 1, and sequence analysis revealed two mutant alleles, one with a 3-bp deletion and another with a 10-bp deletion. Figure 3D represents an example showing that knockout of Rfg1 at the site 2 also led to the formation of root nodules on a transgenic root; sequence analysis identified two mutant alleles, one with a 4-bp deletion and another with a 26-bp deletion when compared with the wild-type allele. Taken together, we conclude that Rfg1 is responsible for nodulation restriction by S. fredii USDA193 in Williams 82.
FIGURE 3.
CRISPR/Cas9-mediated knockout of Rfg1 in the Williams 82 (Nod–) background. (A) Gene structure of Rfg1. The exons and introns are indicated by boxes and lines, respectively. Arrow indicates the transcription direction. (B) Two targeted sites on the fourth exon. The protospacer adjacent motif (PAM) is ‘TGG’ for the site 1 and ‘AGG’ for the site 2. (C) An example showing that knockout of Rfg1 at the site 1 led to the formation of root nodules on a transgenic root (boxed). Sequence analysis revealed that this root contained two mutant alleles, one with a 3-bp deletion and another with a 10-bp deletion (indicated by arrows). (D) An example showing that knockout of Rfg1 at the site 2 also resulted in the formation of root nodules on a transgenic root (boxed). Sequence analysis of the DNA from this root identified two mutant alleles, one with a 4-bp deletion and another with a 26-bp deletion (indicated by arrows).
Functional Analysis of a Chimeric Gene of Rfg1 and Rj2
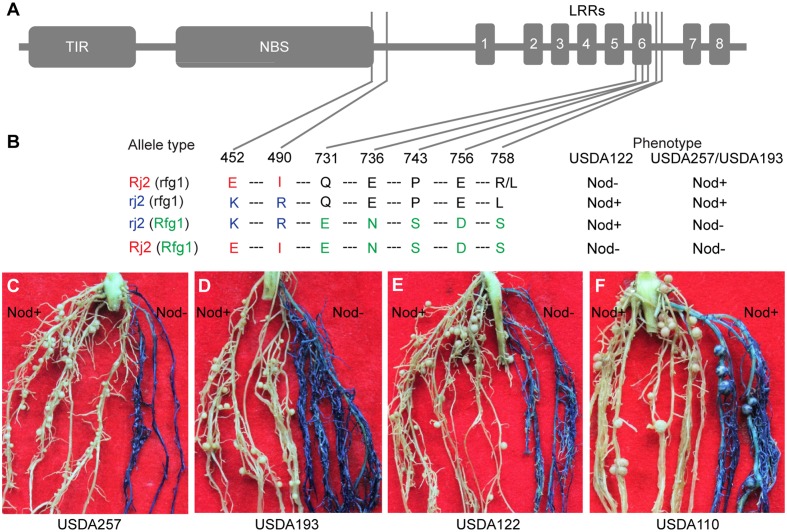
Rfg1 and Rj2 are allelic genes, each encoding a TIR-NBS-LRR protein of 1052 amino acids (Yang et al., 2010). A survey of a group of soybean lines identified three types of naturally occurring alleles, namely Rj2 (rfg1), rj2 (Rfg1), and rj2 (rfg1) (Yang et al., 2010). The Rj2 (rfg1) allele restricts nodulation with USDA122 but not with USDA257; the rj2 (Rfg1) allele restricts nodulation with USDA257 but not with USDA122; and the rj2 (rfg1) allele allows nodulation with both strains. These allelic specificities are determined by seven amino acid substitutions occurring around the C-terminus of the NBS domain and the sixth LRR repeat (Figures 4A,B). Comparing between Rj2 (rfg1) and rj2 (rfg1) and between rj2 (Rfg1), and rj2 (rfg1) suggests that E452 and I490 are required for the Rj2-mediated nodulation restriction against USDA122, while E731, N736, S743, D756, and S758 are essential for Rfg1-mediated resistance against USDA257 and USDA193. If this inference is true, then the chimeric gene of Rj2 and Rfg1, called Rj2 (Rfg1), encoding a protein with E452, I490, E731, N736, S743, D756, and S758, would prevent nodulation with both strains. We generated multiple transgenic roots (from >100 independent plants) expressing the chimeric gene in Peking that carries an rj2 (rfg1) allele and forms nodules with USDA257, USDA193, and USDA122. In consistence with our hypothesis, the transgenic roots restrict nodulation with both USDA257 (Figure 4C), USDA193 (Figure 4D), and USDA122 (Figure 4E). The transgenic roots retained their ability to nodulate with B. japonicum USDA110, a strain that nodulates both Rj2 and Rfg1 genotypes (Figure 4F).
FIGURE 4.
Functional analysis of a chimeric gene of Rfg1 and Rj2. (A,B) Domain structure of the TIR-NBS-LRR protein (A) showing the seven substitution sites and amino acid polymorphisms that distinguish between the Rj2 (rfg1), rj2 (rfg1), and rj2 (Rfg1) protein isoforms (B). Rj2 (Rfg1) represents a protein isoform resulting from expressing the chimeric gene. (C–F) Transgenic roots expressing the chimeric gene (blue) in the Peking background restricted nodulation by USDA257 (C), USDA193 (D), and USDA122 (E) but allowed nodulation with B. japonicum USDA110 (F). In all cases, the nodulation was normal on the non-transgenic roots (white).
Discussion
Previous studies showed that the soybean Rfg1 gene restricts nodulation with the fast-growing S. fredii strains USDA257 and USDA205 (Devine and Kuykendall, 1996; Yang et al., 2010). The Rfg1-mediated resistance to USDA257 was dependent on the bacterial type III secretion system and presumably resulted from the effector-triggered plant immunity, even though the cognate effector(s) has not yet been identified (Krishnan et al., 2003; Yang et al., 2010). In this paper, we demonstrate that the soybean Rfg1 allele also conditions nodulation restriction with S. fredii USDA193, suggesting that Rfg1 likely provide broad-spectrum resistance against a proportion of S. fredii strains (Keyser et al., 1982).
The soybean Rj2/Rfg1 locus confers nodulation specificity toward many naturally occurring B. japonicum and B. fredii strains (Caldwell, 1966; Devine and Kuykendall, 1994; Trese, 1995; Yang et al., 2010). A survey of 847 soybean genotypes from Asian countries revealed an Rj2 allele frequency of 0.02, mainly originated from southeast China (Devine and Breithaupt, 1981). In contrast, the Rfg1 allele occurred with a much higher frequency of 0.44 in a similar population consisting of 285 plant introductions from Asian countries (Devine, 1985). The distribution of the Rfg1 (0.44) and rfg1 (0.56) allele frequencies suggests that these alleles has not been subjected to strong natural selection. However, a screen of 197 soybean lines from the Midwestern United States revealed an Rfg1 allele frequency of 0.83, suggesting that the compatibility with S. fredii strains may be eroded during the breeding process, likely due to the narrow genetic basis of germplasms used in the soybean breeding programs (Balatti and Pueppke, 1992). The diversification of the Rfg1 and rfg1 alleles appears to predate that of the Rj2 and rj2 alleles. The Rj2 (rfg1) allele likely have derived from the rj2 (rfg1) allele through gain-of-function mutations. The rj2 (rfg1) allelic form is most promiscuous, conferring an unrestricted nodulation phenotype with many strains that are restricted by the Rj2 or Rfg1 alleles. Therefore, the rj2 (rfg1) genotype should be prioritized in soybean breeding programs if the goal is to ensure the cultivar to be able to nodulate with indigenous bacterial strains.
The polymorphic Rj2/Rfg1 locus encodes a TIR-NB-LRR gene with three allelic variants that show differing specificities to yet unknown bacterial effectors. E452 and I490 in or near the C-terminal NBS domain of Rj2 are required for the recognition of the cognate effector from B. japonicum USDA122, while E731, N736, S743, D756, and S758 in the LRR domain of Rfg1 are essential for the recognition of the corresponding effectors secreted by USDA257 and USDA193. This situation is similar to the previous studies showing that both TIR, NBS, and LRR domains could be involved in the determination of resistance specificity (Ellis et al., 1999; Dodds et al., 2001; Mondragón-Palomino et al., 2002; Ellis et al., 2007). The chimeric Rj2(Rfg1) allele composed of E452, I490, E731, N736, S743, D756, and S758 could recognize corresponding effectors of all these strains. Such an allele could exist in natural population if intragenic recombination events occur between the Rj2(rfg1) and rj2(Rfg1) alleles. It is possible that these polymorphic sites play a role in direct effector binding or intra- or intermolecular interactions in protein complexes during recognition and signaling. Further analysis of the mechanisms underlying the recognitions mediated by the Rj2/Rfg1 locus awaits the identification of the cognate bacterial effectors.
Author Contributions
YF, JL, SL, and HZ: conception and design of the work. YF, JL, QW, SY, and SL: performed the work and analyzed data. HZ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DR and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Patrick Elia (US Department of Agriculture-Agricultural Research Service, Beltsville, MD, United States) for providing the rhizobial strains and Dr. Qijun Chen (China Agricultural University, Beijing, China) for providing the CRISPR/Cas9 binary vectors.
Footnotes
Funding. This work was supported by the Kentucky Soybean Promotion Board to HZ and by the National Natural Science Foundation of China (grant number 31271751) to SL and YF. YF and SL received support from the Shandong Provincial Education Association of China to study at University of Kentucky.
References
- Balatti P. A., Pueppke S. G. (1992). Identification of North American soybean lines that form nitrogen-fixing nodules with Rhizobium fredii USDA257. Can. J. Plant Sci. 72 49–55. 10.4141/cjps92-006 [DOI] [Google Scholar]
- Bras C. P., Jordá M. A., Wijfjes A. H., Harteveld M., Stuurman N., Thomas-Oates J. E., et al. (2000). A Lotus japonicus nodulation system based on heterologous expression of the fucosyl transferase NodZ and the acetyl transferase NoIL in Rhizobium leguminosarum. Mol. Plant Microbe Interact. 13 475–479. 10.1094/MPMI.2000.13.4.475 [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Jabbouri S., Perret X. (2000). Keys to symbiotic harmony. J. Bacteriol. 182 5641–5652. 10.1128/JB.182.20.5641-5652.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell B. E. (1966). Inheritance of a strain specific ineffective nodulation in soybeans. Crop Sci. 6 427–428. 10.2135/cropsci1966.0011183X000600050010x [DOI] [Google Scholar]
- Deakin W. J., Broughton W. J. (2009). Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat. Rev. Microbiol. 7 312–320. 10.1038/nrmicro2091 [DOI] [PubMed] [Google Scholar]
- Devine T. E. (1985). Nodulation of soybean plant introduction lines with the fast-growing rhizobial strain USDA 205. Crop Sci. 25 354–356. 10.2135/cropsci1985.0011183X002500020037x [DOI] [Google Scholar]
- Devine T. E., Breithaupt B. H. (1981). Frequencies of Nodulation Response Alleles, Rj2 and Rj4 in Soybean Plant Introductions and Breeding Lines. Washington, DC: USDA. [Google Scholar]
- Devine T. E., Kuykendall L. D. (1994). Genetic allelism and linkage tests of a soybean gene, Rfg1, a soybean gene controlling nodulation with fast-growing Rhizobium fredii strain 205. Plant Soil 158 47–51. 10.1007/BF00007916 [DOI] [Google Scholar]
- Devine T. E., Kuykendall L. D. (1996). Host genetic control of symbiosis in soybean (Glycine max L.). Plant Soil 186 173–187. 10.1007/BF00035072 [DOI] [Google Scholar]
- D’Haeze W., Holsters M. (2004). Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 12 555–561. 10.1016/j.tim.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Lawrence G. J., Ellis J. G. (2001). Six amino acid changes confined to the leucine-rich repeat beta-strand/beta-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13 163–178. 10.1105/tpc.13.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Downie J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34 150–170. 10.1111/j.1574-6976.2009.00205.x [DOI] [PubMed] [Google Scholar]
- Ellis J. G., Dodds P. N., Lawrence G. J. (2007). Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol. 45 289–306. 10.1146/annurev.phyto.45.062806.094331 [DOI] [PubMed] [Google Scholar]
- Ellis J. G., Lawrence G. J., Luck J. E., Dodds P. N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. 10.1105/tpc.11.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque O. M., Miwa H., Yasuda M., Fujii Y., Kaneko T., Sato S., et al. (2015). Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 81 6710–6717. 10.1128/AEM.01942-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvart M., Michiels J. (2008). Rhizobial secreted proteins as determinants of host specificity in the rhizobium-legume symbiosis. FEMS Microbiol. Lett. 285 1–9. 10.1111/j.1574-6968.2008.01254.x [DOI] [PubMed] [Google Scholar]
- Geurts R., Heidstra R., Hadri A. E., Downie J. A., Franssen H., Van Kammen A., et al. (1997). Sym2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol. 115 351–359. 10.1104/pp.115.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5 619–633. 10.1038/nrmicro1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y., Kelly S., Nielsen M. W., Hjuler C. T., Gysel K., Muszyñski A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523 308–312. 10.1038/nature14611 [DOI] [PubMed] [Google Scholar]
- Kereszt A., Li D., Indrasumunar A., Nguyen C. D., Nontachaiyapoom S., Kinkema M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2 948–952. 10.1038/nprot.2007.141 [DOI] [PubMed] [Google Scholar]
- Keyser H. H., Bohlool B. B., Hu T. S., Weber D. F. (1982). Fast-growing rhizobia isolated from root nodules of soybean. Science 215 1631–1632. 10.1126/science.215.4540.1631 [DOI] [PubMed] [Google Scholar]
- Keyser H. H., Li F. (1992). Potential for increasing biological nitrogen fixation in soybeans. Plant Soil 141 119–135. 10.1007/BF00011313 [DOI] [Google Scholar]
- Kiers E. T., Hutton M. G., Denison R. F. (2007). Human selection and the relaxation of legume defences against ineffective rhizobia. Proc. Biol. Sci. 274 3119–3126. 10.1098/rspb.2007.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Kaashyap M., Rathore A., Das R. R., Parupalli S., Upadhyaya H. D., et al. (2014). Phylogenetic diversity of Mesorhizobium in chickpea. J. Biosci. 39 513–517. 10.1007/s12038-014-9429-9 [DOI] [PubMed] [Google Scholar]
- Krishnan H. B., Lorio J., Kim W. S., Jiang G., Kim K. Y., DeBoer M., et al. (2003). Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 16 617–625. 10.1094/MPMI.2003.16.7.617 [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344 781–784. 10.1038/344781a0 [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633. 10.1126/science.1090074 [DOI] [PubMed] [Google Scholar]
- Liu J., Yang S., Zheng Q., Zhu H. (2014). Identification of a dominant gene in Medicago truncatula that restricts nodulation by Sinorhizobium meliloti strain Rm41. BMC Plant Biol. 14:167 10.1186/1471-2229-14-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L. F., Shoemaker R. C. (1996). Construction and size characterization of a bacterial artificial chromosomal (BAC) library from soybean. Soybean Genet. Newslett. 23 126–129. [Google Scholar]
- Mondragón-Palomino M., Meyers B. C., Michelmore R. W., Gaut B. S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12 1305–1315. 10.1101/gr.159402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch L. A., Young J. P. (2004). Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol. Ecol. 13 2435–2444. 10.1111/j.1365-294X.2004.02259.x [DOI] [PubMed] [Google Scholar]
- Nakano Y., Yamakawa T., Ikeda M., Ishizuka J. (1997). Nodulation of Rj-soybean varieties with Rhizobium fredii USDA 193 under limited supply of nutrients. Soil Sci. Plant Nutri. 43 929–932. 10.1080/00380768.1997.10414659 [DOI] [Google Scholar]
- Okazaki S., Zehner S., Hempel J., Lang K., Göttfert M. (2009). Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 295 88–95. 10.1111/j.1574-6968.2009.01593.x [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Perret X., Staehelin C., Broughton W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64 180–201. 10.1128/MMBR.64.1.180-201.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Gronlund M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. 10.1038/nature02039 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L. H., Madsen E. B., Jurkiewicz A., Fukai E., Quistgaard E. M., et al. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26 3923–3935. 10.1038/sj.emboj.7601826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Cregan P. B., Rodriguez-Quinones F., Keyser H. H. (1990). Microbial influence on gene-for-gene interactions in legume-Rhizobium symbioses. Plant Soil 129 53–60. 10.1007/BF00011691 [DOI] [Google Scholar]
- Soto M. J., Dominguez-Ferreras A., Perez-Mendoza D., Sanjuan J., Olivares J. (2009). Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 11 381–388. 10.1111/j.1462-5822.2008.01282.x [DOI] [PubMed] [Google Scholar]
- Tang F., Yang S., Liu J., Zhu H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 170 26–32. 10.1104/pp.15.01661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trese A. T. (1995). A single dominant gene in McCall soybean prevents effective nodulation with Rhizobium fredii USDA257. Euphytica 81 279–282. 10.1007/BF00025618 [DOI] [Google Scholar]
- Tsukui T., Eda S., Kaneko T., Sato S., Okazaki S., Kakizaki-Chiba K., et al. (2013). The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 79 1048–1051. 10.1128/AEM.03297-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumaru H., Hashimoto S., Okizaki K., Kanesaki Y., Yoshikawa H., Yamakawa T. (2015). A putative type III secretion system effector encoded by the MA20_12780 gene in Bradyrhizobium japonicum Is-34 causes incompatibility with Rj4 genotype soybeans. Appl. Environ. Microbiol. 81 5812–5819. 10.1128/AEM.00823-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yang S., Tang F., Zhu H. (2012). Symbiosis specificity in the legume-rhizobial mutualism. Cell Microbiol. 14 334–342. 10.1111/j.1462-5822.2011.01736.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Yang S., Liu J., Terecskei K., Ábrahám E., Gombár A., et al. (2017). Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114 6854–6859. 10.1073/pnas.1700715114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H. L., Dong L., Wang Z. P., Zhang H. Y., Han C. Y., Liu B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Tang F., Gao M., Krishnan H. B., Zhu H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107 18735–18740. 10.1073/pnas.1011957107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang Q., Fedorova E., Liu J., Qin Q., Zheng Q., et al. (2017). Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114 6848–6853. 10.1073/pnas.1700460114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M., Miwa H., Masuda S., Takebayashi Y., Sakakibara H., Okazaki S. (2016). Effector-triggered immunity determines host genotype-specific incompatibility in legume-Rhizobium symbiosis. Plant Cell Physiol. 57 1791–1800. 10.1093/pcp/pcw104 [DOI] [PubMed] [Google Scholar]