Abstract
Human glioblastoma is one of the most malignant types of brain tumor in the world. In the present study, the functional mechanisms of microRNA-141 (miR-141) were assessed, and the potential role of miR-141 as a prognostic biomarker in glioblastoma was examined. The gene expression of miR-141 in glioblastoma cell lines and glioblastoma tumors was assessed by reverse transcription-quantitative polymerase chain reaction. Glioblastoma LN229 and U89 cell lines were transfected with synthetic miR-141 mimics to upregulate endogenous miR-141. The subsequent effect on glioblastoma proliferation was assessed by MTT assay. In human glioblastoma, miR-141 expression was compared between patients with tumors of different pathological grades. Statistical analyses were performed to assess the correlation between miR-141 and the clinicopathological properties and overall survival rates (OS) of the patients. In addition, a Cox regression model was used to examine whether miR-141 was a potential biomarker of glioblastoma. miR-141 was aberrantly downregulated in glioblastoma cell lines and human glioblastoma tumors. Forced miR-141 upregulation in glioblastoma LN229 and U89 cell lines suppressed cancer proliferation. In patients with glioblastoma, miR-141 downregulation was closely associated with an advanced disease stage, poor clinicopathological properties and a shorter OS time. The multivariate Cox regression model demonstrated that low miR-141 expression was an effective prognostic biomarker for patients with glioblastoma. Overall, the present study showed that miR-141 may be a functional cancer regulator and a prognostic biomarker for glioblastoma.
Keywords: glioblastoma, microRNA-141, cancer proliferation, biomarker, overall survival
Introduction
Glioblastoma is a heterogeneous and highly aggressive brain tumor in the central nervous system. Patients with glioblastoma often experience high relapse rates and poor prognoses (1–3). Although efforts have been made toward early diagnosis and targeted therapy, there are currently no effective treatments to completely cure patients with glioblastoma, particularly those at advanced clinical stages (4–6). Therefore, it is important to identify novel molecular targets that are effective regulators and prognostic biomarkers for glioblastoma.
MicroRNAs (miRNAs/miRs) are formed from several hundred single-stranded short RNAs that endogenously bind to the 3′-untranslated region (3′UTR) of target genes to post-transcriptionally suppress gene production or induce protein degradation, thus serving important roles in regulating cell and tissue development and pathology in human and animals (7–10). In human cancer types, miRNAs have been demonstrated to be aberrantly upregulated or downregulated in cancerous tissues, thus serving as either oncogenes or tumor suppressors to modulate cancer development (10–12). In addition, cancerous miRNAs have been identified to be effective biomarkers to predict prognosis in patients with cancer (13–15). In human glioblastoma, various miRNAs have been demonstrated to be either upregulated or downregulated in glioma tumors, and played critical roles in regulating glioblastoma proliferation, migration and chemosensitivity (16).
Amongst numerous other cancer-associated miRNAs, miR-141 has been demonstrated to be an effective tumor regulatory gene in various human cancer types, including ovarian cancer, hepatocellular carcinoma, non-small cell lung cancer and renal cancer (17–20). However, the expression pattern or functional role of miR-141 in human glioblastoma remains unknown.
In the present study, the expression pattern of miR-141 was assessed in glioblastoma cell lines and clinical samples from patients with glioblastoma. Secondly, synthetic miRNA mimics were applied to ectopically overexpress miR-141 in glioblastoma cell lines to examine whether miR-141 exhibited functional mechanisms in regulating glioblastoma proliferation. Thirdly, the correlation between cancerous miR-141 expression and clinicopathological factors and overall survival rates (OS) of patients with glioblastoma was analyzed. The purpose of the present study was to investigate whether miR-141 may act as a cancer regulator or as a prognostic biomarker for glioblastoma.
Patients and methods
Ethics, consent and permissions
In the present study, the clinical and experimental procedures were reviewed and approved by the Ethic Committees at Suining Central Hospital (Suining, China) and Chongqing Fourth People's Hospital (Chongqing, China). All experiments were conducted in accordance with the Declaration of Helsinki. Consent forms were obtained from all patients enrolled.
Glioblastoma cell lines
Immortal glioblastoma T98G, U251, A172, LN229 and U89 cell lines, and normal human astrocytes (NHA), were all purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma Aldrich; Millipore, Darmstadt, Germany) in a tissue culturing environment with 5% CO2 at 37°C.
Clinical glioblastoma samples
Between September 2007 and September 2011, 91 patients (mean age, 56±6.3 years; 52 male and 39 female) with glioblastoma were enrolled in the study at Suining Central Hospital and Chongqing Fourth People's Hospital. Clinical samples of glioblastoma tumors and adjacent non-tumorous brain tissues were obtained during surgical resection. The pathological grade of tumors, based on magnetic resonance imaging scans and histological examination, were evaluated by a joint-team of histologists, pathologists and radiologists according to the World Health Organization (WHO) classification of brain tumors (21). All tumorous or non-tumorous brain samples, once obtained from patients, were immediately snap-frozen in liquid nitrogen and stored at −80°C for future RNA extraction.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from clinical samples using a TRIzol kit (Thermo Fisher Scientific, Inc.) and purified using a QiaQuick PCR Purification kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers' protocols. The quantity of extracted RNA was examined by a NanoDrop-3000 spectrophotometer (Thermo Fisher Scientific, Inc.). A total of 5 µg RNA from each sample was reverse synthesized to complementary DNA using a TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Gene expression of human miR-141 (hsa-miR-141) was quantified through RT-qPCR using a TaqMan MicroRNA assay kit (Thermo Fisher Scientific, Inc.) on an ABI Prism 7000 Sequence Detection system (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The thermocycling conditions used were as follows: 30 sec at 95°C; 30 sec at 60°C; and 30 sec at 72°C for 35 cycles. The following primers were used: hsa-miR-141 forward, 5′-CGCTAACACTGTCTGGTAAAG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 snRNA forward, 5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse, 5′-GGAACGCTTCACGAATTTG-3′. The relative expression of hsa-miR-141 was measured as a fold-change and was normalized to U6 small nuclear RNA expression in control samples using the 2−ΔΔCq method (22).
miR-141 upregulation in glioblastoma
Synthetic human miR-141 mimics, miR-141-mimic, and its non-specific control miRNA, miR-Ctrl, were purchased from Sunbiotech Co., Ltd. (Beijing, China). The glioblastoma LN229 and U89 cell lines were transfected for 24 h at 37°C with 100 nM miR-141-mimic or 100 nM miR-Ctrl using the Oligofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The cultures were replenished with fresh RPMI-1640 medium without synthetic miRNAs at 48 h post-transfection. Subsequent experiments, such as RT-qPCR using the same conditions as stated above and a proliferation assay were performed.
Cancer proliferation assay
The in vitro proliferation of glioblastoma cells was performed using an MTT assay (Applied Science, Grass Valley, CA, USA) according to the manufacturer's protocols. Subsequent to transfection with synthetic miRNAs, LN229 and U89 cells were maintained in 96-well plates at a density of 5,000 cells/well for 5 days. Every 24 h, 20 µl 1X MTT medium was added into 96-well plates for 4 h, followed by 20 min application of dimethyl sulphoxide. The 96-well plates were then examined using a Synergy 2 multi-mode microplate reader (BioTek Instruments, Inc., Winooshi, VT, USA) at an absorbance of 490 nm according to the manufacturer's protocol.
Statistical analysis
All experiments were conducted in triplicate and the data are presented as the mean ± standard deviation. All statistical analyses were conducted using SPSS software v11.0 (SPSS, Inc., Chicago, IL, USA). An unpaired two-tailed Student's t-test was used to compare paired or grouped samples. The associations between miR-141 and the clinicopathological features of the patients were analyzed using rank sum and χ2 tests. The associations between the clinicopathological properties of the patients with OS were analyzed using forward univariate or multivariate Cox proportional hazards model with 95% confidence intervals (CIs). The OS of the patients was estimated using a Kaplan-Meier model and compared by log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-141 is aberrantly downregulated in glioblastoma cell lines and human tumors
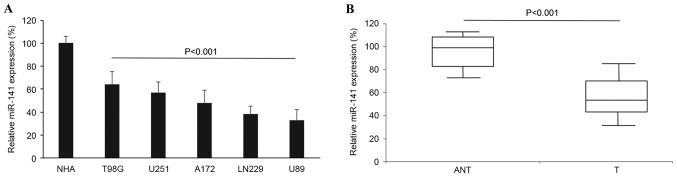
The gene expression pattern of miR-141 among glioblastoma cell lines and human tumors was examined. In 5 glioblastoma cell lines, T98G, U251, A172, LN229 and U89, miR-141 expression was quantified by RT-qPCR and analyzed against miR-141 expression in NHAs. It was revealed that miR-141 was significantly downregulated in the glioblastoma cell lines compared with the level in the NHAs (Fig. 1A; P<0.001). Secondly, miR-141 expression was examined in human samples. In 91 patients with glioblastoma, glioblastoma tumors and adjacent non-tumorous brain tissues were surgically sampled and their endogenous miR-141 expression levels were compared using RT-qPCR. Similar to the result in glioblastoma cell lines, it was demonstrated that miR-141 was also aberrantly downregulated in human glioblastoma tumors compared with the expression in the non-tumorous brain tissues (Fig. 1B; P<0.001).
Figure 1.
miR-141 expression in glioblastoma cells and clinical samples. (A) Endogenous miR-141 expression was measured by RT-qPCR in glioblastoma cell lines T98G, U251, A172, LN229 and U89, and compared with miR-141 expression in NHAs. (B) Endogenous miR-141 expression was compared between clinical samples in 42 patients with glioblastoma. NHAs, normal human astrocytes; ANT, non-tumorous brain tissues; T, human glioblastoma tumors; miR/miRNA, microRNA.
Induced miR-141 overexpression inhibits cancer proliferation in glioblastoma
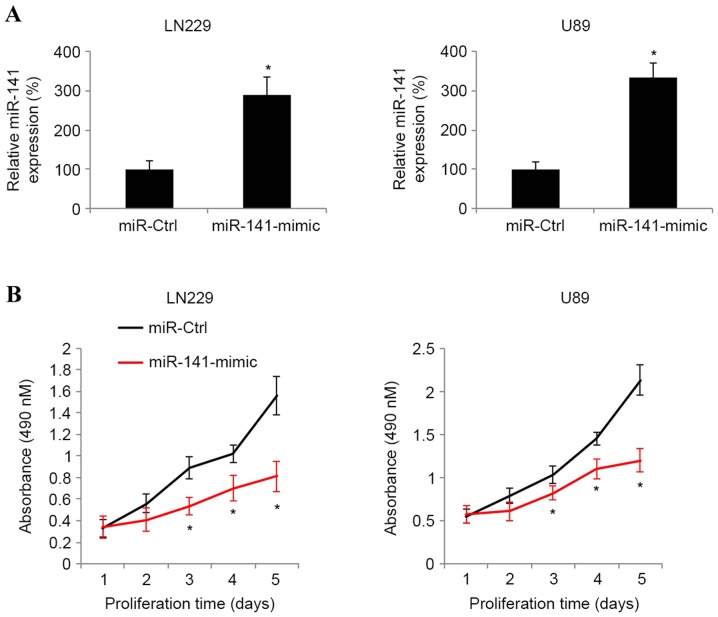
The present study next assessed whether miR-141 exhibited a functional role in glioblastoma. The glioblastoma LN229 and U89 cell lines were transfected with synthetic miR-141-mimic miRNAs to induce endogenous overexpression of miR-141. Control LN229 and U89 cells were transfected with a non-specific control synthetic miRNA, miR-Ctrl. RT-qPCR analysis indicated that at 2 days post-transfection, endogenous miR-141 was significantly upregulated in the glioblastoma cells transfected with miR-141-mimic compared with the glioblastoma cells transfected with miR-Ctrl (Fig. 2A; P<0.05).
Figure 2.
Effect of miR-141 upregulation on glioblastoma proliferation. (A) Glioblastoma LN229 and U89 cell lines were transfected with 100 nM synthetic miR-141 mimics, miR-141-mimic, or 100 nM non-specific control miRNA, miR-Ctrl for 48 h. The efficiency of miR-141 upregulation was assessed by RT-qPCR (*P<0.05 vs. miR-Ctrl). (B) Subsequent to transfection, LN229 and U89 cells were plated into 96-well plates and an MTT assay was conducted for 5 days to assess cancer proliferation (*P<0.05 vs. miR-Ctrl; n=6). miR/miRNA, microRNA.
The regulatory effect of miR-141 upregulation on glioblastoma growth was then examined. Transfected LN229 and U89 cells were seeded into 96-well plates and maintained for 5 days. An MTT assay was performed every 24 h during a 5-day period to compare cancer proliferation between glioblastoma cells. It was revealed that in the LN229 and U89 cells lines, miR-141 upregulation markedly suppressed cancer cell proliferation (Fig. 2B; P<0.05).
miR-141 is correlated with clinicopathological properties of patients with glioblastoma
In 91 patients with glioblastoma, 25 exhibited WHO grade I pilocytic astrocytomas, 23 exhibited WHO grade II diffuse astrocytomas, 21 exhibited WHO grade III anaplasia astrocytomas and 22 exhibited WHO grade IV primary glioblastomas. The miR-141 expression levels were compared based on the tumor grades. It was demonstrated that miR-141 expression was downregulated in grade III or IV tumors compared with non-tumorous brain tissues (Fig. 3; P<0.05). It was also demonstrated that miR-141 expression was significantly downregulated in grade III or IV tumors compared with grade I or grade II tumors (Fig. 3; P<0.05).
Figure 3.
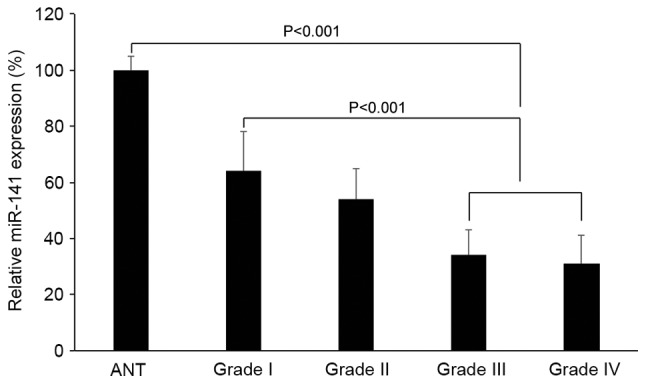
miR-141 expression in glioblastoma tumors with different pathological grades. Glioblastoma tumors were characterized based on World Health Organization grades. The endogenous miR-141 expression of grade I, II, III and IV tumors were compared with miR-141 expression in normal human astrocytes. ANT, adjacent non-tumorous brain tissues; miR, microRNA.
To understand the correlation between miR-141 expression and the prognoses of patients with glioblastoma, patients were divided into two subgroups. One group of 47 patients was characterized to exhibit miR-141 expression levels lower than the median value. The other group of 44 patients exhibited miR-141 expression levels higher than the median value. The clinicopathological properties of the patients were statistically analyzed based on miR-141 expression levels, and it was identified that miR-141 expression demonstrated no association with patient gender, age or tumor size (Table I). Conversely, miR-141 expression was revealed to be significantly correlated with the pathological grade and Karnofsky Performance Scale (KPS) of the patients (Table I; P<0.05).
Table I.
Association of tumor miR-141 expression with clinicopathological properties of the patients (n=91).
| miR-141 expression, n (%) | |||
|---|---|---|---|
| Clinicopathol-ogicalproperty | Low (n=47) | High (n=44) | P-value |
| Gender | |||
| Male | 31 (65.96) | 30 (68.18) | N.S.S. |
| Female | 16 (34.04) | 14 (31.82) | |
| Age, years | |||
| <55 | 12 (25.53) | 7 (15.91) | N.S.S. |
| ≥55 | 35 (74.47) | 37 (84.09) | |
| WHO grade | |||
| I | 7 (14.84) | 18 (40.91) | 0.002 |
| II | 8 (17.02) | 15 (34.09) | |
| III | 14 (29.79) | 7 (15.91) | |
| IV | 18 (38.30) | 4 (9.09) | |
| KPS | |||
| <80 | 35 (74.47) | 14 (31.82) | 0.013 |
| ≥80 | 12 (25.53) | 30 (68.18) | |
| Tumor size, cm | |||
| <6 | 22 (46.81) | 27 (61.36) | N.S.S. |
| ≥6 | 25 (53.19) | 17 (38.64) | |
WHO, World Health Organization; KPS, Karnofsky Performance Scale; N.S.S., not statistically significant; miR, microRNA.
miR-141 is a prognostic biomarker for survival of patients with glioblastoma
A Kaplan-Meier model was applied to separately assess the OS of patients with glioblastoma and high endogenous miR-141 expression levels, and those with low endogenous miR-141 expression levels. Using a log-rank test, it was identified that the two OS curves were significantly different. Patients with low endogenous miR-141 expression levels exhibited much poorer OS compared with patients with high endogenous miR-141 expression levels (Fig. 4; P<0.001). A Cox regression model was applied to assess the association of the clinicopathological properties of the patients with OS. Through the univariate analysis, it was revealed that the WHO grade (95% CI, 3.89–5.40; P=0.020), KPS (95% CI, 1.04–4.78; P=0.241) and endogenous miR-141 expression (95% CI, 4.83–9.63; P=0.003) of the patients were significantly associated with OS (Table II). In addition, the multivariate analysis demonstrated that the WHO grade (95% CI, 2.89–5.92; P=0.107), and endogenous miR-141 expression (95% CI, 3.24–6.36; P=0.007) of the patients were independent prognostic factors for OS in patients with glioblastoma (Table II).
Figure 4.
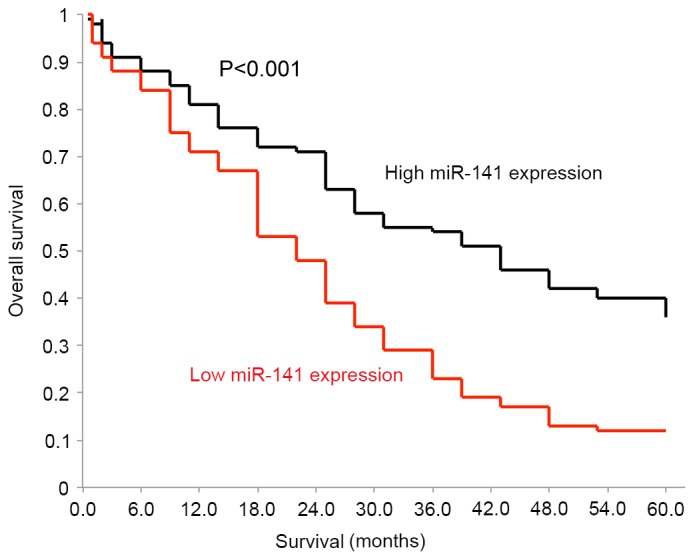
OS of glioblastoma patients with low or high endogenous miR-141 expression. OS was assessed by a Kaplan-Meier model for patients with high endogenous miR-141 expression and for patients with low endogenous miR-141 expression (P<0.001). OS, overall survival.
Table II.
Univariate and multivariate Cox regression model of prognostic properties in patients with glioblastoma.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Prognostic features | P-value | HR (95% CI) | P-value | HR (95% CI) |
| Gender | N.S.S. | 0.87 (0.63–1.41) | ||
| Age, years | N.S.S. | 1.21 (0.89–4.29) | ||
| WHO grade | 0.029 | 4.33 (3.89–5.40) | 0.107 | 3.58 (2.89–5.92) |
| KPS | 0.241 | 1.93 (1.04–4.78) | N.S.S. | 1.28 (0.67–2.71) |
| Tumor size, cm | N.S.S. | 2.04 (1.18–5.31) | ||
| miR-141 expression | 0.003 | 6.73 (4.83–9.63) | 0.007 | 4.12 (3.24–6.36) |
HR, hazard ratio; CI, confidence interval; N.S.S., not statistically significant; WHO, World Health Organization; KPS, Karnofsky Performance Scale; miR, microRNA.
Discussion
miR-141 has been demonstrated to be an active onco-regulator in various types of human cancer (17–20). However, the expression pattern or mechanistic role of miR-141 in human glioblastoma remains unknown. In the present study, quantitative methods were used to compare miR-141 expression between 5 in vitro glioblastoma cell lines and their adjacent non-tumorous brain tissues. The results of the RT-qPCR analysis clearly demonstrated that miR-141 was markedly downregulated in in vitro glioblastoma cell lines and in vivo glioblastoma tumors. These data are concurrent with previous studies demonstrating decreased miR-141 expression in hepatocellular carcinoma, and renal and gastric cancer (17,20,23), suggesting that miR-141 downregulation or dysregulation may be the predominant expression pattern of miR-141 within various types of cancer.
Secondly, the functional role of miR-141 in regulating glioblastoma was investigated in the present study. It was revealed that induced miR-141 overexpression may effectively inhibit glioblastoma proliferation in vitro. A tumor suppressive role of miR-141 has also been observed in other types of human cancer. For example, miR-141 overexpression was demonstrated to induce cell-cycle arrest in renal cell carcinoma (20). Notably, miR-141 may also act as an oncogenic factor, with actions such as increasing cisplatin chemoresistance in epithelial ovarian cancer (18). Thus, although miR-141 may be predominantly downregulated in human cancer, the functional mechanisms of miR-141 may be more complex, including roles as a tumor suppressor or an oncogene, depending on the different downstream signaling pathways associated with miR-141 regulation in different types of cancer (24).
It was previously demonstrated that plasma miR-141 may be an effective biomarker in colon and colorectal cancer (25,26). In addition to the RT-qPCR analysis demonstrating miR-141 downregulation in glioblastoma cell lines and human tumors, the present study also identified that miR-141 downregulation was associated with glioblastoma tumors of advanced stages (Fig. 3). These data indicated that cancerous miR-141 may be a potential cancer biomarker for patients with glioblastoma. The clinicopathological properties and OS of patients with glioblastoma were analyzed to evaluate the significance of miR-141 correlation. It was revealed that low cancerous miR-141 expression was likely to be associated with advanced clinicopathological features in patients with glioblastoma. Also, it was demonstrated that low cancerous miR-141 expression was significantly correlated with poor survival in patients with glioblastoma. Additionally, multivariate Cox regression analysis demonstrated that cancerous miR-141 expression may be an independent predicting factor for glioblastoma. Thus, the analysis of the clinical data of the present suggested that miR-141 may act as a tumor suppressor in regulating cancer proliferation and may serve as a potential biomarker for glioblastoma.
Overall, the present study provided clear molecular and clinical implications for miR-141 in glioblastoma. miR-141 is significantly downregulated in glioblastoma cell lines and human tumor samples. miR-141 may act as a functional tumor suppressor to inhibit cancer proliferation in glioblastoma. Additionally, miR-141 is closely associated with the clinicopathological properties of patients with glioblastoma, and may be a prognostic biomarker for glioblastoma.
Acknowledgements
The present study was supported by the Health and Family Planning Commission Research of Sichuan Province (grant no. 150245).
References
- 1.Alifieris C, Trafalis DT. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol Ther. 2015;152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Ellis HP, Greenslade M, Powell B, Spiteri I, Sottoriva A, Kurian KM. Current challenges in glioblastoma: Intratumour heterogeneity, residual disease, and models to predict disease recurrence. Front Oncol. 2015;5:251. doi: 10.3389/fonc.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jovčevska I, Kočevar N, Komel R. Glioma and glioblastoma - how much do we (not) know? Mol Clin Oncol. 2013;1:935–941. doi: 10.3892/mco.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Mohindra S, Singla N, Sodhi HB, Chatterjee D, Gupta SK. Multiple glioblastoma: A diagnostic challenge and controversies in management. Neurol India. 2015;63:449–452. doi: 10.4103/0028-3886.158267. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Ye G, Li J, Wang Y. Recent advance in molecular angiogenesis in glioblastoma: The challenge and hope for anti-angiogenic therapy. Brain Tumor Pathol. 2015;32:229–236. doi: 10.1007/s10014-015-0233-5. [DOI] [PubMed] [Google Scholar]
- 6.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 7.Guarnieri DJ, DiLeone RJ. MicroRNAs: A new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 8.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Liu MF. Mechanisms of microRNA-mediated gene regulation. Sci China C Life Sci. 2009;52:1111–1116. doi: 10.1007/s11427-009-0152-y. [DOI] [PubMed] [Google Scholar]
- 10.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 11.Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 2015;34:5857–5868. doi: 10.1038/onc.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47:131–144. doi: 10.1007/s12035-012-8349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue J, Niu YF, Huang J, Peng G, Wang LX, Yang YH, Li YQ. miR-141 suppresses the growth and metastasis of HCC cells by targeting E2F3. Tumour Biol. 2014;35:12103–12107. doi: 10.1007/s13277-014-2513-9. [DOI] [PubMed] [Google Scholar]
- 18.van Jaarsveld MT, Helleman J, Boersma AW, van Kuijk PF, van Ijcken WF, Despierre E, Vergote I, Mathijssen RH, Berns EM, Verweij J, et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32:4284–4293. doi: 10.1038/onc.2012.433. [DOI] [PubMed] [Google Scholar]
- 19.Tejero R, Navarro A, Campayo M, Vinolas N, Marrades RM, Cordeiro A, Ruiz-Martinez M, Santasusagna S, Molins L, Ramirez J, Monzó M. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu X, Xiao P, Shi H, Wang R, Chen L, et al. miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res. 2014;20:2617–2630. doi: 10.1158/1078-0432.CCR-13-3224. [DOI] [PubMed] [Google Scholar]
- 21.Feiden S, Feiden W. WHO classification of tumours of the CNS: Revised edition of 2007 with critical comments on the typing und grading of common-type diffuse gliomas. Pathologe. 2008;29:411–421. doi: 10.1007/s00292-008-1064-5. (In German) [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T, Si J. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol. 2009;44:556–561. doi: 10.1007/s00535-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 24.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]