Perspectives piece
Histone post-translational modifications (PTMs) have been implicated in guiding a multitude of diverse cellular processes, including gene expression, DNA replication, chromosome segregation and DNA repair. In particular, acetylation of histone lysine (K) residues – a chromatin mark first discovered by Vincent Allfrey in 1964 – is known to regulate gene expression (Verdin and Ott 2015). In the last few years, our understanding of how histone acetylation contributes to chromatin-based functions has greatly expanded, in part through the identification of reader domains that recognize K acetylation. The first identified K acetylation reader domain was the bromodomain, an ~100 amino acid module found in many chromatin-associated proteins (Dhalluin, Carlson et al. 1999). For years, bromodomains remained in the spotlight as the only known reader of K acetylation. However, the list of K acetylation readers has grown with the discoveries that the double PHD finger (DPF) domain and the YEATS domain are also K acetylation readers (Zeng, Zhang et al. 2010, Sabari, Zhang et al. 2017). Bromo, DPF and YEATS domains occur in diverse chromatin-associated proteins, including K acetyltransferases (KATs), ATP-dependent chromatin remodelers, and K methyltransferases. In this issue of Structure, Kutateladze and co-authors unveil how MORF, a double PHD-containing KAT, is a preferential reader of H3K14 acylation (Klein, Simithy et al. 2017), thereby expanding our knowledge of the types of PTM landscapes reader domains interact with.
New technologies in mass spectrometry have dramatically enhanced our capabilities to detect novel, yet related PTMs and to determine their locations in histones (Huang, Lin et al. 2015). Over the last few years, our knowledge regarding the locations and types of histone PTMs has rapidly expanded. Of the newly discovered PTM types, K acylations that include propionylation (pr), butyrylation (bu), crotonylation (cr), β-hydroxybutyrylation, succinylation, and malonylation, are garnering increasing attention, partly due to recent findings that several of these PTMs contribute to chromatin-based gene transcription (Sabari, Zhang et al. 2017). K butyrylation (Kbu) and crotonylation (Kcr) particularly have been linked to the stimulation of gene transcription in yeast and mammals, and these PTMs exhibit similar genome-wide occupancy patterns with enrichment at promoter regions and enhancers (Sabari, Zhang et al. 2017).
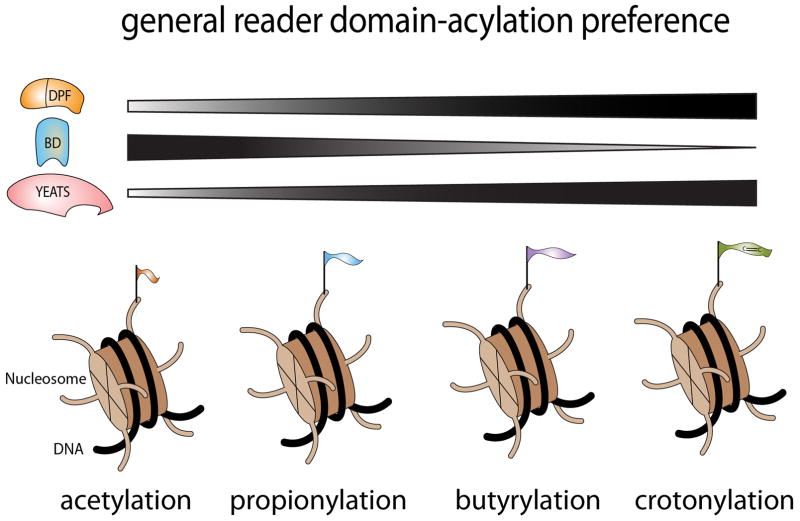
The advances mentioned above raise an intriguing question: How do histone acylations, such as Kbu and Kcr, cause downstream effects in chromatin? Although bromodomains might be likely suspects to bind these PTMs, these domains outside of a few exceptions prefer Kac and Kpr (Figure 1 and Table 1) (Flynn, Huang et al. 2015, Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016). The answer to this puzzle lies in the discovery that the YEATS domain possesses acyl selectivity (Figure 1, Table 1). Structural studies revealed that the YEATS domains of Taf14, YEATS2, and AF9 bind to K acylation through an aromatic cage, with the highest affinity towards crotonylation followed by the other acyl marks approximately in order of the length of the fatty acid chain (Table 1) (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016). Although these studies cracked one piece of the K acyl puzzle, the Li lab has added another K acyl reader to the list with a recent report demonstrating that the DPF domains of MOZ and DPF2 are also selective readers of H3K14 acylation, notably H3K14cr (Xiong, Panchenko et al. 2016). In this issue of Structure, Kutateladze and co-authors now expand our basic understanding of K acyl reading by showing that the DPF module of the MORF KAT is also a selective reader of H3K14 acylation, with a preference for H3K14Kbu (Klein, Simithy et al. 2017).
Figure 1. General preferences of histone readers for the distinct histone lysine acylation states.
Recent studies have uncovered the presence of additional lysine acylation states in histones, including lysine propionylation, butyrylation, crotonylation, β-hydroxybutyrylation, succinylation and malonylation. Investigators are now unveiling how these distinct acylation “flavors” are “read” or interpreted in the context of chromatin. New studies from the Li and Kutateladze groups have added to the list of reader domains that bind the bulkier histone acyl forms that are largely “off limits” to most bromodomains (BD). In this issue of Structure, Kutateladze et al. define the structural basis of the interaction of H3K14bu with the double PHD finger (DPF) of MORF, thus contributing to the finding that the DPF domain is a novel reader of lysine acylation. DPF domains, similar to YEATS domains, largely prefer longer acyl forms of lysines. It will be of great interest to determine how DPF domains contribute to transcriptional regulation through their ability to be recruited to lysine acylation events in chromatin.
Table 1.
Readers of Histone Acylation
| Protein | Domain | Residue(s) | Acylation preference | Reference |
|---|---|---|---|---|
| BRD9 | Bromo | H4 K5, K8 | Pr>Bu>Ac | (Flynn, Huang et al. 2015) |
| CECR2 | Bromo | H4 K5, K8 | Bu>Ac>Pr | (Flynn, Huang et al. 2015) |
| TAF1 | Bromo(2) | H4 K5, K8 | Ac>Pr>Cr>Bu | (Flynn, Huang et al. 2015, Andrews, Shinsky et al. 2016) |
| AF9 | YEATS | H3 K9 | Cr>Pr>Bu>Ac | (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016) |
| YEATS2 | YEATS | H3 K27 | Cr>Bu>Pr>Ac | (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016) |
| ENL | YEATS | H3 K9, 27 | Cr>Pr>Bu>Ac | (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016) |
| GAS41 | YEATS | H3 K9, 27 | Cr>Pr>Bu>Ac | (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016) |
| TAF14 | YEATS | H3 K9 | Cr>Pr>Bu>Ac | (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016) |
| MOZ | DPF | H3 K14 | Cr>Bu>Pr>Ac | (Xiong, Panchenko et al. 2016) |
| DPF2 | DPF | H3 K14 | Cr>Bu>Pr>Ac | (Xiong, Panchenko et al. 2016) |
| MORF | DPF | H3 K14 | Bu>Ac | (Klein, Simithy et al. 2017) |
Similar to the DPF domains of MOZ and DPF2, the DPF domain of MORF shows a slight preference for the longer butyryl-lysine compared with the well-characterized acetyl moiety at the same residue. A hydrophobic binding pocket in the first PHD finger that encapsulates the lipophilic butyryl-lysine dictates the specificity of the DPF domain for longer acylated lysine residues. This mechanism of recognition is distinct from that of the YEATS domain, which utilizes an aromatic cage to form a unique π-stacking interaction with the amide and alkene groups of the crotonyl-lysine, providing an increased affinity for crotonyl-lysine compared with other acyl-lysines. Another feature of the YEATS binding pocket is its openness, which prevents steric clashes with the longer acyl carbon chains (Andrews, Shinsky et al. 2016, Li, Sabari et al. 2016).
As mentioned above, a select number of bromodomains can accommodate butyrl- and/or crotonyl-lysine (Flynn, Huang et al. 2015). BRD9 and CECR2 are two such bromodomains that can read butyryl-lysine; these bromodomains are characterized by a “tyrosine gatekeeper” and “methionine core” that provide increased flexibility in the binding pocket to accommodate the bulkier butyryl moiety. The second bromodomain of TAF1 can bind both butyryl- and crotonyl-lysine. Crotonyl recognition in the TAF1-2 bromodomain displaces water molecules in the binding pocket to allow a coplanar orientation of the crotonyl double bond with the amide group.
The distinct ways in which longer K acyl chains are bound by the different reader domain types argues for distinct physiological functions for the different acyl forms. Intriguingly, Klein et al. also showed that the bulkier K acyl forms (forms larger than acetylation and propionylation) are present at very low levels in vivo (Klein, Simithy et al. 2017). Significantly, it has not been fully elucidated how these various acyl-CoAs end up on chromatin. Klein et al. began to address this knowledge gap by performing histone acyl-transferase assays in vitro with several well-studied KATs in the presence of the various acyl-CoAs. GCN5 and PCAF indiscriminately catalyzed the addition of acetyl, propionyl, and butyryl groups to H3K14; however, they did not add the other assayed acyl groups. MOF catalyzed butyrylation of H3K14, but to a lesser extent than it catalyzed acetylation of the same residue. Interestingly, none of the KATs tested robustly catalyzed the crotonylation of H3K14, although prior evidence has shown that p300 could catalyze crotonylation at H3K18 (Sabari, Tang et al. 2015). Recent studies of the DPF domains of MOZ and DPF2 indicate a DPF domain preference for H3K14cr (Xiong, Panchenko et al. 2016), but it has not been established which KAT installs this modification.
The discovery that the MORF DPF domain is an H3K14 acylation reader raises many exciting questions. For example, what is the biological relevance of MORF DPF binding in gene transcription? Why do the preferred acyl forms of MORF exist at such low levels in vivo? Perhaps answers to these questions will emerge by exploring the metabolism of the acyl-CoA forms. Under different nutrient conditions, humans possess vastly different levels of acyl-CoA (Sabari, Zhang et al. 2017). During fasting when glucose levels are low, fatty acids are oxidized in the liver to generate an alternative source of acetyl-CoA. Propionyl-, butyryl-, and crotonyl-CoA are all generated during this process (Sabari, Zhang et al. 2017). A portion of the acetyl-CoA derived from fatty acid oxidation is converted to β-hydroxybutyrate via ketogenesis. When mice were subjected to similar starvation conditions, the level of histone K-β-hyrodxybutyrylation was dramatically increased, and it correlated with activated transcription of starvation-induced stress response genes (Sabari, Zhang et al. 2017). Clearly, future studies must examine the importance of the various acyl readers, writers, and erasers under different metabolic conditions and disease states, such as diabetic ketoacidosis in which levels of β-hyrodxybutyrate in blood can increase into the mM range (Sabari, Zhang et al. 2017). The stoichiometry of the various histone K-acylations has also been demonstrated to change during cellular differentiation and development, such as in spermatogenesis (Sabari, Zhang et al. 2017). The dynamic environment of histone acylation offers the possibility of an “acyl code” in which, during different metabolic or developmental states, readers of the bulkier acyl groups, such as YEATS and DPF domain-containing proteins, stay bound to chromatin, whereas most bromodomain-containing proteins are excluded.
Acknowledgments
We thank Krzysztof Krajewski for help on Figure 1, Tanya Kutatelazde for helpful discussions, and Howard Fried for editorial suggestions. B.D.S is supported by grants from the NIH (GM068088 and GM110058).
References
- Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, Kutateladze TG. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12(6):396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Flynn EM, Huang OW, Poy F, Oppikofer M, Bellon SF, Tang Y, Cochran AG. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure. 2015;23(10):1801–1814. doi: 10.1016/j.str.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115(6):2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Simithy J, Wang X, Ahn J, Andrews FH, Zhang Y, Cote J, Shi X, Garcia BA, Kutateladze TG. Recognition of Histone H3K14 Acylation by MORF. Structure. 2017 doi: 10.1016/j.str.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, Roeder RG, Shi X, Allis CD, Li H. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol Cell. 2016;62(2):181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, Allis CD. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2017;18(2):90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Xiong X, Panchenko T, Yang S, Zhao S, Yan P, Zhang W, Xie W, Li Y, Zhao Y, Allis CD, Li H. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol. 2016;12(12):1111–1118. doi: 10.1038/nchembio.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466(7303):258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]