Abstract

The emergence of Mycobacterium tuberculosis (MTB) strains that are resistant to most or all available antibiotics has created a severe problem for treating tuberculosis and has spurred a quest for new antibiotic targets. Here, we demonstrate that trans-translation is essential for growth of MTB and is a viable target for development of antituberculosis drugs. We also show that an inhibitor of trans-translation, KKL-35, is bactericidal against MTB under both aerobic and anoxic conditions. Biochemical experiments show that this compound targets helix 89 of the 23S rRNA. In silico molecular docking predicts a binding pocket for KKL-35 adjacent to the peptidyl-transfer center in a region not targeted by conventional antibiotics. Computational solvent mapping suggests that this pocket is a druggable hot spot for small molecule binding. Collectively, our findings reveal a new target for antituberculosis drug development and provide critical insight on the mechanism of antibacterial action for KKL-35 and related 1,3,4-oxadiazole benzamides.
Keywords: Mycobacterium tuberculosis; antibiotics; 1,3,4-oxadiazoles; ribosome rescue
Over 1.8 billion people are infected with Mycobacterium tuberculosis (MTB) worldwide, 10% of whom are predicted to develop the active disease.1 These infections produce 1.5 million deaths annually. Antibiotic treatment has reduced the mortality rate of MTB, but the rise of multidrug resistant (MDR-TB) and extensively drug resistant (XDR-TB) strains has raised an urgent need for new antibiotics.2 Drugs with new chemical scaffolds and new molecular targets are particularly desirable because they are less likely to be counteracted by existing resistance mechanisms in clinical strains. The trans-translation pathway for rescue of nonstop ribosomes presents a potential target for antibiotics because it is required for viability or virulence in many pathogens and is not found in metazoans.3,4trans-Translation is used to rescue ribosomes that are trapped at the 3′ end of an mRNA that has no in-frame stop codon to allow termination. During trans-translation, a specialized RNA molecule, tmRNA, and a small protein, SmpB, recognize these nonstop translation complexes.4 tmRNA acts first like a tRNA to accept the nascent polypeptide, and then, a reading frame within tmRNA is inserted into the mRNA channel. Translation resumes using tmRNA as a message and terminates at a stop codon within tmRNA, releasing the ribosome and a protein with the tmRNA-encoded peptide sequence at its C terminus.4−6 Multiple proteases recognize the tmRNA-encoded peptide and rapidly degrade the protein, thereby clearing both the stalled ribosome and the incomplete polypeptide.7,8 Nonstop translation complexes occur frequently in bacteria because they arise both from damaged mRNAs that lack a stop codon (nonstop mRNA) and from cleavage of mRNAs before or during translation.9 In some bacteria, trans-translation is the only mechanism known to rescue nonstop translation complexes, and both tmRNA and SmpB are essential for viability.10 Other species have the ArfA or ArfB backup systems that can release ribosomes from nonstop translation complexes in the absence of trans-translation.11,12 The MTB genome does not encode ArfA or ArfB, suggesting that trans-translation is likely to be essential and, therefore, a good candidate for target-based drug development. Despite a report that the antituberculosis drug pyrazinamide targets trans-translation,13 careful experiments have shown that trans-translation is not inhibited by pyrazinamide or its active metabolite, pyrazinoic acid, in vitro or in vivo.14 Therefore, there are currently no antibiotics that target this pathway.
Results and Discussion
SmpB Is Essential in M. tuberculosis
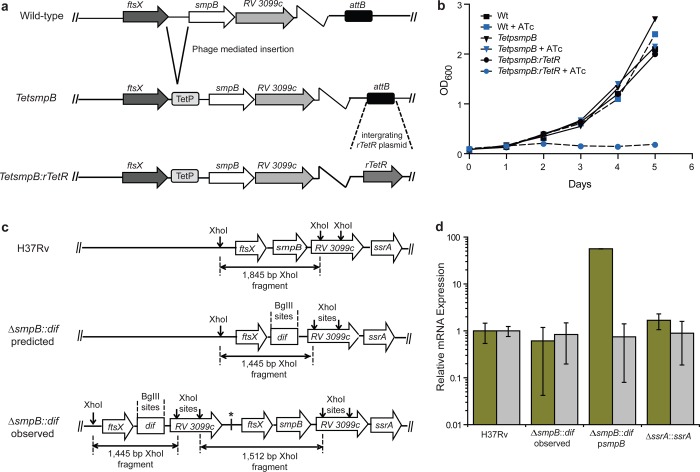
To assess the importance of trans-translation in MTB, we first attempted to delete the genes encoding tmRNA (ssrA) and SmpB (smpB) from the MTB chromosome using allelic exchange, but we could not obtain a deletion of either gene. To rigorously determine if trans-translation is essential in MTB, we engineered a strain (TetpsmpB:rTetR) in which the expression of smpB at its chromosomal locus is controlled by the tet repressor (TetR), such that addition of anhydrotetracycline (ATc) shuts off SmpB production (Figure 1a). TetpsmpB:rTetR cells grew at a similar rate to wild-type cells in the absence of ATc, but addition of ATc severely inhibited growth (Figure 1b). Addition of ATc had no effect on growth of wild-type cells or control strains lacking tetR (Figure 1b). These data indicate that SmpB is required for growth of MTB in culture. This conclusion is consistent with data from saturating transposon mutagenesis screens that failed to recover insertions in ssrA or smpB15 and with data demonstrating that the chromosomal copy of ssrA could only be deleted in the presence of an additional copy of the gene.16 A MTB strain deleted for smpB has been reported,16 but whole-genome sequencing of this strain showed that the smpB coding sequence was present (Figure 1c; GenBank accession numbers: SAMN05907893 and SAMN05907849). qRT-PCR to detect the SmpB mRNA in this deletion strain, ΔsmpB::dif, revealed that the gene is expressed (Figure 1d). Taken together, these results demonstrate that trans-translation is essential for growth in MTB.
Figure 1.
SmpB is essential in MTB. (a) Schematic illustration for the design of the smpB depletion constructs in MTB. (b) Growth curves for the SmpB depletion and control strains. (c) Schematic diagram of the smpB locus in the parental H37Rv strain, the reported ΔsmpB::dif,16 and ΔsmpB::dif observed from whole genome sequencing showing that the ΔsmpB::dif strain has a copy of smpB. (d) qRT-PCR analysis showing that both ssrA and smpB are expressed in the ΔsmpB::dif strain. smpB (yellow) and ssrA (gray) mRNA levels in midexponential phase MTB cells were quantified by qRTPCR and normalized to the housekeeping gene sigA. Mean values from 3 technical replicates of one biological sample are shown with error bars indicating the standard deviation.
KKL-35 Kills Growing and Nonreplicating Persister Cells of M. tuberculosis
KKL-35 (Figure 2a) and related 1,3,4-oxadiazole benzamides were identified by cell-based screening for inhibitors of trans-translation and were found to have broad-spectrum antibacterial activity.17,18 To assess the ability of KKL-35 to inhibit growth of MTB, MIC and plating assays were performed. KKL-35 inhibited growth of MTB cultures with a MIC of 1.6 μg/mL, and plating assays showed that 8.0 μg/mL KKL-35 killed >90% of MTB cells within 7 days (Figure 2b,c). Tuberculosis infections can be difficult to treat in part because MTB cells can enter a nonreplicating persister state in which they are not sensitive to most antibiotics. We used a hypoxia persistence model19 to evaluate the activity of KKL-35 against nonreplicating persister bacilli. 1.6 μg/mL KKL-35 killed >90% of nonreplicating MTB cells under these conditions, demonstrating that KKL-35 was equally active against nonreplicating MTB cells as it was against actively growing cells (Figure 2d). The observed activity against persister cells suggests that trans-translation is required for survival in this state and indicates that trans-translation inhibitors may be effective against multiple physiological states of MTB during infection. Despite its potency against MTB, KKL-35 and its analogs displayed no cytotoxic activity against HeLa cells (Table 1) or HepG2 cells18 at concentrations >20-fold MIC. The combination of significant antibiotic activity against MTB and low cytotoxic activity for KKL-35 indicates that this compound is a promising antitubercular agent.
Figure 2.
Compound structures and activity for KKL-35 against MTB. (a) Structures of the 1,3,4-oxadiazole benzamides KKL-35, KKL-40, the photolabile click probe KKL-2098, and the trifunctional fluorescent molecule, KKL-2107. (b) Growth inhibitory profiles for MTB cultures treated with KKL-35 and monitored by luminescence. (c) CFU counts for MTB liquid cultures treated with KKL-35. (d) CFUs recovered from MTB cells grown using the hypoxia model for nonreplicating persisters treated with KKL-35. The median from two replicates is shown with error bars indicating the standard deviation.
Table 1. Comparison of the Antibacterial Activity for KKL-35, KKL-40, and KKL-2098a.
| MICb (μg/mL) |
cytotoxicityh | |||||
|---|---|---|---|---|---|---|
| compound ID | B. anthracisc | E. coli ΔtolCd | S. flexnerie | M. smegmatisf | MTBg | HeLa (μg/mL) |
| KKL-35 | 0.3 (0.1) | 0.5 (0.1) | 1.6 (0) | 0.4 (0) | 1.6 (0) | >31.8 |
| KKL-40 | 0.1 (0) | 0.2 (0) | 1.8 (0) | 0.3 (0) | 1.8 (0) | >31.8 |
| KKL-2098 | 0.3 (0.1) | 0.5 (0.1) | 1.7 (0) | 0.4 (0) | ND | ND |
For footnotes c to f, MICs for KKL-35 and KKL-40 for these strains have been previously reported.17 ND: not determined.
Data are averages from three independent assays each performed in triplicate (SD).
Sterne strain 34F2.
Strain MG1655.
Strain 2a 2457T.
Strain mc2155 (ATCC 700084).
Erdman strain.
From >3 independent determinations.
KKL-35 Targets Helix 89 of 23S rRNA
To determine the molecular target for KKL-35, we designed and synthesized an analog, KKL-2098, incorporating a photoreactive azide group and a terminal alkyne moiety (Figure 2a, Scheme 1). The MICs for KKL-2098 against Mycobacterium smegmatis and other bacterial species were similar to those for KKL-35 (Table 1). The similarity in activity suggests that the structural modifications in this analog did not significantly alter antibiotic properties or target binding of the compound. We therefore used M. smegmatis for the KKL-35 target identification. Intracellular photoaffinity labeling followed by click bioconjugation was used in the molecular target identification process (Figure 3).20,21
Scheme 1. Synthesis of the Dual Function Photo-Reactive Click Probe: 4-Azido-N-(5-(4-ethynylphenyl)-1,3,4-oxadiazol-2-yl)benzamide (KKL-2098).
Reagent conditions: (a) NaNO2, HCl, H2O, 0 °C, 1 h; (b) NaN3, HCl, H2O, 0 °C, 1 h; (c) SOCl2, reflux 12 h; (d) NaOH, MeOH, rt, 4 h; (e) 1 N HCl, pH 2; (f) POCl3, reflux, 12 h; (g) pyridine, 50 °C, 12 h.
Figure 3.
Target identification workflow. The photolabile probe KKL-2098 was added to a growing bacterial culture. Cells were irradiated with UV light to activate the probe and enable cross-linking. Cells were lysed, and protein was denatured and subjected to click chemistry with the fluorescent affinity compound KKL-2107 and analyzed by SDS-PAGE. Alternatively, total RNA was purified and used in click conjugation assays with KKL-2107 or primer extension assays to detect RNA modification. Agarose or polyacrylamide gel electrophoresis was used to visualize and identify the probe-linked macromolecule.
To facilitate isolation and visualization of the target, we also synthesized a trifunctional probe, KKL-2107 (Figure 2a, Scheme 2), that incorporated an azide group, an affinity conjugate, and a fluorescent moiety. The target was identified by incubating KKL-2098 with growing M. smegmatis cells and irradiating the culture with UV light to initiate cross-linking (Figure 3). Following cross-linking, the cells were lysed and click conjugation was used to attach the fluorescent molecule (KKL-2107) to the alkyne moiety of KKL-2098, facilitating purification and visualization of cross-linked molecules. Analysis of proteins using SDS-PAGE showed no fluorescent bands, indicating that KKL-2098 was not cross-linked to a protein (Figure S1). However, analysis of RNA preparations from KKL-2098-treated cells revealed a fluorescent band that comigrated with 23S rRNA on agarose gels (Figure 4a). Similar results were obtained when cross-linking was repeated with RNA extracts from M. tuberculosis and E. coli (Figure S2). Primer extension assays were used to confirm that KKL-2098 was cross-linked to 23S rRNA. Assays using RNA from KKL-2098-treated M. smegmatis cells reproducibly showed a prominent band that was not present in control reactions using RNA from cells treated with KKL-35 instead of KKL-2098 (KKL-35 will not cross-link but causes the same physiological response in the cells) (Figure 4b, Figure S3). This band indicated that reverse transcriptase activity was terminated after nucleotide 2505 (E. coli numbering), suggesting KKL-2098 was cross-linked to nucleotide Ψ2504 (Figure 4b). Primer extension on 23S rRNA from E. coli after cross-linking with KKL-2098 indicated modification of nucleotide C2452 (Figure S4), which base pairs with Ψ2504.22 Ψ2504 and C2452 are positioned at the base of H89, a structure that extends from the PTC to the factor binding site (Figure 4c,d).23 Nucleotides that border the PTC and those located at the base of H89 adjacent to the PTC (Figure 5) are highly conserved and essential for efficient ribosome function in bacteria.23,24 Mutation of Ψ2504, C2452, or other nearby nucleotides, as well as conformational changes in H89, have been shown to have moderate to severe effects on translation fidelity, ribosome function, and/or cell growth.24−27 Collectively, these data indicate that the base of H89 is the target for KKL-35 and related 1,3,4-oxadiazole benzamides.
Scheme 2. Synthesis of the Tri-Functional Fluorescent Reporter N-(6-((6-Azidohexyl)(6-(5-(dimethyl amino) Naphthalene-1-sulfonamido)hexyl)amino)hexyl)-5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl) Pentanamide (KKL-2107).
Reagent conditions: (a) Et3N, CH2Cl2, 0 °C, rt, 12 h; (b) DIPEA, CH3CN, reflux 12 h; (c) DIPEA, CH3CN, reflux 12 h; (d) TFA/CH2Cl2 (1:1), 0 °C, rt, 3 h; (e) Et3N, MeOH, rt, 12 h.
Figure 4.
KKL-2098 binds 23S rRNA. (a) Agarose gel analysis for the click conjugation and control reactions with total RNA preparations from M. smegmatis cells. (b) Autoradiogram showing primer extension results using RNA prepared from cells treated with KKL-35 or KKL-2098. The arrow indicates the extension product seen only in the cross-linked sample treated with KKL-2098 (see Figure S3 for the full gel and experiments using other primers). (c) Structure of the E. coli ribosome (PDB ID 4V69) showing the location of H89 (magenta) extending from the PTC (green) to the factor binding site (purple). (d) Cartoon and surface structures of H89 showing the location of Ψ2504 and C2452.
Figure 5.
Nucleotides that cross-link to KKL-2098 are highly conserved. Comparison of the nucleotide sequence and secondary structures of H89. The KKL-2098 cross-link sites are indicated by asterisks. Mutation of the nucleotides highlighted in red is known to impair peptidyl-transferase activity, ribosome fidelity/integrity, or cell growth in bacteria.24−27
Docking and Solvent Mapping Predict Binding Site for 1,3,4-Oxadiazole Benzamides
Guided by results from the cross-linking experiments, we performed in silico molecular docking studies using the Autodock-Vina program.28 KKL-35 was docked to a region of the 50S ribosome encompassing the PTC and the full-length H89 to the edge of the factor-binding site (Figure S5). The 6 lowest energy structures had KKL-35 in the same pocket at the base of H89 near nucleotides Ψ2504 and C2452 (Figure 6). The conformation of KKL-35 within this pocket varied, but in all cases, the main contributors to the binding energy based on the docked conformation were the predicted polar interactions between KKL-35 and H89 originating from the carbonyl oxygen atom and the oxadiazole core (Figure 6). The latter contribution is in agreement with experimental evidence showing favorable electron donor capabilities for the 1,3,4-oxadiazole ring as a result of a large dipole moment generated by the nitrogen atoms.29 Docking experiments using KKL-2098 and KKL-40 localized these compounds to the same pocket, with similar docked conformations and binding energies as KKL-35 (ΔG = −8.8 kcal/mol for KKL-35, −9.0 kcal/mol for KKL-2098, and −9.5 kcal/mol for KKL-40) (Figure S5). In some structures, the oxadiazole-amide core was oriented in a conformation that would place the azide group of KKL-2098 in position to cross-link with Ψ2504 and C2452, but in others, the orientation of the oxadiazole-amide was reversed (Figure S5). The similar docked energies of the forward and reverse conformations are a result of partial symmetry in the molecules based on the location H-donor and acceptor atoms in the oxadiazole-amide core and the phenyl-rings (Figure S5). Therefore, the docking studies predict a binding pocket for 1,3,4-oxadiazole benzamides at the base of H89 but do not specify the conformation of the molecules within this pocket. Because the ribosome structure bound by 1,3,4-oxadiazole benzamides may be subtly different from the available crystal and cryo-EM structures, we cannot exclude the possibility that these molecules bind in the PTC where they could contact the other face of the Ψ2504–C2452 base pair. Other antibiotics, including linezolid, bind in the PTC close to Ψ2504 and C2452.30 Unlike linezolid, KKL-35 and other oxadiazoles do not inhibit translation,17 so they would have to bind within the PTC in a manner that inhibited trans-translation but not translation. Ongoing structural studies should provide more insight on the binding site for 1,3,4-oxadiazole benzamides.
Figure 6.
KKL-35 docks to a predicted binding hot spot in H89. (a) Docked KKL-35 (cyan) in a pocket at the base of H89 adjacent to the PTC (PDB ID 4ABR). (b) Lateral view (from the loop region) of the docking site for KKL-35 illustrating potential polar interactions (dashed lines). (c) Reversed view (from the PTC side) of the binding site. (d) Solvent mapping results for H89 showing probe clustering. The dotted region indicates a hot spot located within the predicted binding site for KKL-35. (e) Close-up of the highlighted hot spot in “d” showing the docked KKL-35 and FTMap probe clusters. Probes are color-coded to distinguish between different consensus sites (CSs): CS1-(9), CS2-(12), CS3-(11), and CS4-(4) (number of probe clusters in each CS in parentheses).
We used a computational solvent mapping algorithm (FTMap) to assess solvent accessibility and druggability of H89.31 The FTMap server performs solvent mapping using standard probes with variable chemical structures to identify probable drug binding hot spots within a macromolecule.31 These hot spots, typically located within probe-cluster consensus sites (CS), are regions capable of binding a variety of chemically diverse probes and are predicted to contribute significantly to the binding free energy.31,32 Probe clustering was observed at several sites on H89 (Figure 6d), but the base region had one major hot spot which comprised four CSs (Figure 6d, Figure S6). Structural alignment of docked KKL-35 with solvent mapping revealed that the predicted binding pocket for KKL-35 superimposes with the solvent-mapped hot spot with probe cluster coverage along the entire KKL-35 structure (Figure 6e). These mapping data together with the docking results for KKL-35 support the presence of a druggable binding pocket for 1,3,4-oxadiazole benzamides at the base of H89 adjacent to the PTC.
KKL-35 Selectivity Suggests Structural Changes in the Ribosome during Rescue
The data presented here indicate that KKL-35 bind to a highly conserved region of the ribosome, and previous results showed that KKL-35 inhibits trans-translation but not translation initiation, elongation, or termination.17 Binding of a drug near the PTC might be expected to interfere with translation and mutation of nucleotides that form the KKL-35 binding site (for example, U2460, U2492, and U2493) (Figure 5b), leading to significantly decreased PTC activity and impaired cell growth.24−27 However, KKL-35 did not inhibit translation of mRNAs containing an in-frame stop codon when tested at concentrations >100-fold above the IC50 for inhibiting trans-translation in vitro.17 KKL-2098 cross-linked to 23S rRNA during in vitro translation of mRNAs containing an in-frame stop codon as well as translation of nonstop mRNAs (Figure S2), indicating that the binding site may be accessible during normal translation. No cross-linking was observed in reactions that did not contain mRNA (Figure S2), suggesting that the inhibitors bind a structure that is only present during translation. How could binding of KKL-35 to H89 inhibit trans-translation but not translation? One possible explanation is that binding of KKL-35 could introduce polar interactions that limit flexibility of H89, preventing structural changes that are required for ribosome rescue but not for translation. This selectivity might also explain the ability KKL-35 to inhibit ribosome rescue by ArfA and ArfB in E. coli and C. crescentus: these alternative rescue pathways recognize nonstop translation complexes in the absence of trans-translation.11,12
In summary, the 1,3,4-oxadiazole benzamides present a unique chemical scaffold that is distinct both in structure and mechanism of action from existing antituberculosis drugs.33,34 KKL-35 and its analogs display promising bactericidal activity against actively growing and nonreplicating persister cells of MTB while exhibiting minimal cytotoxicity against eukaryotic cells (Table 1).17 Collectively, these properties make the 1,3,4-oxadiazole benzamides good antitubercular drug candidates. In an effort to circumvent current resistance trends, the druggable state of trans-translation in MTB presents an excellent opportunity to develop novel antituberculosis drugs.
Methods
Bacterial Strains and Growth Conditions
M. tuberculosis H37Rv, ΔsmpB::dif, ΔsmpB::dif::smpB, and ΔssrA::ssrA (gifts from Prof. Tanya Parish)16 and the Erdman TMC 107 (ATCC 35801) strain were cultured at 37 °C in 7H9 media (Difco, Becton Dickinson, Franklin lakes, NJ) supplemented with 10% OADC (Middlebrook), 0.5% glycerol, and 0.05% TWEEN 80. Solid medium plates were prepared using 7H10 agar (Difco) supplemented with 10% OADC (Middlebrook) and 0.5% glycerol. E. coli ΔtolC (MG1655) was cultured in LB growth medium at 37 °C.
MIC and MBC Determination for KKL-35 against M. tuberculosis
One milliliter of culture was grown in 5 mL bottles to OD600 = 0.0125 in 5 mL ink wells, and KKL-35 was added at concentrations ranging from 0 to 1 mM. Cultures were incubated at 37 °C, and the MIC was recorded as the minimum concentration of drug that inhibited visible growth. The MBC was obtained through CFUs, which were determined by plating serial dilutions of cultures onto 7H10 agar plates. Plates were incubated for 3–4 weeks at 37 °C prior to enumeration of CFUs.
Assessment of Growth Inhibition
Autoluminescent M. tuberculosis (LuxTB) was generated by transforming the wild-type Erdman strain with the pMlux plasmid, encoding the mycobacterial MOPS promoter driving expression of a synthetic GC-rich luxCDABE operon from P. luminescens.35,36 LuxTB was cultured in 7H9 medium to OD600 = 0.0125. KKL-35 was added, and the cultures were incubated at 37 °C. Luminescence readings were recorded every 24 h for 12 days using an Infinite M200 plate reader (Tecan Trading AG, Mannedorf, Switzerland).
M. tuberculosis SmpB Depletion Assays
The TetpsmpB:rTetR mutant was constructed by replacing 500 bp upstream of the smpB ATG start site with a tet operator (tetO)-containing mycobacterial promoter (Psymc).37 This mutation was made by homologous recombination using a specialized mycobacterium phage system as previously described.38 After the addition of the Psymc promoter, the strain was transformed with a plasmid that integrates at the attB site and expresses the reverse repressor TetR.37 Repression of SmpB was achieved by incubation of cells with 300 ng/mL of anhydrotetracycline (Sigma-Aldrich, St. Louis, MO).
M. tuberculosis Hypoxia Assay
For growth under hypoxia, MTB was grown in 17 mL glass test tubes in triplicate and gradual hypoxia was generated using the Wayne Model.19,39 KKL-35 was added to hypoxic cultures at various concentrations, and the cultures were incubated for 1 week. These cultures were then spread on 7H10 agar plates and incubated at 37 °C for 3–4 weeks before enumeration of the CFUs.
Genome Sequencing
Full genome sequencing of strain ΔsmpB::dif was performed by generating a library from randomly sheared 350 bp genomic DNA fragments using a TruSeq DNA Kit (Illumina Inc., San Diego, CA) following the manufacturer’s protocol. Paired-end sequencing was performed for 100 cycles using an Illumina HiSeq 2500 by the University of Minnesota Genomics Center. Approximately 1.3 GB of data was obtained representing >300-fold sequence coverage (NCBI BioProject accession number PRJNA343132). High-quality paired-end reads were trimmed using Cutadapt (http://cutadapt.readthedocs.io/en/stable/guide.html#trimming-paired-end-reads) and mapped to the H37Rv reference genome sequence40 using Geneious 6.0 (Biomatters Ltd., Auckland, New Zealand). Sequence of the smpB region was independently verified by sequencing of PCR amplicons covering open reading frames from Rv3098c to Rv3102c.
qRT-PCR Analysis of smpB Expression
Midexponential phase cultures of strains H37Rv, ΔsmpB::dif, ΔsmpB::dif::smpB, and ΔssrA::ssrA were harvested by centrifugation; cell pellets were resuspended in buffer containing 10 mM Tris-HCl, 1 mM EDTA, and 15 mg/mL lysozyme and incubated at 37 °C for 16 h. RNA was extracted using the E.Z.N.A. bacterial RNA kit (Omega Biotek, Norcross, GA). Residual DNA was removed using the TURBO DNA-free kit (Life Technologies Corp., Grand Island, NY). qRT-PCR was performed with the QuantiFast SYBR Green RT-PCR kit (Qiagen). qRT-PCR reactions were prepared with 2X QuantiFast SYBR Green RT-PCR master mix, 10 μM primers, 0.1 μL of QuantiFast RT Mix, and 1 ng of RNA and were run on a LightCycler480 with the following cycle conditions: 50 °C for 10 min, 95 °C for 5 min, 35 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s with fluorescence quantification for each cycle. A melting curve cycle of 95 °C for 15 s, 60 °C for 15 s, and 95 °C with 2% ramp rate was used to determine product specificity. A no reverse transcriptase qRT-PCR control reaction was performed to test for contaminating DNA. Primers to express the mature tmRNA were used for these studies.41 tmRNA primer sequences were as follows: MSTSSRA-5 TGCAGGCAAGAGACCACCGTA; MTSSRA-6 CCGGTCACGCGAACTAGCCGAGA.
Bioorthogonal Photoaffinity Labeling with KKL-2098
Intracellular photolabeling was performed by adding either KKL-35 or KKL-2098 at the MIC to midexponential cultures of M. smegmatis, M. tuberculosis, or E. coli ΔtolC. These cultures were grown for 1 h, and cells were harvested by centrifugation at 2716g and resuspended in phosphate buffered saline solution (1.2 g of Na2HPO4, 0.22 g of NaH2PO4, 8.5 g of NaCl in 1 L, pH 7.5). This suspension was irradiated with 312 nm UV light for 10 min, and cells were recovered by centrifugation. RNA extracts were prepared using the Norgen total RNA preparation kit (Norgen Biotek Corp., Thorold, ON, Canada) according to the manufacturer’s protocols, and the purity of the isolated RNA was assessed by agarose gel electrophoresis.
Primer Extension Assays
DNA oligonucleotides covering 23S rRNA were end-labeled with [32]P using polynucleotide kinase (NEB, Ipswich, MA) according to the manufacturer’s instructions. Primer extension assays42 were performed using RNA from KKL-35 and KKL-2098-treated cells with Superscript II reverse transcriptase (Thermo Fisher Scientific, Bellefonte, PA) according to the manufacturer’s instructions. The products were separated on an 8% polyacrylamide urea gel and visualized using a Typhoon 9410 imager (GE Healthcare, Tyrone, PA). These experiments were repeated using Superscript IV and Sunscript reverse transcriptase for confirmation of modified sites.
Oligonucleotide sequences used in the primers extension assays:
Copper Catalyzed Azide–Alkyne Huisgen Cycloaddition (AAHC) Click Conjugation Assay
RNA
Click conjugation reactions were performed in a 20 μL scale by combining 8 μL of acetonitrile (final 40% v/v), KKL-2107 (1 mM final), 2 μL of 1 M Hepes (pH 7.4, final 100 μM), 8.6 μL of RNA solution, 2 μL of premixed solution of CuSO4/THPTA (final concentrations of 0.1 and 1 mM, respectively), and NH2NH2 (final concentration of 0.1 mM). Samples were mixed, and the reaction was incubated at room temperature for 15 min. Fifteen μL of 2× formamide loading buffer was added, and the sample was incubated at 65 °C for 10 min. Samples were analyzed by gel electrophoresis on a 1% agarose TAE gel.
Protein
KKL-35- or KKL-2098-treated cell pellets were resuspended in lysis buffer (100 mM NaH2PO4 (pH 7.5), 100 mM NaCl, 0.1% SDS, 2 mM BME), lysed by sonication, and clarified by centrifugation at 22 000g for 10 min. The lysate was then subjected to click conjugation by mixing 100 μL of acetonitrile, KKL-2107 (final concentration of 1 mM), 172 μL of clarified lysate, a premixed solution of CuSO4/THPTA (final concentrations of 0.4 and 2 mM, respectively), and NH2NH2 (final concentration of 0.1 mM). Reactions were incubated at room temperature for 3 h with gentle agitation, and protein was precipitated by addition of acetone. The recovered protein pellet was air-dried and redissolved in binding buffer (100 mM Na3PO4, 100 mM NaCl, 0.1% SDS, 2 mM BME). For affinity chromatography, NeutrAvidin (Thermo Fisher Scientific, Bellefonte, PA) agarose resin was equilibrated in binding buffer according to the manufacturer’s protocols. A mixture of the resin and lysate was incubated for 1 h at room temperature with gentle agitation and then transferred to a column. The column was washed with 10 volumes of binding buffer; the resin was transferred to a clean tube, and protein was eluted by addition of 1× SDS sample buffer (pH 6.8, 34.2 mM Tris, 13.1 mM glycerol (w/v), 1% SDS, 0.01% bromophenol blue) and incubation at 95 °C for ∼5 min. Samples were analyzed by SDS PAGE.
In Vitro Photolabeling and Click Conjugation
Assays were set up using the PURExpress in vitro protein synthesis kit (NEB, Ipswich, MA) according to the manufacturer’s protocols. The reactions were performed with no DNA template, a nonstop DHFR template, the full length DHFR gene, full length DHFR gene with 0 bases after the stop codon, or full length DHFR gene with 33 bases after the stop codon.16,43 KKL-2098 (final concentration of 1 μM) was added to a mixture of assay components, and the samples were incubated at room temperature for 1 h. Samples were placed on ice, irradiated with 312 nm UV light for 10 min, and used to set up click conjugation assays in the presence of KKL-2107 (final concentration of 0.5 mM). After incubating for 30 min, an equal volume of 2× formamide loading buffer was added, and the tubes were incubated at 65 °C for 10 min. The samples were resolved on a 1% agarose gel. The gel was first scanned for fluorescence (to visualize the conjugated probe) and then stained with ethidium bromide to visualize the RNA.
In Silico Molecular Docking and Solvent Mapping
Molecular docking studies were performed with the AutoDock Vina program28 utilizing the AutoDock tools graphical interface. Energy minimizations for KKL-35 and KKL-2098 were performed using the Open Babel module. Modeling, structural manipulation, and visualization were performed using PyMOL (Schrödinger). Receptor grid maps were generated using the AutoGrid module and KKL-35 or KKL-2098 docked using the Lamarckian genetic algorithm. Docking for KKL-35 and KKL-2098 to the 70S ribosome was guided by results from the cross-linking experiments with KKL-2098. The dimensions of the dock-search space were adjusted to encompass the peptidyl-transfer center and helix 89 of the 50S ribosomal crystal structure. These studies were performed with multiple crystal structures: PDB ID: 4ABR, 4V69, 3DLL, and 4V7T. The best binding conformations were selected from the 10 generated from the docking event based on a criterion combining the highest dock score and the lowest root-mean-square deviation values (RMSD < 1).
Computational Solvent Mapping
The H89 structure was extracted from the protein databank ribosome structure (PDB ID: 4ABR). All water molecules and ions were removed, and mapping was performed using the FTMap algorithm31,32 (Boston University, MA) remotely through its servers online (http://ftmap.bu.edu). The entire surface of H89 was scanned with a mini probe library of 16 organic small molecules with variable hydrophobic and hydrogen bonding properties. The program utilized CHARMM energy minimized conformations for all the probes to scope from potential binding sites on H89. The algorithm retained six bound clusters with the lowest mean interaction energies for each probe. Probe-cluster consensus sites (CS) were then identified from congregated groups of structurally diverse probe. Each of the CSs represented a potential binding hot spot within H89 and was ranked on the basis of the number of probe clusters it contained. The CS with the largest number of probe clusters characterized the most probable small molecule binding sites.
Acknowledgments
The authors thank Yusuke Minato for assistance with MTB MIC determinations, Teresa Repasy for assistance with MTB studies, Tanya Parish for providing strains, and Amber Miller for performing toxicity screens against HeLa cells. J.N.A. is grateful to Steven J. Benkovic for the use of the chemical synthesis facilities. This work was supported by NIH grants GM068720 and AI132275 to K.C.K., DP1AI124619 to J.S.C., and NIH grant AI123146 and the Bill and Melinda Gates Foundation Grand Challenges Explorations grant to A.D.B.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00028.
Results for additional in vitro biochemical assays, in silico molecular modeling, and experimental details for the chemical synthesis (PDF)
Accession Codes
Whole genome sequencing results for the MTB SmpB deletion strains are accessible via GenBank using the accession numbers SAMN05907893 and SAMN05907849.
Author Present Address
‡ P.S.M.: Genentech, Immunology Department, San Francisco, California, USA.
Author Present Address
⊥ N.D.P.: University of Massachusetts Medical School, Worcester, Massachusetts, USA.
Author Present Address
∥ T.L.: Department of Cellular and Molecular Pharmacology, University of California, San Francisco, California, USA.
Author Contributions
J.N.A., K.C.K., and J.S.C. conceived the study. J.N.A., K.C.K, A.D.B., and J.S.C. designed the experiments. J.N.A. performed chemical synthesis and compound characterization, in vitro biochemical analysis, target identification, and in silico modeling studies. J.N.A. and K.C.K. performed the primer extension assays. P.S.M. and T.L. screened for activity against MTB, performed smpB knockout experiments, and carried out the MTB hypoxia model assays. N.D.P. and A.D.B. generated, analyzed, and interpreted full genome sequencing data and qRT-PCR data for MTB strains H37Rv, ΔsmpB::dif, ΔsmpB::dif::smpB, and ΔssrA::ssrA. J.N.A., K.C.K., J.S.C., and A.D.B. prepared and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- World Health Organization. (2016) Global tuberculosis report, World Health Organization, Geneva, Switzerland, http://www.who.int/tb/publications/global_report/en/. [Google Scholar]
- Kurz S. G.; Furin J. J.; Bark C. M. (2016) Drug-Resistant Tuberculosis: Challenges and Progress. Infect. Dis. Clin. North Am. 30, 509–522. 10.1016/j.idc.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C.; Alumasa J. N. (2013) The potential of trans-translation inhibitors as antibiotics. Future Microbiol. 8, 1235–1237. 10.2217/fmb.13.110. [DOI] [PubMed] [Google Scholar]
- Keiler K. C. (2008) Biology of trans-translation. Annu. Rev. Microbiol. 62, 133–151. 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler K. C.; Waller P. R.; Sauer R. T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Giudice E.; Macé K.; Gillet R. (2014) Trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front. Microbiol. 5, 113. 10.3389/fmicb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H.; Kurita D.; Muto A. (2014) tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front. Genet. 5, 66. 10.3389/fgene.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H.; Nameki N.; Kurita D.; Muto A.; Abo T. (2015) Ribosome rescue systems in bacteria. Biochimie 114, 102–112. 10.1016/j.biochi.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Ito K.; Chadani Y.; Nakamori K.; Chiba S.; Akiyama Y.; Abo T. (2011) Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One 6, e28413. 10.1371/journal.pone.0028413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C. (2015) Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297. 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- Chadani Y.; Ono K.; Kutsukake K.; Abo T. (2011) Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from ssrA and ArfA-mediated pathways. Mol. Microbiol. 80, 772–785. 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- Chadani Y.; Ito K.; Kutsukake K.; Abo T. (2012) ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl–tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 86, 37–50. 10.1111/j.1365-2958.2012.08190.x. [DOI] [PubMed] [Google Scholar]
- Shi W.; Zhang X.; Jiang X.; Yuan H.; Lee J. S.; Barry C. E.; Wang H.; Zhang W.; Zhang Y. (2011) Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333, 1630–2. 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N. A.; Peterson N. D.; Feaga H. A.; Keiler K. C.; Baughn A. D. (2017) Anti-tubercular activity of pyrazinamide is independent of trans-Translation and RpsA. Sci. Rep. 7, 6135. 10.1038/s41598-017-06415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J.; Ioerger T. R.; Huttenhower C.; Long J. E.; Sassetti C. M.; Sacchettini J. C.; Rubin E. J. (2012) Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 8, e1002946. 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personne Y.; Parish T. (2014) Mycobacterium tuberculosis possesses an unusual tmRNA rescue system. Tuberculosis 94, 34–42. 10.1016/j.tube.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Ramadoss N. S.; Alumasa J. N.; Cheng L.; Wang Y.; Li S.; Chambers B. S.; Chang H.; Chatterjee A. K.; Brinker A.; Engels I. H.; Keiler K. C. (2013) Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. U. S. A. 110, 10282–10287. 10.1073/pnas.1302816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralski T. D.; Dewan K. K.; Alumasa J. N.; Avanzato V.; Place D. E.; Markley R. L.; Katkere B.; Rabadi S. M.; Bakshi C. S.; Keiler K. C.; Kirimanjeswara G. S. (2016) Inhibitors of Ribosome Rescue Arrest Growth of Francisella tularensis at All Stages of Intracellular Replication. Antimicrob. Agents Chemother. 60, 3276–3282. 10.1128/AAC.03089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey C. D.; Voskuil M. I. (2015) In vitro models that utilize hypoxia to induce non-replicating persistence in Mycobacteria. Methods Mol. Biol. 1285, 201–213. 10.1007/978-1-4939-2450-9_11. [DOI] [PubMed] [Google Scholar]
- Sumranjit J.; Chung S. J. (2013) Recent Advances in Target Characterization and Identification by Photoaffinity Probes. Molecules 18, 10425–10451. 10.3390/molecules180910425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alumasa J. N.; Keiler K. C. (2015) Clicking on trans-translation drug targets. Front. Microbiol. 6, 498. 10.3389/fmicb.2015.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Z.; Matzov D.; Krupkin M.; Paukner S.; Riedl R.; Rozenberg H.; Zimmerman E.; Bashan A.; Yonath A. (2016) A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism. Sci. Rep. 6, 39004. 10.1038/srep39004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek N.; Mankin A. S. (2005) The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 40, 285–311. 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- Burakovsky D. E.; Sergiev P. V.; Steblyanko M. A.; Konevega A. L.; Bogdanov A. A.; Dontsova O. A. (2011) The structure of helix 89 of 23S rRNA is important for peptidyl transferase function of Escherichia coli ribosome. FEBS Lett. 585, 3073–3078. 10.1016/j.febslet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- Porse B. T.; Garrett R. A. (1995) Mapping important nucleotides in the peptidyl transferase centre of 23S rRNA using a random mutagenesis approach. J. Mol. Biol. 249, 1–10. 10.1006/jmbi.1995.0276. [DOI] [PubMed] [Google Scholar]
- O’Connor M.; Dahlberg A. E. (1995) The involvement of two distinct regions of 23S ribosomal RNA in tRNA selection. J. Mol. Biol. 254, 838–847. 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- Long K. S.; Poehlsgaard J.; Hansen L. H.; Hobbie S. N.; Böttger E. C.; Vester B. (2009) Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol. Microbiol. 71, 1218–1227. 10.1111/j.1365-2958.2009.06596.x. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström J.; Hogner A.; Llinàs A.; Wellner E.; Plowright A. T. (2012) Oxadiazoles in medicinal chemistry. J. Med. Chem. 55, 1817–1830. 10.1021/jm2013248. [DOI] [PubMed] [Google Scholar]
- Belousoff M. J.; Eyal Z.; Radjainia M.; Ahmed T.; Bamert R. S.; Matzov D.; Bashan A.; Zimmerman E.; Mishra S.; Cameron D.; Elmlund H.; Peleg A. Y.; Bhushan S.; Lithgow T.; Yonath A. (2017) Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome. mBio 8, e00395-17. 10.1128/mBio.00395-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D.; Grove L. E.; Hall D. R.; Bohnuud T.; Mottarella S. E.; Luo L.; Xia B.; Beglov D.; Vajda S. (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 10, 733–755. 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D.; Hall D. R.; Chuang G. Y.; Cencic R.; Brenke R.; Grove L. E.; Beglov D.; Pelletier J.; Whitty A.; Vajda S. (2011) Structural conservation of druggable hot spots in protein–protein interfaces. Proc. Natl. Acad. Sci. U. S. A. 108, 13528–13533. 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. T.; Liu J.; Lee R. B.; Lee R. E. (2016) New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv. Drug Delivery Rev. 102, 55–72. 10.1016/j.addr.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J. P.; Appelberg R.; Gama F. M. (2016) Antimicrobial peptides as novel anti-tuberculosis therapeutics. Biotechnol. Adv. 34, 924–940. 10.1016/j.biotechadv.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Pledger G. W.; Laxer K. D.; Sahlroot J. T.; Taylor M. R.; Cereghino J. J.; McCormick C.; Whitley L.; Manning L. W. (1992) Pharmacokinetic and dose tolerability study of ADD 94057 in comedicated patients with partial seizures. Epilepsia 33, 112–118. 10.1111/j.1528-1157.1992.tb02292.x. [DOI] [PubMed] [Google Scholar]
- Eisele N. A.; Ruby T.; Jacobson A.; Manzanillo P. S.; Cox J. S.; Lam L.; Mukundan L.; Chawla A.; Monack D. M. (2013) Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14, 171–182. 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsche M.; Ehrt S.; Schnappinger D. (2009) Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 37, 1778–1788. 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M. S.; Cox J. S.; Jacobs W. R. Jr. (2000) A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5, 717–727. 10.1016/S1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- Wayne L. G.; Hayes L. G. (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T.; Brosch R.; Parkhill J.; Garnier T.; Churcher C.; Harris D.; Gordon S. V.; Eiglmeier K.; Gas S.; Barry C. E.; Tekaia F.; Badcock K.; Basham D.; Brown D.; Chillingworth T.; Connor R.; Davies R.; Devlin K.; Feltwell T.; Gentles S.; Hamlin N.; Holroyd S.; Hornsby T.; Jagels K.; Krogh A.; McLean J.; Moule S.; Murphy L.; Oliver K.; Osborne J.; Quail M. A.; Rajandream M. A.; Rogers J.; Rutte r S.; Seeger K.; Skelton J.; Squares R.; Squares S.; Sulston J. E.; Taylo K.; Whitehead S.; Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Andini N.; Nash K. A. (2011) Expression of tmRNA in mycobacteria is increased by antimicrobial agents that target the ribosome. FEMS Microbiol. Lett. 322, 172–179. 10.1111/j.1574-6968.2011.02350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y.; Muller S.; Behm-Ansmant I.; Branlant C. (2007) Identification of Modified Residues in RNAs by Reverse Transcription-Based Methods. Methods Enzymol. 425, 21–53. 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- Feaga H. A.; Quickel M. D.; Hankey-Giblin P. A.; Keiler K. C. (2016) Human Cells Require Non-stop Ribosome Rescue Activity in Mitochondria. PLoS Genet. 12, e1005964. 10.1371/journal.pgen.1005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.