Abstract
Background
Pancreatic β-cells require a constant supply of zinc to maintain normal insulin secretory function. Following co-exocytosis with insulin, zinc is replenished via the Zrt- and Irt-like (ZIP; SLC39A) family of transporters. However the ZIP paralogues of particular importance for zinc uptake, and associations with β-cell function and Type 2 Diabetes remain largely unexplored. We retrieved and statistically analysed publically available microarray and RNA-seq datasets to perform a systematic review on the expression of β-cell SLC39A paralogues. We complemented results with experimental data on expression profiling of human islets and mouse β-cell derived MIN6 cells, and compared transcriptomic and proteomic sequence conservation between human, mouse and rat.
Results
The 14 ZIP paralogues have 73–98% amino sequence conservation between human and rodents. We identified 18 datasets for β-cell SLC39A analysis, which compared relative expression to non-β-cells, and expression in response to PDX-1 activity, cytokines, glucose and type 2 diabetic status. Published expression data demonstrate enrichment of transcripts for ZIP7 and ZIP9 transporters within rodent β-cells and of ZIP6, ZIP7 and ZIP14 within human β-cells, with ZIP1 most differentially expressed in response to cytokines and PDX-1 within rodent, and ZIP6 in response to diabetic status in human and glucose in rat. Our qPCR expression profiling data indicate that SLC39A6, −9, −13, and − 14 are the highest expressed paralogues in human β-cells and Slc39a6 and −7 in MIN6 cells.
Conclusions
Our systematic review, expression profiling and sequence alignment reveal similarities and potentially important differences in ZIP complements between human and rodent β-cells. We identify ZIP6, ZIP7, ZIP9, ZIP13 and ZIP14 in human and rodent and ZIP1 in rodent as potentially biologically important for β-cell zinc trafficking. We propose ZIP6 and ZIP7 are key functional orthologues in human and rodent β-cells and highlight these zinc importers as important targets for exploring associations between zinc status and normal physiology of β-cells and their decline in Type 2 Diabetes.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4119-2) contains supplementary material, which is available to authorized users.
Keywords: Type 2 diabetes, Zinc, ZIP, SLC39A, Systematic review, Expression data, Microarray, RNA-seq
Background
Pancreatic β-cells require a constant supply of zinc for normal function in maintaining glycaemic control [1, 2]. Zinc acts at multiple stages within the insulin secretory pathway [3, 4]. Zinc ions (Zn2+) are loaded into insulin granules via the predominantly β-cell specific zinc transporter 8 (ZnT8) [5], where two ions co-crystallise with insulin hexamers [6], important for proper insulin processing, protection of insulin from proteolytic degradation [7] and for maintaining granule osmotic stability [8]. Zinc is subsequently co-released with mature insulin upon exocytosis where it is proposed to fulfil additional roles in glycaemic control [9–11].
Significant amounts of Zn2+ are lost from β-cells during insulin secretion and coordinated replenishment is required. The Zrt- and Irt-like (ZIP; SLC39A) family of zinc importer proteins, of which 14 paralogues are present within both humans and rodents [12, 13], tightly control cellular Zn2+ influx into the cytosol and are thought responsible for restoring β-cell zinc content [14]. ZIP paralogues exhibit differing Zn2+ affinities (K0.5) and transporting efficiencies, and show cell- and condition-dependent expression [12, 15], thus it is expected that the β-cell ZIP profile closely reflects the unique cellular demand for Zn2+ and ability to adapt to stresses such as hyperglycaemia and inflammatory cytokines. Since both hyperzincemic and hypozincemic Zn2+statuses are observed in diabetic patients [16–18] and animal models of diabetes [19, 20], one can hypothesize that altered ZIP expression profiles are associated with disease state. However exploration of the β-cell SLC39A transcriptome, and therefore the liable transporters, has been limited to a few studies [4, 14, 21–23], where an importance of ZIP4 [23], ZIP6 [21, 22], ZIP7 [14, 21, 22], ZIP8 [22], and ZIP14 [14, 24] has been suggested.
Type 2 Diabetes is rapidly evolving into a major public health crisis. The disease pathogenesis generally results from an increasingly inadequate insulin response due to enhanced insulin resistance and a compensatory demand on insulin production that eventually leads to β-cell failure. Multiple studies have associated diabetes with hypozincemia, likely caused by hyperzincuria, and a negative correlation between the glycated haemoglobin percentage and plasma zinc [16–18]. Accordingly, there is a positive effect of adequate plasma zinc levels on glycemic control [18], suggesting a compromised zinc status in diabetes [25].
Since zinc plays an integral role within β-cells, understanding its regulation may prove central for targeting loss of secretory function during Type 2 Diabetes. Much of our understanding of β-cell physiology has derived from studies on rodents due to very limited accessibility of human islets [26]. However, differences in physiology between humans and rodents remain often unacknowledged when interpreting rodent studies. We hypothesised that the ZIP transporters most important to β-cells should be robustly expressed and show enrichment relative to other cell types [27], with changes in expression influenced by cellular stresses associated with compromised insulin secretion. We thereby aimed to identify and evaluate the complement of ZIP transporters most important within human and rodent (mouse and rat) β-cells for regulating zinc influx and accumulation.
Here we show through systematic review of microarray and RNA-seq studies [28, 29] that transcripts for multiple ZIP paralogues are enriched in β-cells and/or show transcriptional regulation in response to cytokines, hyperglycaemia, Type 2 Diabetes status, and pancreatic and duodenal homeobox 1 (PDX-1) activity, the major transcription factor for β-cells. We used quantitative PCR (qPCR) to verify the relative expression of these paralogues within human islets and/or murine MIN6 β-cells. Furthermore, we computationally aligned human, mouse and rat SLC39A mRNA and protein sequences to demonstrate high cross-species conservation of the paralogues identified as key for β-cell zinc homeostasis within our systematic review. We highlight ZIP6, ZIP7, ZIP9, ZIP13 and ZIP14 in human and rodent, and ZIP1 in rodent as biologically important candidates for mediating β-cell Zn2+ influx and zinc-signalling processes, such as cell proliferation. In addition to normal physiology, we suggest ZIP6, ZIP7 and ZIP14 downregulation is associated with diabetic status; however the relationship to zinc content in the β-cells/pancreas remains unknown. Critically, our review highlights potentially important differences between human islets and rodent cells in their complements of zinc importers, again demonstrating the limitations of rodent models for human diabetes.
Methods
Systematic review
Identification of eligible expression datasets
This systematic review was conducted in accordance with the guidelines provided in the PRISMA statement. Microarray and RNA-seq expression profiling studies were identified through searching the NCBI PubMed database and the Gene Expression Omnibus (GEO) database [30] to April 2016, using combinations of the following key terms: “β-cell, islet” and “diabetes, gene expression, microarray, RNA-seq”, and compiled studies screened for duplicates. Eligibility was independently assessed through first screening by title and abstract, and then by the full text, based on the following inclusion criteria: original research article published in English, RNA-seq or microarray platform, expression profiling of mature β-cells, islets and/or β-cell line, and human or rodent genome. The eligibility was finally confirmed through verifying the presence of accessible expression data for ZIP transporter transcripts (SLC39A/Slc39a). Included datasets explored: (a) expression within β-cells compared to non-β-cells, (b) expression in response to extracellular cytokines, (c) expression in response to PDX-1 activity, (d) expression in response to extracellular glucose, and (e) expression within human diabetic islets. From each identified dataset, the accession number (if appropriate), platform, species, sample types and sizes, and gene expression data were extracted. This pipeline is depicted in Fig. 1.
Fig. 1.
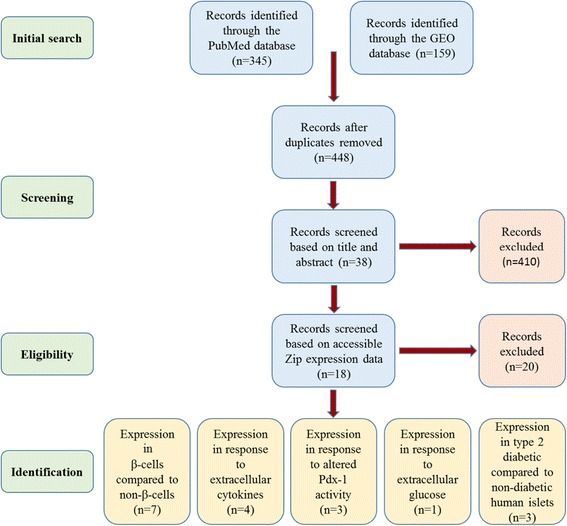
The pipeline followed for identification of suitable microarray and RNA-seq datasets
Data pre-processing
The heterogeneity of different platforms, gene nomenclature and control samples can cause difficulties when comparing datasets from different sources. Normalisation is therefore critical to reduce the chance of skewing the results and enhances credibility of individual expression changes. To minimise inconsistency, a standardised normalisation method was performed within datasets [31] using Qlucore Omics Explorer (version 3.2; Qlucore AB, Lund, Sweden). Raw data was log (base = 2; log2) transformed and normalised through applying a standard score (Z-score) transformation, which calculates normalised expression intensities (y_i) of each probe as follows:
Where x_i represents raw intensity values (x_i, i = 0…N-1) for each gene, m represents average gene intensity for the experiment, and δ represents the standard deviation of all measured intensities.
Statistical analysis
Statistical analysis was undertaken using the Qlucore Omics Explorer (version 3.2; Qlucore AB, Lund, Sweden) bioinformatics software. Fold differences (FD) in expression between relevant conditions were derived, and significance calculated on the global transcriptomic data set through unpaired t-tests, adjusted using the Benjamini-Hochberg False Discovery Rate (FDR) procedure [32]. Genes were considered differentially expressed in comparisons at an FDR of 15%. Arbitrary FD cut offs of ≥1.5-fold on significantly regulated genes were chosen to indicate biologically relevant differential expression. Full results from analysis are provided in Additional file 1.
Datasets with inaccessible raw data
Where the full raw datasets were not available for download, normalised data were extracted from supplementary data tables and log2 transformed FD (Log2FD) values and significance extracted/calculated using Excel as appropriate. Data analysed in this way are annotated.
Experimental analysis
Human islet cDNA
Human islet cDNA originating from healthy cadaver donors was obtained via the Human Islet Isolation Unit at King’s College Hospital.
Cell line and RNA extraction
The adherent insulinoma β-cell line MIN6 (Mus musculus) was maintained within Dulbecco’s Modified Eagle’s Medium supplemented with 15% fetal bovine serum, 4 mM L-glutamine, 50 μM β-mercaptoethanol, 100 μg/ml streptomycin and 100 units/ml penicillin (both Sigma-Aldrich), at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Total RNA was extracted using TRIzol Reagent (ThermoFisher), reverse transcribed to cDNA using the high capacity RNA-to-cDNA kit (ThermoFisher), and diluted ≥1:10 prior to experimentation.
Quantitative PCR
Quantitative PCR (qPCR) assays were designed using the online Universal Probe Library (UPL) assay design tool (Roche). Assay designs are provided within Additional files 2 and 3: Tables S1 and S2. Primer Blast [33] was used to predict the binding of our primers to mouse and human RNA. The mouse primers bind all respective ZIP transporter isoforms. The human primers bind all isoforms for ZIP2, ZIP4, ZIP5, ZIP6, ZIP8, ZIP10, ZIP11, ZIP12 and ZIP13. Primers for ZIP1, ZIP3, ZIP7, ZIP9 and ZIP14 covered either all the isoforms expressed within human islets, or those predominantly expressed (Additional files 2 and 3: Tables S1 and S2, Additional file 4). PCR plates were loaded using the Biomek FX liquid handling robot (Beckman Coulter) and reactions [20-40 μg cDNA, 0.1 μM UPL probe, 0.2 μM forward primer, 0.2 μM reverse primer and 1X TaqMan Fast Advanced Mastermix (Applied Biosystems)] amplified using the Prism7900HT sequence detection system, Applied Biosystems, and analysed with SDS (sequence detection systems) 2.4 software. All gene expression values were normalised to the housekeeping gene ubiquitin C (UBC), and relative expression calculated using the ΔΔCT method. The efficiencies for primers targeting genes with appreciable expression (mouse: ZIP1, ZIP2, ZIP3, ZIP4, ZIP6, ZIP7, ZIP8, ZIP9, ZIP10, ZIP11, ZIP13, ZIP14, UBC, GADPH; human: ZIP1, ZIP3, ZIP4, ZIP5, ZIP6, ZIP7, ZIP8, ZIP9, ZIP10, ZIP13, ZIP14, UBC, GADPH) ranged from 88 to 111% for mouse and 75–106% for human. Data show an average of two biological repeats for human islets and three biological repeats for MIN6 cells.
Exploration of zinc transporter heterogeneity
Human and mouse ZIP orthologues were aligned to assess transcriptomic and proteomic similarities using MUSCLE (3.8) [34, 35] and percent similarity values recorded.
Results
Overview of included datasets
A systematic review allows integrated analysis of multiple high throughput gene expression datasets. Following the pre-defined criteria, 18 appropriate β-cell/islet expression profiling studies were identified. These studies are summarised in Table 1. Seven studies compared expression within β-cells to non-β-cells, four investigated expression in response to extracellular cytokines, three studied expression in response to PDX-1 activity, one explored expression in response to extracellular glucose, and three measured expression within human diabetic islets.
Table 1.
Overview of the datasets identified for analysis
| Dataset ID | Platform | Species | Sample | Number of samples | Reference |
|---|---|---|---|---|---|
| E-MTAB-463 and E-MTAB-465 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | Homo sapiens | β-cells and α-cells | β-cells: 4 α-cells: 4 |
Dorrell et al., 2014 |
| – | RNA-seq | Homo sapiens | Sorted β-cells, α-cells and exocrine cells (duct and acinar) | β-cells: 3 α-cells: 3 Exocrine cells: 2 |
Bramswig et al., 2013 |
| EGAS00001000442 | RNA-seq | Homo sapiens | Sorted β-cells, α-cells and non-β-cells | 7 DNA libraries, pooled | Nica et al., 2013 |
| GSE30803 | [HG-U133A] Affymetrix Human Genome U133A Array | Homo sapiens | Islets (and 16 other primary cell types) | Islets: 3 | Martens et al., 2011 |
| – | RNA-seq | Mus musculus | β-cells and islets | Unknown. Data for 5 additional cell types downloaded from NCBI | Ku et al., 2012 |
| GSE13381 | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | β-cells and α-cells | β-cells: 2 Non-β-cells: 2 |
Kutlu et al., 2009 |
| GSE10785 | Rosetta/Merck Mouse 44 k 1.0 microarray | Mus musculus | Islets (and 5 other primary cell types) | Islets: 40 | Keller et al., 2008 |
| GSE35296 | RNA-seq | Homo sapiens | Islets | Control: 5 IL-1β and IFN-γ: 5 |
Eizirik et al., 2012 |
| – | [HG-U133A] Affymetrix Human Genome U133A Array | Homo sapiens | Islets | Control: 3 IL-1β and IFN-γ: 3 |
Ylipaasto et al., 2005 |
| – | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | FACS purified β-cells | For control and cytokine stimulation at each time point: 3 | Ortis et al., 2010 |
| – | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | INS-1E rat insulinoma cell line | For control and cytokine stimulation at each time point: 3 | Moore et al., 2011 |
| GSE40642 | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | INS-1ab rat insulinoma cell line | Control: 8 IL-1β: 20 PDX-1 overexpression: 8 PDX-1 overexpression and IL-1β: 20 |
Hansen et al., 2012 |
| GSE49786 | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | Islets | Untreated: 5 Pdx-1 overexpression: 5 |
Hayes et al., 2013 |
| E-MTAB-127 | A-CBIL-10-UPenn Mouse PancChip 6.1 | Mus musculus | Islets and MIN6 murine insulinoma cell line | Islets Pdx-1+/−: 3 Islets Pdx-1+/+: 3 MIN6 treated: 4 MIN6 control: 4 |
Sachdeva et al., 2009 |
| GSE12817 | [Rat230_2] Affymetrix Rat Genome 230 2.0 Array | Rattus norvegicus | Islets | 2 mM Glucose: 4 5 mM Glucose: 4 10 mM Glucose: 4 30 mM Glucose: 4 |
Bensellam et al., 2009 |
| GSE25724 | [HG-U133A] Affymetrix Human Genome U133A Array | Homo sapiens | Islets | Non-diabetic: 7 Diabetic: 6 |
Dominguez et al., 2011 |
| GSE20966 | [U133_X3P] Affymetrix Human X3P Array | Homo sapiens | β-cell-enriched pancreatic tissue | Non-diabetic: 10 Diabetic: 10 |
Marselli et al., 2010 |
| GSE38642 | [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] | Homo sapiens | Islets | Non-diabetic: 54 Diabetic: 9 |
Taneera et al., 2012 |
Specificity of ZIP transporter expression within β-cells
Multiple microarray and RNA-seq studies have sought to assess β-cell gene expression relative to other pancreatic cells and additional tissues. Since ZIP paralogues exhibit cell-specific profiles reflecting function [12, 15], β-cell enrichment may indicate important cell-specific roles. Analysis of human islet cell transcriptomics datasets uncovered SLC39A13 and SLC39A14 as enriched within β-cells compared to α-cells [2- to 3-fold], and SLC39A1, SLC39A10 and SLC39A11 as ≥1.5-fold depleted [36, 37]. However, when β-cell expression was compared to sorted pancreatic exocrine cell populations (human duct and acinar cells), enrichment of SLC39A7 and SLC39A9 was observed (1.7- and 1.6-fold respectively) alongside relative depletion of SLC39A5 (11-fold), SLC39A8 (4.3-fold), SLC39A10 (1.8-fold) and SLC39A11 (1.5-fold) (data calculated from supplementary tables). Similarly RNA-seq data from Nica et al. uncovered depletion of SLC39A5 and SLC39A10 within sorted human β-cells over both total islets (2- and 6.8-fold, respectively) and non-β-cells [islet cell populations considered depleted of β-cells (2.8- and 4-fold, respectively)], accompanied by depletion of SLC39A2 (2-fold over total islets and 4-fold over non-β-cells) and SLC39A3 (1.7-fold over both total islets and non-β-cells), with enrichment of SLC39A1 (2.4-fold over total islets and 2.1-fold over non-β-cells), and of SLC39A14 (1.9-fold, only over non-β-cell preparations) [38].
Analysis of microarray datasets of human β-cell-enriched pancreatic samples and 15 other tissues [β-cell-enriched pancreas, pancreatic duct cells, cerebrum, colon, foetal brain, kidney, liver, lung, myocardial, skeletal muscle, prostate, small intestine, spleen, stomach, testis and thymus (dataset GSE30803)] revealed ≥1.5-fold enrichment of SLC39A1, SLC39A6, SLC39A7 and SLC39A14, however without statistical significance [39]. Further investigation of probe-specific expression revealed that relative enrichment was biased by elevated expression of specific paralogues within other tissues (ZIP6 within the brain [40] and prostate [41], ZIP7 within the colon [42] and ZIP14 within the liver [43]). Omitting these tissues indicated ≥3-fold enrichment of SLC39A6 (q < 0.1) and SLC39A14 (q < 0.15), and 1.6-fold SLC39A7 (q < 0.05) enrichment within β-cells compared to the remaining tissues analysed (Fig. 2a).
Fig. 2.
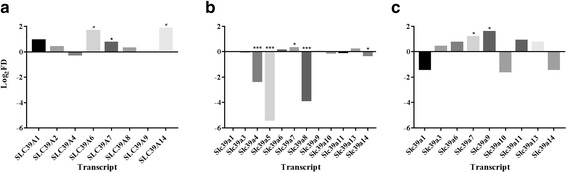
Enrichment of SLC39A paralogues within human and murine β-cells. a Expression within human β-cell-enriched pancreatic samples compared to 11 other tissues (pancreatic duct cells, cerebrum, kidney, lung, myocardial, skeletal muscle, small intestine, spleen, stomach, testis and thymus). b Expression within sorted mouse β-cells compared to mouse islets. c Expression within sorted mouse β-cells compared to six other cell types (brain, liver, lung fibroblasts, NPC, skeletal muscle, islets), after exclusion of β-cell depleted paralogues. Data for (a) analysed from dataset GSE30803 and (b-c) analysed from supplementary tables within [44]. # P < 0.15, *P < 0.05, ***P < 0.001. NPC = neural progenitor cells
Analysis of a mouse RNA-seq dataset [44] suggested Slc39a4, Slc39a5 and Slc39a8 are ≥4-fold depleted within sorted β-cells over islets (Fig. 2b). Further investigation of non-β-cell depleted paralogues compared to total islets and six other cell types [brain, liver, lung fibroblasts, neural progenitor cells (NPC), skeletal muscle, total islet (Fig. 2c)] revealed Slc39a7 and Slc39a9 as the most β-cell enriched Zip paralogues in mouse with 2.3- and 3-fold elevated expression, respectively. Analysis of a further rat dataset comparing expression of β-cells over α-cells (dataset GSE13381) displayed ≥1.5-fold enrichment of Slc39a3 and Slc39a6, and ≥2-fold enrichment of Slc39a7 and Slc39a14, but without statistical significance. However, there was no differential Slc39a expression between murine islets and five other tissues (adipose, gastrocnemius muscle, hypothalamus, liver, and soleus muscle) from 10-week old lean and obese C57BL/6 and BTBR mice (dataset GSE10785).
Cytokine stimulation and ZIP transporter expression
Pro-inflammatory cytokines profoundly affect cellular metabolism and utilisation of nutrients such as metal ions [45]. Chronic exposure of islets to the inflammatory cytokines interleukin-1 beta (IL-1β), tumor necrosis factor-alpha (TNF-α) and interferon-gamma (IFN-γ) is associated with β-cell destruction and decreased secretory parameters in both Type 1 and Type 2 Diabetes [46]. Cytokine-dependent expression may indicate ZIP paralogues important for maintaining normal β-cell parameters when adapting to extracellular cytokine stress. RNA-seq dataset analysis of human islets exposed to IL-1β and IFN-γ for two days (dataset GSE35296) revealed 1.4- and 2.0-fold upregulation of SLC39A8 and SLC39A14 transcripts, respectively, and 2-fold downregulation of SLC39A10 [47]. However an additional microarray study using human islets [48] did not show any ZIP transporter transcripts differentially expressed following 48 h incubation with IL-1β and IFN-γ.
Within independent studies, both fluorescence assisted cell sorting (FACS) purified rat β-cells [49] and the rat INS-1E β-cell line [50] were cultured with IL-1β and IFN-γ before microarray analysis (data from both studies calculated from supplementary data). Within rat β-cells [49] differential regulation of Slc39a1 (2.5-fold) was observed at 2 h, and of Slc39a1 and Slc39a10 at both 12 h (2.0- and −3.5-fold, respectively) and 24 h (1.5-fold and −1.6-fold, respectively). Similarly, INS-1E cells [50] displayed upregulation of Slc39a1 at both 6 and 24 h (2.6- and 2.1-fold, respectively) and of Slc39a6 at 24 h (1.7-fold), alongside downregulation of Slc39a13 at 6 h (−1.5-fold). INS-1E cells were additionally analysed after 6 and 24 h incubation with IFN-γ and TNF-α to show Slc39a1 upregulation (2.8- and 2.4-fold, respectively) and Slc39a14 downregulation (2.8- and 2.4-fold, respectively).
PDX-1 and ZIP transporter expression
PDX-1 is the key transcription factor mediating β-cell-specific gene expression within developing and mature β-cells [51]. Changes in ZIP expression as a consequence of PDX-1 activity may indicate roles of respective transporters in maintaining normal β-cell parameters. Critically, constitutive overexpression of PDX-1 sensitises β-cells to cytokine-induced apoptosis [52, 53]. Overexpression of PDX-1 within rat INS-1ab cells (dataset GSE40642) resulted in downregulation of Slc39a6 and Slc39a14 (Fig. 3). Stimulation of PDX-1 overexpressing cells with cytokine IL-1β further exacerbated these effects and also upregulated Slc39a1 ≥ 2.5-fold [54] (Fig. 3). Whereas within rat islets (dataset GSE49786), PDX-1 overexpression upregulated Slc39a8 (2.1-fold, P < 0.01) [55]; however Slc39a5 and Slc39a8 were up- and downregulated 1.5- and −2.6-fold, respectively, in mouse MIN6 cells (dataset E-MTAB-127) [56]. Data analysis from E-MTAB-127 additionally showed 2.8-fold downregulation of Slc39a5 in PDX+/− mouse islets compared to PDX+/+ control mouse islets.
Fig. 3.
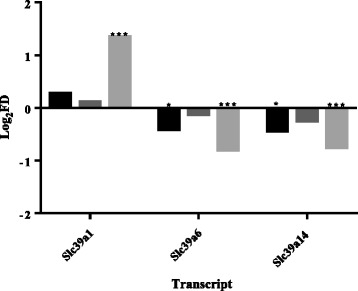
Slc39a transcripts differentially expressed in rat INS-1ab cells following PDX-1 overexpression and IL-1β stimulation. Black bars: PDX-1 overexpression; dark grey bars: IL-1β stimulation; light grey bars: PDX-1 overexpression and IL-1β stimulation. Data analysed from dataset GSE40642. *P < 0.05, ***P < 0.001
Expression in response to glucose stimulation
Hyperglycaemia is universal within all prediabetic and diabetic cases, and glucose-responsive expression may indicate genes and pathways important for adapting to an enhanced demand for insulin secretion. Examination of microarray datasets of isolated rat islets cultured with 2 mM, 5 mM, 10 mM and 30 mM glucose (dataset GSE12817) uncovered ≥1.5-fold upregulation of Slc39a2, Slc39a4 and Slc39a6 and ≥1.5-fold downregulation of Slc39a3 and Slc39a5 when glucose increases [57].
Expression within islets from type 2 diabetic patients
Transcriptomic datasets of islets derived from normoglycaemic and Type 2 diabetic patients were next analysed to explore relevance to the human disease. Three paralogues (SLC39A2, SLC39A5 and SLC39A8) showed ≥1.5-fold upregulation and four paralogues (SLC39A6, SLC39A7, SLC39A8 and SLC39A14) ≥1.5-fold downregulation in diabetic compared to non-diabetic individuals [results combined from studies GSE25724 [58] and GSE20966 [59], Fig. 4]. However in a cohort of Nordic patients (dataset GSE38642), no ZIP paralogues were found differentially (≥1.5-fold) expressed between diabetic and non-diabetic islets [60].
Fig. 4.
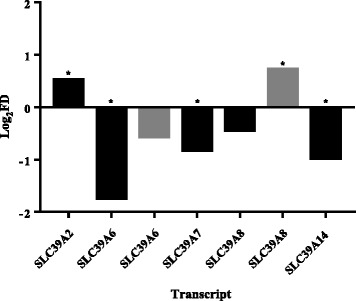
SLC39A transcripts differentially expressed in human type 2 diabetic islets compared to non-diabetic islets. Probes targeting the same transcript(s) are averaged for each assay. Black bars: data analysed from dataset GSE25724; grey bars: data analysed from dataset GSE20966. *P < 0.05
SLC39A Paralogues identified within our systematic review are experimentally verified to show high β-cell/islet expression and sequence conservation
Enhanced relative expression may suggest a biological relevance of respective putative ZIP orthologues in maintaining intracellular Zn2+ homeostasis in β-cells/islets. To verify the biological relevance of the ZIP orthologues we identified in our systematic review in terms of β-cell function, we performed qPCR expression profiling of human and mouse SLC39A mRNA transcripts. There was notably a wider range of mRNA for ZIP transporters expressed in human islets, compared with murine MIN6 cells. We observed highest expression of SLC39A6, SLC39A9, SLC39A13 and SLC39A14 in human islets and of Slc39a6 and Slc39a7 in mouse MIN6 cells (Fig. 5).
Fig. 5.
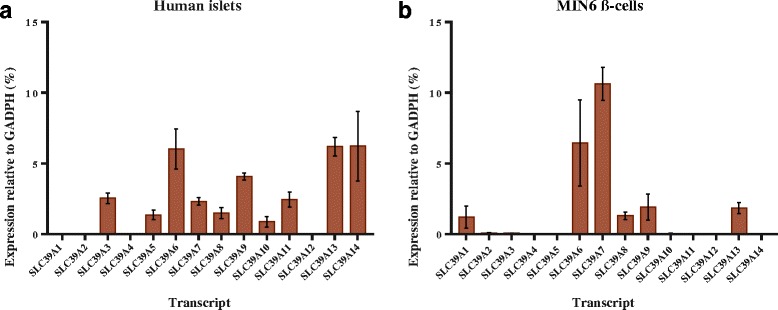
Expression profiles of SLC39A mRNA transcripts in human islets and murine MIN6 cells. a The human SLC39A transcriptome. N = 2 and error bars show ± range. b The MIN6 β-cell Slc39a transcriptome. N = 3 and error bars show ±SEM
We next assessed cross-species homologies between human and putative rodent ZIP orthologues. Enhanced similarity increases the likelihood of inferring molecular function [61], and increases confidence when applying results encompassing data derived from multiple species. We calculated the transcriptomic and proteomic homologies between human and mouse ZIP orthologues, and between mouse and rat ZIP orthologues through bioinformatics. We showed all respective orthologues have high similarities, with lowest protein homology between human and mouse observed for ZIP4 (73%) and highest for ZIP1 (94%). All rat and mouse orthologues showed protein similarities of ≥90% aside from ZIP4 (89.8%) and ZIP14 (89.5%) (Table 2). The high sequence similarities observed indicate that all ZIP paralogues identified as important in our systematic review have the potential to substitute functionally in human, mouse and rat β-cells.
Table 2.
Percentage similarity between human, mouse and rat ZIP/SLC39A protein and mRNA sequences
| Gene | Refseq transcript ID | Entrez protein ID | Percentage similarities (human:mouse) |
Percentage similarities (mouse:rat) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | Human | Mouse | Rat | Transcript | Protein | Transcript | Protein | |
| SLC39A1 | NM_014437.4 | NM_013901.2 | NM_001134577.1 | Q9NY26 | Q9QZ03 | B5DEF5 | 80.77 | 93.83 | 91.60 | 98.46 |
| SLC39A2 | NM_014579.3 | NM_001039676.2 | NM_001107260.1 | Q9NP94 | Q2HIZ9 | D3ZIN1 | 78.51 | 77.99 | 91.87 | 94.82 |
| SLC39A3 | NM_144564.4 | NM_134135.1 | NM_001008356.1 | Q9BRY0 | Q99K24 | Q5U1X7 | 80.99 | 84.04 | 91.23 | 97.16 |
| SLC39A4 | NM_017767.2 | NM_028064.2 | NM_001077669.1 | Q6P5W5 | Q78IQ7 | A0JPN2 | 74.55 | 73.17 | 91.98 | 89.79 |
| SLC39A5 | NM_173596.2 | NM_028051.3 | NM_001108728.1 | Q6ZMH5 | Q9D856 | D3ZSF7 | 83.42 | 84.30 | 92.24 | 94.00 |
| SLC39A6 | NM_012319.3 | NM_139143.3 | NM_001024745.1 | Q13433 | Q8C145 | Q4V887 | 80.04 | 88.20 | 92.01 | 95.28 |
| SLC39A7 | NM_006979.2 | NM_008202.2 | NM_001164744.1 | Q92504 | Q31125 | Q6MGB4 | 81.72 | 85.90 | 90.62 | 93.72 |
| SLC39A8 | NM_022154.5 | NM_001135150.1 | NM_001011952.1 | Q9C0K1 | Q91W10 | Q5FVQ0 | 76.15 | 89.35 | 90.74 | 96.10 |
| SLC39A9 | NM_018375.4 | NM_026244.2 | NM_001034929.1 | Q9NUM3 | Q8BFU1 | Q3KR82 | 78.04 | 93.49 | 93.35 | 92.33 |
| SLC39A10 | NM_001127257.1 | NM_172653.2 | NM_001108796.2 | Q9ULF5 | Q6P5F6 | D4A517 | 84.50 | 87.36 | 92.25 | 96.16 |
| SLC39A11 | NM_001159770.1 | NM_001166503.1 | NM_001013042.1 | Q8N1S5 | Q8BWY7 | Q6P6S2 | 77.62 | 90.32 | 91.32 | 95.22 |
| SLC39A12 | NM_001145195.1 | NM_001012305.2 | XM_006254285.3 | Q504Y0 | Q5FWH7 | D4A8R5 | 78.31 | 78.17 | 91.15 | 90.41 |
| SLC39A13 | NM_001128225.2 | NM_001290765.1 | NM_001039196.1 | Q96H72 | Q8BZH0 | Q2M1K6 | 81.54 | 90.58 | 93.42 | 93.84 |
| SLC39A14 | NM_001128431.2 | NM_001135151.1 | NM_001107275.1 | Q15043 | Q75N73 | D3ZZM0 | 75.84 | 86.91 | 90.11 | 89.53 |
Discussion
Associations between Zn2+ status and β-cell function have been extensively described in independent studies [1, 2, 62]. ZnT8 expression is positively correlated with granule Zn2+ release and glucose tolerance in mice [63], and high glucose stimulation increases free Zn2+ content within mouse islets [22] and hamster HIT-T15 cells [64]. Intracellular Zn2+ exhibits roles in protection against oxidative stress-induced apoptosis [65] whereas chronic elevation contributes to β-cell dysfunction [22]. The ZIP importer paralogues responsible for maintaining β-cell Zn2+ homeostasis remain largely unexplored and are important to investigate for understanding β-cell function in health and diabetic disease.
A systematic review allows integrated analysis of relative consistencies in differential expression from high throughput gene expression techniques, despite heterogeneities between studies involving experimental design and platform used. It has the capacity to identify consistent but modest variations, important for genes involved in processes where small expression changes can have amplified effects. Through this systematic review we re-analysed raw microarray and RNA-seq data in parallel with unannotated high-throughput datasets to compare and contrast β-cell ZIP complemets in human, mouse and rat β-cells/islets. We show enrichment of mRNA for ZIP7 and ZIP9 within rodent and ZIP6, ZIP7 and ZIP14 within human, with mRNA for ZIP1, ZIP6 and ZIP14 differentially expressed in response to cytokines and PDX-1 within rodent, and ZIP6 in response to diabetic status in human and glucose in rat. To query the biological relevance of our data, we carried out experimental expression profiling of human islet and MIN6 β-cell cDNA, and computationally aligned human, mouse and rat mRNA and protein sequences. Highest expression was observed for mRNA corresponding to ZIP6, ZIP9, ZIP13 and ZIP14 in human islets and ZIP6 and ZIP7 in mouse MIN6 cells, which is in agreement with previous observations [21]. The mRNA profile for ZIPs generated through our qPCR analyses also corresponds well to expression data on specific isoforms in human islets as produced by RNA-seq (Additional file 4). All ZIP orthologues displayed high sequence conservation between species. Surprisingly ZIP4, which is essential for intestinal zinc uptake in both mouse and human [27, 66], shows the lowest homology (73%) between these two species and it also does not appear to play a major role in β-cells. Based on their expression levels, relative enrichment in β-cells/islets (compared with other cells/tissues), and regulation in response to conditions relevant to diabetes, we propose that ZIP6, ZIP7 and ZIP14 in human, and ZIP6 and ZIP7 in rodent may be of particular importance for β-cell Zn2+ uptake and/or homeostasis. This conclusion is similar to that of Liu et al. [21], who highlighted the roles of ZIP6 and ZIP7 in β-cell zinc transport and viability. Our study also indentifies ZIP1, ZIP9 and ZIP13 as being of potential additional significance for β-cell function.
The abundance of ZIP transporters varies substantially between tissues and cells, allowing those with differing Zn2+ affinities, cellular localisations and regulatory mechanisms to tightly maintain the homeostatic balance [67]. We found significant differences in ZIP mRNA abundance between β-cells and non-pancreatic tissues; specifically, enrichment of ZIP7 and ZIP9 within mouse β-cells, and ZIP6, ZIP7 and ZIP14 within human islets. With the exception for ZIP7, which is found in the endoplasmic reticulum (ER) and in some cells in the Golgi apparatus, these zinc channels are operating at the plasma membrane [13, 68]. ZIP6 and ZIP7 enrichment is consistent with a report [21], suggesting that ZIP6 and ZIP7 mediate influx of zinc into the β-cell cytosol in tandem from the plasma membrane and the ER. In addition to their roles in transporting zinc, ZIP6, ZIP7 and ZIP14 strongly stimulate cell proliferation, drastically increasing the number of cells in G2/M phase, and their expression changes in cancers [27, 69–73]. Also of potential importance is that ZIP14 mediates import of both zinc and non-heme iron [74, 75] and that ZIP9 has been identified as a plasma membrane androgen receptor [76]. Interestingly, transcripts of ZIP9 and ZIP14, which were both found expressed at comparable abundances to ZIP6 and ZIP7 within human islets by ourselves and others [21], were additionally enriched within β-cells. ZIP9 and ZIP14 both show predicted localisation at the plasma membrane (with localisation of ZIP9 at the Golgi and trans-Golgi network additionally described) [77, 78], and currently remain unexplored in this context. Our expression profiling further identified ZIP13 as highly expressed in both human islets and MIN6 cells. ZIP13 is phylogenically grouped with ZIP7 [79] and studies have suggested ZIP13 localises at the ER, Golgi [80, 81] and intracellular vesicles [82]. However, to our knowledge ZIP13 has not been studied in β-cells. ZIP9, ZIP13 and ZIP14 may represent novel targets for understanding β-cell zinc uptake and homeostasis.
PDX-1 is the major transcriptional regulator in mature β-cells and mediates expression of key β-cell genes, with homozygous mutations linked to Type 2 Diabetes development [83]. Furthermore, PDX-1 drives β-cell (re)generation from neurogenin-3 positive endocrine precursors and pancreatic α-cells [84, 85], and β-cell-specific recovery of activity within Ins2Akita mice (βPdx1; Ins2Akita mice) promotes significantly improved glucose tolerance compared to control littermates [86]. Of interest, PDX-1 binds enhancers (cis elements) of the ZnT8 gene SLC30A8 [87], indicating a role of PDX-1 in β-cell zinc homeostasis parallel to its role in insulin gene regulation [88]. Our analysis suggests PDX-1 activity sensitizes the β-cell zinc response to cytokines through ZIP6 and ZIP14 downregulation and ZIP1 upregulation within rat INS-1ab cells. We additionally established ZIP1 to be consistently upregulated following stimulation with IL-1β and IFN-γ, and IFN-γ and TNF-α within rat β-cells [49] and INS-1E cells [50], highlighting ZIP1 as potentially important in the adaptive response to cytokines. Interestingly, ZIP1 and ZIP6 abundances have been negatively correlated with the obesity-associated inflammatory state [89]. In contrast to the data in rodents, our review further identified ZIP8 and ZIP14 upregulation in response to the inflammatory cytokines IL-1β and IFN-γ in human islets [47]. Inflammatory mediators such as lipopolysaccharides (LPS) and TNF-α upregulate SLC39A8 within human lung epithelia [90] and Slc39a14 is upregulated in response to LPS-initiated inflammation within the mouse pancreas and liver [43] and shows an acute-phase gene response to IL-6 [91].
Hyperglycaemia is well recognised as a universal driver in the pathogenesis of Type 2 Diabetes [92]. Our analysis showed high glucose stimulation of rat islets significantly enhanced ZIP6 mRNA expression, consistent with glucose-dependent increases of additional ZIP7 and ZIP8 upregulation [22]. Similarly, analysis of islets from human type 2 diabetic donors displayed ZIP6, ZIP7, ZIP8 and ZIP14 mRNA downregulation compared to normoglycaemic controls [58, 59]. Decreased transcript expression supports a disease relevance of these paralogues for mediating β-cell zinc accumulation, indicating abnormally low zinc uptake may occur within diabetic β-cells as a result of disrupted ZIP6, ZIP7, ZIP8 and/or ZIP14 expression.
At a proteomic level no significant differences in protein abundances were observed for any ZIP paralogue within human islets incubated with high or low glucose [93–95]. Though in one of these studies non-significant trends for enrichment of ZIP6 (2.6-fold) and ZIP14 (1.6-fold) in human islets were observed following culture in high compared to low glucose [95]. However, these proteomic studies likely bias towards proteins with higher abundances [96], and accurately evaluating less abundant species away from central pathways and those in complexes remains challenging, with membrane proteins imposing further challenges [97]. Although it is acknowledged that mRNA abundances often poorly correlate with protein abundances and functional activity [98], transcriptomic analysis remains important for pinpointing expression control and pathways of disruption during disease states.
This systematic review provides an overview of ZIP transcript expression in the context of β-cell specificity, cytokine stimulation, PDX-1 activity, glucose status and Type 2 Diabetes. It has allowed us to collectively analyse ZIP expression within multiple high throughput datasets, complemented by experimental work, providing evidence for differential regulation as a consequence of β-cell stresses associated with decreased insulin secretion. The study’s limitations should nevertheless be acknowledged. Firstly, although all raw datasets (if appropriate) were subjected to the same normalisation process to minimise inconsistencies, the platform and genomic heterogeneities and differing probe hybridization efficiencies could skew global interpretation, and the analysis used may not have been equally suited to all datasets. Secondly, RNA-seq offers unbiased analysis of sequences present however microarray datasets are limited by hybridization efficiencies and the probes present [99], such that in multiple studies probes did not target all ZIP paralogues. Multiple datasets analysed and our qPCR expression data utilised islets incorporating non-β-cells. Relative SLC39A abundances may be impacted by additional cell populations, such as SLC39A14 enrichment in α-cells [100]. Furthermore, although we have shown high conservation of ZIP mRNA and protein sequences between human and mouse, results may not be entirely translatable across species. Finally, several microarray studies identified within the systematic review search criteria were excluded during the final screening due to the absence of available experimental data for download and analysis. Despite these limitations, our systematic review distinguishes specific SLC39A paralogues as important within each human and rodent β-cells. The results are strongly supported by our experimental expression profiling of human islet and MIN6 β-cell cDNA through confirming relative enrichment and a biological relevance.
Conclusions
We have used a systematic approach to identify key ZIP complements in human, mouse and rat β-cells. We have verified a biological importance of these paralogues through proving high relative expression in human islets and/or murine MIN6 β-cells, and have demonstrated their potential to serve as functional orthologues in human and rodent through verifying high sequence similarities. Importantly, our results highlight similarities and potentially biologically relevant differences in zinc regulation between human and rodent ZIP orthologues which may prove critical when evaluating rodent β-cell models of disease. We propose ZIP6 and ZIP7 serve as key functional rodent-human orthologues in β-cells. We further identify ZIP9, ZIP13 and ZIP14 in human and rodent, and ZIP1 in rodent as potentially biologically important for β-cell function (Fig. 6). These paralogues represent interesting targets for future investigation into zinc regulation and homeostasis in β-cell failure and Type 2 Diabetes.
Fig. 6.
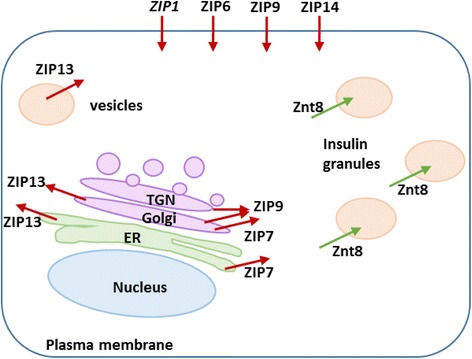
Predicted subcellular localisation of the identified ZIP transporters within human and rodent β-cells. Based on the data analysed in the present study ZIP6, ZIP7, ZIP9, ZIP13 and ZIP14 in human, and the same transporters in addition to ZIP1 in rodent appear to be of particular importance for β-cell biology and pathology. Zinc Transporter 8 (ZnT8) which transports zinc into the insulin granules is also shown. ER = endoplasmic reticulum; TGN = trans-Golgi network
Additional files
Analysed datasets. (XLSX 61 kb)
Designs for human qPCR assays undertaken. (DOCX 12 kb)
Designs for mouse qPCR assays undertaken. (DOCX 12 kb)
ZIP isoforms in human islets. (DOCX 56 kb)
Acknowledgments
Human islet material was obtained from Dr. Pratik Choudhary and Dr. Guo-Cai Wang of the Human Islet Isolation Unit at King’s College Hospital. We thank Dr. Matthew Arno (King’s College London Genomics Centre) for technical support and Dr. Christine Baldwin (King’s College London) for advice on presenting the systematic review methodology. The authors wish to thank Dr. Theodora Stewart for proofreading the final manuscript.
Funding
RL was supported by the King’s Bioscience Institute and the Guy’s and St Thomas’ Charity Prize PhD Programme in Biomedical and Translational Science. The funding body was not involved in the design of the study, collection, analysis or interpretation of data, or in writing the manuscript.
Availability of data and materials
Datasets analysed during the current study are available in either the GEO repository [https://www.ncbi.nlm.nih.gov/geo/] or the ArrayExpress repository [https://www.ebi.ac.uk/arrayexpress/]. Respective dataset ID’s are provided in Table 1.
Abbreviations
- ER
Endoplasmic reticulum
- FACS
Fluorescence assisted cell sorting
- FD
Fold difference
- FDR
False Discovery Rate
- GEO
Gene Expression Omnibus
- IFN-γ
Interferon-gamma
- IL-1β
Interleukin-1 beta
- Log2FD
Log2 transformed FD
- LPS
Lipopolysaccharides
- NPC
Neural progenitor cells
- PDX-1
Pancreatic and duodenal homeobox 1
- qPCR
quantitative PCR
- TNF-α
Tumor necrosis factor-alpha
- UPL
Universal Probe Library
- ZIP
Zrt- and Irt-like protein
- Zn2+
Zinc ions
- ZnT8
Zinc transporter 8
Authors’ contributions
RL performed the systematic review, all experiments, analysis and drafted the manuscript. CH and WM supervised the study. All the authors reviewed, approved and contributed to the final version of this manuscript.
Ethics approval and consent to participate
The King’s College Hospital Research Ethics Committee has approved human islet isolation and use for research (Protocol number 01–082, Human Islet Isolation and Research), and an assent form is completed by a relative of the cadaver pancreas donor for all islets used for research. The MIN6 cell line was a kind gift from Dr. Jun-ichi Miyazaki [101], distributed to WM under the appropriate materials transfer agreement.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4119-2) contains supplementary material, which is available to authorized users.
Contributor Information
Rebecca Lawson, Email: rebecca.tarrant@kcl.ac.uk.
Wolfgang Maret, Email: wolfgang.maret@kcl.ac.uk.
Christer Hogstrand, Email: christer.hogstrand@kcl.ac.uk.
References
- 1.Li YV. Zinc and insulin in pancreatic beta-cells. Endocrine. 2014;45(2):178–189. doi: 10.1007/s12020-013-0032-x. [DOI] [PubMed] [Google Scholar]
- 2.Chabosseau P, Rutter GA. Zinc and diabetes. Arch Biochem Biophys. 2016;611:79-85. [DOI] [PubMed]
- 3.Maret W. Zinc in pancreatic islet biology, insulin sensitivity, and diabetes. Prev Nutr Food Sci. 2017;22(1):1–8. doi: 10.3746/pnf.2017.22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L. Zinc and its transporters, pancreatic β-cells, and insulin metabolism. Vitam Horm. 2014;95:365–390. doi: 10.1016/B978-0-12-800174-5.00014-4. [DOI] [PubMed] [Google Scholar]
- 5.Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53(9):2330–2337. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
- 6.Chan SJ, Seino S, Gruppuso PA, Schwartz R, Steiner DF. A mutation in the B chain coding region is associated with impaired proinsulin conversion in a family with hyperproinsulinemia. Proc Natl Acad Sci U S A. 1987;84(8):2194–2197. doi: 10.1073/pnas.84.8.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu FY, Kildsig DO, Mitra AK. Insulin aggregation in aqueous media and its effect on alpha-chymotrypsin-mediated proteolytic degradation. Pharm Res. 1991;8(7):925–929. doi: 10.1023/A:1015820032648. [DOI] [PubMed] [Google Scholar]
- 8.Noormägi A, Gavrilova J, Smirnova J, Tõugu V, Palumaa P. Zn(II) ions co-secreted with insulin suppress inherent amyloidogenic properties of monomeric insulin. Biochem J. 2010;430(3):511–518. doi: 10.1042/BJ20100627. [DOI] [PubMed] [Google Scholar]
- 9.Tamaki M, Fujitani Y, Hara A, Uchida T, Tamura Y, Takeno K, et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J Clin Invest. 2013;123(10):4513–4524. doi: 10.1172/JCI68807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim BJ, Kim YH, Kim S, Kim JW, Koh JY, Oh SH, et al. Zinc as a paracrine effector in pancreatic islet cell death. Diabetes. 2000;49(3):367–372. doi: 10.2337/diabetes.49.3.367. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer R, Soria B, Dawson CM, Atwater I, Rojas E. Effects of Zn2+ on glucose-induced electrical activity and insulin release from mouse pancreatic islets. Am J Phys. 1984;246(5 Pt 1):C520–C527. doi: 10.1152/ajpcell.1984.246.5.C520. [DOI] [PubMed] [Google Scholar]
- 12.Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14(3–4):251–270. doi: 10.1023/A:1012988914300. [DOI] [PubMed] [Google Scholar]
- 13.Taylor KM, Nicholson RI. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta. 2003;1611(1–2):16–30. doi: 10.1016/S0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 14.Bosco MD, Mohanasundaram DM, Drogemuller CJ, Lang CJ, Zalewski PD, Coates PT. Zinc and zinc transporter regulation in pancreatic islets and the potential role of zinc in islet transplantation. Rev Diabet Stud. 2010;7(4):263–274. doi: 10.1900/RDS.2010.7.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukada T, Kambe T. Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics. 2011;3(7):662–674. doi: 10.1039/c1mt00011j. [DOI] [PubMed] [Google Scholar]
- 16.Pidduck HG, Wren PJ, Evans DA. Hyperzincuria of diabetes mellitus and possible genetical implications of this observation. Diabetes. 1970;19(4):240–247. doi: 10.2337/diab.19.4.240. [DOI] [PubMed] [Google Scholar]
- 17.Garg VK, Gupta R, Goyal RK. Hypozincemia in diabetes mellitus. J Assoc Physicians India. 1994;42(9):720–721. [PubMed] [Google Scholar]
- 18.de Carvalho GB, Brandão-Lima PN, Maia CS, Barbosa KB, Pires LV. Zinc’s role in the glycemic control of patients with type 2 diabetes: a systematic review. Biometals. 2017;30(2):151–62. [DOI] [PubMed]
- 19.Simon SF, Taylor CG. Dietary zinc supplementation attenuates hyperglycemia in db/db mice. Exp Biol Med (Maywood) 2001;226(1):43–51. doi: 10.1177/153537020122600107. [DOI] [PubMed] [Google Scholar]
- 20.Begin-Heick N, Dalpe-Scott M, Rowe J, Heick HM. Zinc supplementation attenuates insulin secretory activity in pancreatic islets of the ob/ob mouse. Diabetes. 1985;34(2):179–184. doi: 10.2337/diab.34.2.179. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Batchuluun B, Ho L, Zhu D, Prentice KJ, Bhattacharjee A, et al. Characterization of Zinc Influx Transporters (ZIPs) in Pancreatic Beta Cells: roles in regulating cytosolic zinc homeostasis and insulin secretion.J Biol Chem. 2015. [DOI] [PMC free article] [PubMed]
- 22.Bellomo EA, Meur G, Rutter GA. Glucose regulates free cytosolic Zn2+ concentration, Slc39 (ZiP), and metallothionein gene expression in primary pancreatic islet β-cells. J Biol Chem. 2011;286(29):25778–25789. doi: 10.1074/jbc.M111.246082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy AB, Prentice KJ, Froese S, Liu Y, Andrews GK, Wheeler MB. Zip4 Mediated zinc influx stimulates insulin secretion in pancreatic beta cells. PLoS One. 2015;10(3):e0119136. doi: 10.1371/journal.pone.0119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohanasundaram D, Drogemuller C, Brealey J, Jessup CF, Milner C, Murgia C, et al. Ultrastructural analysis, zinc transporters, glucose transporters and hormones expression in new world primate (Callithrix Jacchus) and human pancreatic islets. Gen Comp Endocrinol. 2011;174(2):71–79. doi: 10.1016/j.ygcen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 25.S P, Pasula S, Sameera K. Trace elements in diabetes mellitus. J Clin Diagn Res. 2013;7(9):1863–1865. doi: 10.7860/JCDR/2013/5464.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkarni RN, Stewart AF. Summary of the Keystone islet workshop (April 2014): the increasing demand for human islet availability in diabetes research. Diabetes. 2014;63(12):3979–81. [DOI] [PMC free article] [PubMed]
- 27.Hogstrand C, Maret W. Genetics of Human Zinc Deficiencies. In: eLS: John Wiley & Sons, Ltd; 2016. [Google Scholar]
- 28.Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, et al. Erratum to: comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2015;16:261. doi: 10.1186/s13059-015-0813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang M, Cowley AW, Greene AS. High throughput gene expression profiling: a molecular approach to integrative physiology. J Physiol. 2004;554(Pt 1):22–30. doi: 10.1113/jphysiol.2003.049395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y, Li D. Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med Genet. 2014;15:80. doi: 10.1186/1471-2350-15-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57(57):289–300. [Google Scholar]
- 33.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54(11):2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123(3):1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nica AC, Ongen H, Irminger JC, Bosco D, Berney T, Antonarakis SE, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23(9):1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martens GA, Jiang L, Hellemans KH, Stangé G, Heimberg H, Nielsen FC, et al. Clusters of conserved beta cell marker genes for assessment of beta cell phenotype. PLoS One. 2011;6(9):e24134. doi: 10.1371/journal.pone.0024134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowanadisai W, Lönnerdal B, Kelleher SL. Zip6 (LIV-1) regulates zinc uptake in neuroblastoma cells under resting but not depolarizing conditions. Brain Res. 2008;1199:10–19. doi: 10.1016/j.brainres.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Lue HW, Yang X, Wang R, Qian W, Xu RZ, Lyles R, et al. LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PLoS One. 2011;6(11):e27720. doi: 10.1371/journal.pone.0027720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogstrand C, Kille P, Nicholson RI, Taylor KM. Zinc transporters and cancer: a potential role for ZIP7 as a hub for tyrosine kinase activation. Trends Mol Med. 2009;15(3):101–111. doi: 10.1016/j.molmed.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Beker Aydemir T, Chang SM, Guthrie GJ, Maki AB, Ryu MS, Karabiyik A, et al. Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia) PLoS One. 2012;7(10):e48679. doi: 10.1371/journal.pone.0048679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ku GM, Kim H, Vaughn IW, Hangauer MJ, Myung Oh C, German MS, et al. Research resource: RNA-Seq reveals unique features of the pancreatic β-cell transcriptome. Mol Endocrinol. 2012;26(10):1783–1792. doi: 10.1210/me.2012-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moldawer LL, Copeland EM. Proinflammatory cytokines, nutritional support, and the cachexia syndrome: interactions and therapeutic options. Cancer. 1997;79(9):1828–1839. doi: 10.1002/(SICI)1097-0142(19970501)79:9<1828::AID-CNCR28>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 46.Cnop M, Welsh N, Jonas JC, Jörns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(Suppl 2):S97–107. doi: 10.2337/diabetes.54.suppl_2.S97. [DOI] [PubMed] [Google Scholar]
- 47.Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ylipaasto P, Kutlu B, Rasilainen S, Rasschaert J, Salmela K, Teerijoki H, et al. Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia. 2005;48(8):1510–1522. doi: 10.1007/s00125-005-1839-7. [DOI] [PubMed] [Google Scholar]
- 49.Ortis F, Naamane N, Flamez D, Ladrière L, Moore F, Cunha DA, et al. Cytokines interleukin-1beta and tumor necrosis factor-alpha regulate different transcriptional and alternative splicing networks in primary beta-cells. Diabetes. 2010;59(2):358–374. doi: 10.2337/db09-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore F, Naamane N, Colli ML, Bouckenooghe T, Ortis F, Gurzov EN, et al. STAT1 is a master regulator of pancreatic {beta}-cell apoptosis and islet inflammation. J Biol Chem. 2011;286(2):929–41. [DOI] [PMC free article] [PubMed]
- 51.Cerf ME, Muller CJ, Du Toit DF, Louw J, Wolfe-Coote SA. Transcription factors, pancreatic development, and beta-cell maintenance. Biochem Biophys Res Commun. 2005;326(4):699–702. doi: 10.1016/j.bbrc.2004.10.217. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen K, Karlsen AE, Deckert M, Madsen OD, Serup P, Mandrup-Poulsen T, et al. Beta-cell maturation leads to in vitro sensitivity to cytotoxins. Diabetes. 1999;48(12):2324–2332. doi: 10.2337/diabetes.48.12.2324. [DOI] [PubMed] [Google Scholar]
- 53.Nielsen K, Kruhøffer M, Orntoft T, Sparre T, Wang H, Wollheim C, et al. Gene expression profiles during beta cell maturation and after IL-1beta exposure reveal important roles of Pdx-1 and Nkx6.1 For IL-1beta sensitivity. Diabetologia. 2004;47(12):2185–2199. doi: 10.1007/s00125-004-1578-1. [DOI] [PubMed] [Google Scholar]
- 54.Hansen JB, Tonnesen MF, Madsen AN, Hagedorn PH, Friberg J, Grunnet LG, et al. Divalent metal transporter 1 regulates iron-mediated ROS and pancreatic β cell fate in response to cytokines. Cell Metab. 2012;16(4):449–461. doi: 10.1016/j.cmet.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Hayes HL, Moss LG, Schisler JC, Haldeman JM, Zhang Z, Rosenberg PB, et al. Pdx-1 activates islet α- and β-cell proliferation via a mechanism regulated by transient receptor potential cation channels 3 and 6 and extracellular signal-regulated kinases 1 and 2. Mol Cell Biol. 2013;33(20):4017–4029. doi: 10.1128/MCB.00469-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachdeva MM, Claiborn KC, Khoo C, Yang J, Groff DN, Mirmira RG, et al. Pdx1 (MODY4) Regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci U S A. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bensellam M, Van Lommel L, Overbergh L, Schuit FC, Jonas JC. Cluster analysis of rat pancreatic islet gene mRNA levels after culture in low-, intermediate- and high-glucose concentrations. Diabetologia. 2009;52(3):463–476. doi: 10.1007/s00125-008-1245-z. [DOI] [PubMed] [Google Scholar]
- 58.Dominguez V, Raimondi C, Somanath S, Bugliani M, Loder MK, Edling CE, et al. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem. 2011;286(6):4216–4225. doi: 10.1074/jbc.M110.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marselli L, Thorne J, Dahiya S, Sgroi DC, Sharma A, Bonner-Weir S, et al. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, Ahlqvist E, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16(1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Sokolov A, Funk C, Graim K, Verspoor K, Ben-Hur A. Combining heterogeneous data sources for accurate functional annotation of proteins. BMC Bioinformatics. 2013;14(Suppl 3):S10. doi: 10.1186/1471-2105-14-S3-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rutter GA, Chabosseau P, Bellomo EA, Maret W, Mitchell RK, Hodson DJ, et al. Intracellular zinc in insulin secretion and action: a determinant of diabetes risk? Proc Nutr Soc. 2016;75(1):61–72. doi: 10.1017/S0029665115003237. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell RK, Hu M, Chabosseau PL, Cane MC, Meur G, Bellomo EA, et al. Molecular genetic regulation of Slc30a8/ZnT8 reveals a positive association with glucose tolerance. Mol Endocrinol. 2016;30(1):77–91. doi: 10.1210/me.2015-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slepchenko KG, Li YV. Rising intracellular zinc by membrane depolarization and glucose in insulin-secreting clonal HIT-T15 beta cells. Exp Diabetes Res. 2012;2012:190309. doi: 10.1155/2012/190309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes. 1991;40(4):405–412. doi: 10.2337/diab.40.4.405. [DOI] [PubMed] [Google Scholar]
- 66.Kasana S, Din J, Maret W. Genetic causes and gene–nutrient interactions in mammalian zinc deficiencies: acrodermatitis enteropathica and transient neonatal zinc deficiency as examples. J Trace Elem Med Biol. 2015;29:47–62. doi: 10.1016/j.jtemb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 67.Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- 68.Huang L, Kirschke CP, Zhang Y, Yu YY. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J Biol Chem. 2005;280(15):15456–15463. doi: 10.1074/jbc.M412188200. [DOI] [PubMed] [Google Scholar]
- 69.Thorsen K, Mansilla F, Schepeler T, Oster B, Rasmussen MH, Dyrskjot L, et al. Alternative Splicing of SLC39A14 in Colorectal Cancer is Regulated by the Wnt Pathway. Molecular & Cellular Proteomics. 2011;10(1)M110.002998. [DOI] [PMC free article] [PubMed]
- 70.Aydemir TB, Sitren HS, Cousins RJ. The Zinc Transporter Zip14 Influences c-Met Phosphorylation and Hepatocyte Proliferation During Liver Regeneration in Mice. Gastroenterology. 2012;142(7):1536–1546.e1535. [DOI] [PMC free article] [PubMed]
- 71.Taylor KM, Muraina IA, Brethour D, Schmitt-Ulms G, Nimmanon T, Ziliotto S, et al. Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem J. 2016;473:2531–2544. doi: 10.1042/BCJ20160388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hogstrand C, Kille P, Ackland ML, Hiscox S, Taylor KM. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3) Biochem J. 2013;455:229–237. doi: 10.1042/BJ20130483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor KM, Hiscox S, Nicholson RI, Hogstrand C, Kille P. Protein Kinase CK2 Triggers Cytosolic Zinc Signaling Pathways by Phosphorylation of Zinc Channel ZIP7. Sci Signal. 2012;5(210):ra11. doi: 10.1126/scisignal.2002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liuzzi JP, Lichten LA, Lanata T, Cousins RJ. Zip14 Protein is induced in mouse liver by endotoxin injection and functions as a zinc transporter. FASEB J 2005;19(5):A974-A974.
- 75.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. Zip14 (Slc39a14) Mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci U S A. 2006;103(37):13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas P, Pang Y, Dong J. Membrane androgen receptor characteristics of human ZIP9 (SLC39A) zinc transporter in prostate cancer cells: androgen-specific activation and involvement of an inhibitory G protein in zinc and MAP kinase signaling. Mol Cell Endocrinol. 2017;447:23-34. [DOI] [PubMed]
- 77.Matsuura W, Yamazaki T, Yamaguchi-Iwai Y, Masuda S, Nagao M, Andrews GK, et al. SLC39A9 (ZIP9) Regulates zinc homeostasis in the secretory pathway: characterization of the ZIP subfamily I protein in vertebrate cells. Biosci Biotechnol Biochem. 2009;73(5):1142–1148. doi: 10.1271/bbb.80910. [DOI] [PubMed] [Google Scholar]
- 78.Jenkitkasemwong S, Wang CY, Mackenzie B, Knutson MD. Physiologic implications of metal-ion transport by ZIP14 and ZIP8. Biometals. 2012;25(4):643–655. doi: 10.1007/s10534-012-9526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor KM, Morgan HE, Smart K, Zahari NM, Pumford S, Ellis IO, et al. The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol Med. 2007;13(7–8):396–406. doi: 10.2119/2007-00040.Taylor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giunta C, Elçioglu NH, Albrecht B, Eich G, Chambaz C, Janecke AR, et al. Spondylocheiro dysplastic form of the Ehlers-Danlos syndrome--an autosomal-recessive entity caused by mutations in the zinc transporter gene SLC39A13. Am J Hum Genet. 2008;82(6):1290–1305. doi: 10.1016/j.ajhg.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukada T, Civic N, Furuichi T, Shimoda S, Mishima K, Higashiyama H, et al. The zinc transporter SLC39A13/ZIP13 is required for connective tissue development; its involvement in BMP/TGF-beta signaling pathways. PLoS One. 2008;3(11):e3642. doi: 10.1371/journal.pone.0003642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeong J, Walker JM, Wang F, Park JG, Palmer AE, Giunta C, et al. Promotion of vesicular zinc efflux by ZIP13 and its implications for spondylocheiro dysplastic Ehlers-Danlos syndrome. Proc Natl Acad Sci U S A. 2012;109(51):E3530–E3538. doi: 10.1073/pnas.1211775110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clocquet AR, Egan JM, Stoffers DA, Muller DC, Wideman L, Chin GA, et al. Impaired insulin secretion and increased insulin sensitivity in familial maturity-onset diabetes of the young 4 (insulin promoter factor 1 gene) Diabetes. 2000;49(11):1856–1864. doi: 10.2337/diabetes.49.11.1856. [DOI] [PubMed] [Google Scholar]
- 84.Matsuoka TA, Kawashima S, Miyatsuka T, Sasaki S, Shimo N, Katakami N, et al. Mafa enables Pdx1 to effectively convert pancreatic islet progenitors and committed islet α-cells into β-cells in vivo. Diabetes. 2017;66(5)1293-1300. [DOI] [PMC free article] [PubMed]
- 85.Cheng CW, Villani V, Buono R, Wei M, Kumar S, Yilmaz OH et al. Fasting-Mimicking Diet Promotes Ngn3-Driven β-Cell Regeneration to Reverse Diabetes. Cell. 2017;168(5):775–788.e12. [DOI] [PMC free article] [PubMed]
- 86.Yamamoto Y, Miyatsuka T, Sasaki S, Miyashita K, Kubo F, Shimo N, et al. Preserving expression of Pdx1 improves β-cell failure in diabetic mice. Biochem Biophys Res Commun. 2017;483(1):418–424. doi: 10.1016/j.bbrc.2016.12.128. [DOI] [PubMed] [Google Scholar]
- 87.Pound LD, Hang Y, Sarkar SA, Wang Y, Milam LA, Oeser JK, et al. The pancreatic islet β-cell-enriched transcription factor Pdx-1 regulates Slc30a8 gene transcription through an intronic enhancer. Biochem J. 2011;433(1):95–105. doi: 10.1042/BJ20101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12(11):4251–9. [DOI] [PMC free article] [PubMed]
- 89.Noh H, Paik HY, Kim J, Chung J. The alteration of zinc transporter gene expression is associated with inflammatory markers in obese women. Biol Trace Elem Res. 2014;158(1):1–8. doi: 10.1007/s12011-014-9902-1. [DOI] [PubMed] [Google Scholar]
- 90.Besecker B, Bao S, Bohacova B, Papp A, Sadee W, Knoell DL. The human zinc transporter SLC39A8 (Zip8) is critical in zinc-mediated cytoprotection in lung epithelia. Am J Physiol Lung Cell Mol Physiol. 2008;294(6):L1127–L1136. doi: 10.1152/ajplung.00057.2008. [DOI] [PubMed] [Google Scholar]
- 91.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, et al. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc Natl Acad Sci U S A. 2005;102(19):6843–6848. doi: 10.1073/pnas.0502257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aronson D. Hyperglycemia and the pathobiology of diabetic complications. Adv Cardiol. 2008;45:1–16. doi: 10.1159/000115118. [DOI] [PubMed] [Google Scholar]
- 93.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci U S A. 2009;106(45):18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maris M, Ferreira GB, D'Hertog W, Cnop M, Waelkens E, Overbergh L, et al. High glucose induces dysfunction in insulin secretory cells by different pathways: a proteomic approach. J Proteome Res. 2010;9(12):6274–6287. doi: 10.1021/pr100557w. [DOI] [PubMed] [Google Scholar]
- 95.Schrimpe-Rutledge AC, Fontès G, Gritsenko MA, Norbeck AD, Anderson DJ, Waters KM, et al. Discovery of novel glucose-regulated proteins in isolated human pancreatic islets using LC-MS/MS-based proteomics. J Proteome Res. 2012;11(7):3520–3532. doi: 10.1021/pr3002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brackeva B, Kramer G, Vissers JP, Martens GA. Quantitative proteomics of rat and human pancreatic beta cells. Data Brief. 2015;3:234–239. doi: 10.1016/j.dib.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chandramouli K, Qian PY. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009:239204. [DOI] [PMC free article] [PubMed]
- 98.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 99.Steger D, Berry D, Haider S, Horn M, Wagner M, Stocker R, et al. Systematic spatial bias in DNA microarray hybridization is caused by probe spot position-dependent variability in lateral diffusion. PLoS One. 2011;6(8):e23727. doi: 10.1371/journal.pone.0023727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gyulkhandanyan AV, Lu H, Lee SC, Bhattacharjee A, Wijesekara N, Fox JE, et al. Investigation of transport mechanisms and regulation of intracellular Zn2+ in pancreatic alpha-cells. J Biol Chem. 2008;283(15):10184–10197. doi: 10.1074/jbc.M707005200. [DOI] [PubMed] [Google Scholar]
- 101.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysed datasets. (XLSX 61 kb)
Designs for human qPCR assays undertaken. (DOCX 12 kb)
Designs for mouse qPCR assays undertaken. (DOCX 12 kb)
ZIP isoforms in human islets. (DOCX 56 kb)
Data Availability Statement
Datasets analysed during the current study are available in either the GEO repository [https://www.ncbi.nlm.nih.gov/geo/] or the ArrayExpress repository [https://www.ebi.ac.uk/arrayexpress/]. Respective dataset ID’s are provided in Table 1.


