Abstract
Background
Drug resistance limits options for antiretroviral therapy (ART) and results in poorer health outcomes among HIV-infected persons. We sought to characterize resistance patterns and to identify predictors of resistance in Washington, DC.
Methods
We analyzed resistance in the DC Cohort, a longitudinal study of HIV-infected persons in care in Washington, DC. We measured cumulative drug resistance (CDR) among participants with any genotype between 1999 and 2014 (n = 3411), transmitted drug resistance (TDR) in ART-naïve persons (n = 1503), and acquired drug resistance (ADR) in persons with genotypes before and after ART initiation (n = 309). Using logistic regression, we assessed associations between patient characteristics and transmitted resistance to any antiretroviral.
Results
Prevalence of TDR was 20.5%, of ADR 40.5%, and of CDR 45.1% in the respective analysis groups. From 2004 to 2013, TDR prevalence decreased for nucleoside and nucleotide analogue reverse transcriptase inhibitors (15.0 to 5.5%; p = 0.0003) and increased for integrase strand transfer inhibitors (INSTIs) (0.0–1.4%; p = 0.04). In multivariable analysis, TDR was not associated with age, race/ethnicity, HIV risk group, or years from HIV diagnosis.
Conclusions
In this urban cohort of HIV-infected persons, almost half of participants tested had evidence of CDR; and resistance to INSTIs was increasing. If this trend continues, inclusion of the integrase-encoding region in baseline genotype testing should be strongly considered.
Keywords: HIV, Antiretroviral therapy, Drug resistance, Transmitted drug resistance, Acquired drug resistance, Cumulative drug resistance, Prevalence, Washington, DC
Background
Since 1995, the use of combination antiretroviral therapy (ART) has dramatically improved life expectancy and health outcomes for people infected with HIV, but resistance to antiretroviral drugs (ARVs) undermines their effectiveness [1–4]. Drug resistance may be acquired in response to drug pressure (ADR) or transmitted at the time of infection (TDR). In the United States (US), estimates of TDR prevalence range from 4 to 27% [5–28]. Some reports indicate that resistance to nucleoside and nucleotide analogue reverse transcriptase inhibitors (NRTIs) has remained stable or decreased, while resistance to nonnucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs) has remained stable or increased [14, 19, 23, 26–28]. Few data are available on the prevalence of resistance to the newer ARV classes: entry/fusion inhibitors (EIs) and integrase strand transfer inhibitors (INSTIs). Two recent studies found no resistance to INSTIs [29, 30]; however, with INSTI-based regimens featuring prominently in the latest US Department of Health and Human Services treatment guidelines [31], increasing resistance to this class is likely.
While TDR has been fairly well documented, fewer data exist on rates of ADR and of cumulative drug resistance (CDR), a term we use to encompass all resistance, whether transmitted, acquired, or of unknown origin. One study of homeless persons in San Francisco found ADR prevalence of 36% [9]. The same study and one other found CDR prevalence rates of 27 and 45%, respectively [9, 32]. These categories of resistance may provide an indication of how well a city is maintaining treatment and adherence in its infected population. Additionally, Tilghman et al. found that high levels of CDR at specific gene locations predicted TDR in the same locations [33].
In Washington, DC, which has an HIV prevalence of 2.5% [34], recent studies of TDR have found that 17–23% of participants had mutations associated with resistance to at least one drug [26, 28] while two earlier studies reported resistance rates of up to 17%, depending on drug class [35, 36]. These findings suggest resistance is common, yet citywide prevalence is unknown. The DC Cohort, a longitudinal observational study of HIV-infected persons receiving outpatient care at 13 clinics throughout Washington, DC [37], affords a unique opportunity to characterize prevalence in a major urban area with a high burden of HIV. With 6743 people enrolled as of December 2014, including 4969 DC residents, the study aims to provide a representative sample of the 16,423 people estimated to be living with HIV in the city [34]. Additionally, the longitudinal nature of the study makes it possible to distinguish, for some participants, between transmitted and acquired drug resistance.
In this analysis, we aimed to describe the prevalence of and trends in ARV drug resistance among DC Cohort participants by category of resistance (TDR, ADR, and CDR); specifically, to measure prevalence of individual drug resistant mutations and to estimate resistance to individual drugs and drug classes. We further sought to examine associations between patient characteristics and the presence of transmitted drug resistance.
Methods
Data source and study population
Enrollment in the DC Cohort began in January 2011. Data on all consenting participants are electronically exported on a monthly basis. Historical data are manually abstracted including genotype and phenotype tests and date of ART initiation where available [38]. For the present analysis, we included all active participants enrolled through December 2014 and not perinatally infected (n = 6506). Our study population included both recently infected individuals and people who had been living with HIV for many years. Participants with any documented genotype test between 1999 and 2014 were included for the estimates of CDR (n = 3411). Those who were documented treatment-naïve at first genotype test were evaluated for TDR (n = 1503). Among the latter group, those who had one or more additional genotype tests after ART initiation were assessed for ADR (n = 309) (Fig. 1). The DC Cohort study was approved by the George Washington University Institutional Review Board (IRB), and all 13 sites received IRB approval to participate in the study.
Fig. 1.
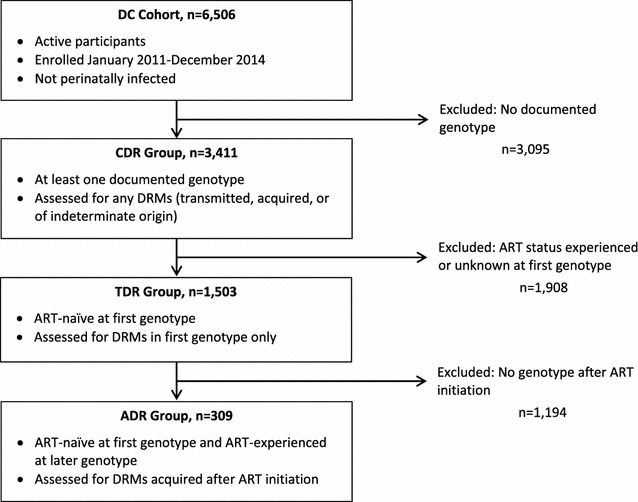
Composition of DC Cohort analysis groups, Washington DC, 1999–2014. CDR, cumulative drug resistance; TDR, transmitted drug resistance; ADR, acquired drug resistance; DRM, drug-resistant mutation; ART, antiretroviral therapy
Measurement of resistance
Multiple commercial assays were used for the genotype testing, some of which occurred prior to study enrollment. A total of 5993 genotypes were analyzed (LabCorp: 3047; TruGene: 1279; Monogram Biosciences: 621; Quest: 467; other: 579) representing all 13 clinical sites. Although major and minor mutations were available for the reverse transcriptase and protease genes, and sometimes for the envelope and integrase genes, full sequences were not available, and we did not have information on which specific genotypes were evaluated for EI and INSTI resistance. We measured the prevalence of individual drug resistance mutations (DRMs) that were included in the WHO Surveillance Drug Resistance Mutations list [39] or in the 2014 International Antiviral Society-USA (IAS) HIV-1 drug mutations classification [40]. From the latter, we included all bolded amino acid substitutions and all mutations at bolded PI locations. We then interpreted resistance to drugs and drug classes using the IAS classification alone; however, for PIs, since the IAS guidelines identified only major locations and not specific amino acid substitutions, we used the 2014 Stanford HIVDB genotypic resistance interpretation algorithm (Version 7.0), including intermediate and high-level resistance mutations at bolded locations [41]. Phenotypic data were not examined.
To determine the prevalence of TDR, we used the first genotype test for each ART-naïve participant (1503 tests). For ADR, we assessed mutations present in tests after ART initiation (557 tests) and absent in the initial test (309 tests). We did not have complete data on the drug regimen for each participant at the time of the test; therefore specific regimen was not taken into account. The CDR analysis group included all participants in the TDR and ADR groups as well as many more for whom we were not able to ascertain whether mutations were transmitted or acquired. To estimate the prevalence of cumulative drug resistance in this group, we included all DRMs on every test, regardless of treatment status at the time of testing (5993 tests). A participant with a given mutation on any test was considered to have that mutation for the remainder of the study period.
Analysis
Characteristics of the DC Cohort at enrollment were assessed as frequencies and proportions for categorical variables and as medians and interquartile ranges for continuous variables. To evaluate trends in resistance from 2004 to 2013, we used the Cochran–Armitage test with 2-sided p values. We performed simple and multivariable logistic regression to examine potential associations between patient characteristics and transmitted resistance to any drug class. In the multivariable model, we included, a priori, age at genotype test, race/ethnicity, transmission risk group, and years from HIV diagnosis along with any variables that proved statistically significant with α = 0.05 in bivariate regression. All analysis was conducted using SAS 9.2 (SAS Institute, Cary, North Carolina).
Results
Demographics
The median age of DC Cohort participants at enrollment was 48 years. Participants were mostly male (73.8%), non-Hispanic black (76.4%) and infected through male-to-male sex (38.7%) or heterosexual sex (30.7%). Nearly two-thirds had public health insurance (64.9%), and roughly equal numbers of participants received care at hospital-based clinics (48.2%) and community-based clinics (51.8%). Most participants had CD4 counts above 500 cells/μl (51.5%) and viral loads below 400 copies/ml (75.7%) at enrollment; 41.6% of participants had been diagnosed with AIDS. The median interval between HIV diagnosis and consent date was 9.3 years (Table 1). Clinical characteristics at enrollment were not reflected in the resistance results, which were based on genotype tests that were often performed years earlier or later.
Table 1.
Characteristics of DC Cohort at enrollment, Washington DC, 1999–2014
| Characteristic at enrollmenta | N (%) |
|---|---|
| All participants | 6506 (100) |
| Age | |
| <18 | 17 (0.26) |
| 18–29 | 693 (10.65) |
| 30–39 | 1134 (17.43) |
| 40–49 | 1945 (29.90) |
| 50–59 | 1948 (29.94) |
| 60+ | 769 (11.82) |
| Sex | |
| Female | 1702 (26.16) |
| Male | 4804 (73.84) |
| Race/ethnicity | |
| Non-Hispanic black | 4972 (76.42) |
| Non-Hispanic white | 904 (13.89) |
| Hispanic | 282 (4.33) |
| Other | 127 (1.95) |
| Unknown | 221 (3.40) |
| Transmission risk group | |
| Male-to-male sexual contact (MMS) | 2515 (38.66) |
| Heterosexual contact | 1995 (30.66) |
| Injection drug use (IDU) | 458 (7.04) |
| MMS/IDU | 80 (1.23) |
| Other | 121 (1.86) |
| Unknown/missing | 1337 (20.55) |
| Insurance | |
| Public | 4222 (64.89) |
| Private | 1767 (27.16) |
| Other | 131 (2.01) |
| Unknown | 386 (5.93) |
| Clinic type | |
| Hospital-based | 3136 (48.20) |
| Community-based | 3370 (51.80) |
| Clinical status | |
| HIV | 3798 (58.38) |
| AIDS | 2708 (41.62) |
| CD4 count (cells/µl) | |
| <50 | 158 (2.43) |
| 50–199 | 525 (8.07) |
| 200–349 | 938 (14.42) |
| 350–499 | 1285 (19.75) |
| ≥500 | 3349 (51.48) |
| Unknown | 251 (3.86) |
| Viral load (copies/ml) | |
| 0–399 | 4922 (75.65) |
| 400–999 | 186 (2.86) |
| 1000–9999 | 366 (5.63) |
| 10,000–49,999 | 362 (5.56) |
| 50,000–99,999 | 155 (2.38) |
| ≥100,000 | 250 (3.84) |
| Unknown | 265 (4.07) |
| Alcohol use | |
| Current | 915 (14.06) |
| Previous | 924 (14.20) |
| Never | 3038 (46.70) |
| Unknown | 1629 (25.04) |
| Recreational drug use | |
| Current | 774 (11.90) |
| Previous | 1416 (21.76) |
| Never | 2147 (33.00) |
| Unknown | 2169 (33.34) |
| Intravenous drug use | |
| Current | 45 (0.69) |
| Previous | 476 (7.32) |
| Never | 3132 (48.14) |
| Unknown | 2853 (43.85) |
| Characteristic at enrollmenta | Median (IQR) |
|---|---|
| HIV diagnosis to consent (years) | 9.3 (4.2–16.3) |
| HIV diagnosis to ART start (years) | 1.2 (0.1–6.2) |
| ART start to consent (years) | 3.6 (1.2–7.8) |
ART, antiretroviral treatment
aClinical characteristics at enrollment do not correspond to resistance results, which are based on tests performed at other times
Prevalence
Among the 5993 genotypes analyzed, 5895 were subtype B, 48 were C, 17 were AG, and 33 were other subtypes. In the TDR group (ART-naïve at genotype), prevalence of TDR to any drug class was 20.5%: 7.9% for NRTIs, 11.7% for NNRTIs, 5.7% for PIs, 1.1% for EIs, and 0.9% for INSTIs (Table 2). In the ADR group (genotypes before and after ART initiation), ADR prevalence was 40.5%; while in the CDR group (all participants tested), CDR prevalence was 45.1%. In terms of specific drugs, all three groups were most resistant to efavirenz (TDR: 10.0%; ADR: 24.6%; CDR 27.2%), and nevirapine (TDR: 10.2%; ADR: 23.9%; CDR 27.1%). The ADR and CDR groups also had high levels of resistance to emtricitabine and lamivudine (TDR: 3.1%; ADR: 20.4%; CDR: 24.3%), and abacavir (TDR: 3.5%; ADR: 19.1%; CDR: 24.2%). Among the protease inhibitors, the highest levels of TDR and CDR were to nelfinavir (TDR: 1.9%; ADR: 0.0%; CDR: 7.2%), and of ADR to atazanavir (TDR: 1.8; ADR: 3.2%; CDR: 5.3). No resistance to darunavir was detected as TDR, ADR, or CDR. Resistance to the fusion inhibitor enfuvirtide was found in a few participants (TDR: 1.1%; ADR: 1.0; CDR: 1.5). Maraviroc resistance was not assessed because tropism determination was not available. Mutations conferring resistance to raltegravir (TDR: 0.6%; ADR: 1.6%; CDR: 1.5%) and elvitegravir (TDR: 0.9%; ADR: 0.6%; CDR: 1.3%) were found in all three analysis groups; while in the CDR group only, 4 participants had evidence of resistance to dolutegravir (TDR: 0.0%; ADR: 0.0; CDR: 0.1%). Resistance to three or more classes was 1.2% for TDR, 1.9% for ADR, and 7.1% for CDR.
Table 2.
Prevalence of resistance to antiretroviral agents, Washington DC, 1999–2014
| Transmitted drug resistance (n = 1503) |
Acquired drug resistance (n = 309) |
Cumulative drug resistance (n = 3411) |
||||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| NRTIsa | ||||||
| Abacavir | 52 | 3.5 | 59 | 19.1 | 826 | 24.2 |
| Didanosine | 14 | 0.9 | 12 | 3.9 | 158 | 4.6 |
| Emtricitabine | 47 | 3.1 | 63 | 20.4 | 828 | 24.3 |
| Lamivudine | 47 | 3.1 | 63 | 20.4 | 828 | 24.3 |
| Stavudine | 88 | 5.9 | 23 | 7.4 | 582 | 17.1 |
| Tenofovir | 7 | 0.5 | 10 | 3.2 | 98 | 2.9 |
| Zidovudine | 82 | 5.5 | 17 | 5.5 | 524 | 15.4 |
| NNRTIsa | ||||||
| Efavirenz | 151 | 10.0 | 76 | 24.6 | 929 | 27.2 |
| Etravirine | 19 | 1.3 | 11 | 3.6 | 212 | 6.2 |
| Nevirapine | 153 | 10.2 | 74 | 23.9 | 926 | 27.1 |
| Rilpivirine | 42 | 2.8 | 24 | 7.8 | 331 | 9.7 |
| PIsb | ||||||
| Atazanavir | 27 | 1.8 | 10 | 3.2 | 181 | 5.3 |
| Darunavir | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Fosamprenavir | 18 | 1.2 | 5 | 1.6 | 118 | 3.5 |
| Indinavir | 17 | 1.1 | 2 | 0.6 | 175 | 5.1 |
| Lopinavir | 22 | 1.5 | 4 | 1.3 | 166 | 4.9 |
| Nelfinavir | 29 | 1.9 | 0 | 0.0 | 247 | 7.2 |
| Saquinavir | 25 | 1.7 | 1 | 0.3 | 195 | 5.7 |
| Tipranavir | 13 | 0.9 | 3 | 1.0 | 117 | 3.4 |
| EIsa,c | ||||||
| Enfuvirtide | 17 | 1.1 | 3 | 1.0 | 52 | 1.5 |
| INSTIsa | ||||||
| Dolutegravir | 0 | 0.0 | 0 | 0.0 | 4 | 0.1 |
| Elvitegravir | 14 | 0.9 | 2 | 0.6 | 45 | 1.3 |
| Raltegravir | 9 | 0.6 | 5 | 1.6 | 50 | 1.5 |
| Any ARV | 308 | 20.5 | 125 | 40.5 | 1538 | 45.1 |
| Any NRTI | 118 | 7.9 | 71 | 23.0 | 1013 | 29.7 |
| Any NNRTI | 176 | 11.7 | 81 | 26.2 | 998 | 29.3 |
| Any PI | 86 | 5.7 | 19 | 6.1 | 498 | 14.6 |
| Any EI | 17 | 1.1 | 3 | 1.0 | 52 | 1.5 |
| Any INSTI | 14 | 0.9 | 6 | 1.9 | 60 | 1.8 |
| Any 2 classes | 67 | 4.5 | 42 | 13.6 | 582 | 17.1 |
| Any 3 classes | 18 | 1.2 | 5 | 1.6 | 228 | 6.7 |
| Any 4 classes | 0 | 0.0 | 1 | 0.3 | 15 | 0.4 |
NRTI, nucleoside/nucleotide analogue reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; EI, entry/fusion inhibitor; INSTI, integrase strand transfer inhibitor
aInterpreted using 2014 International Antiviral Society-USA (IAS) HIV-1 drug mutations classification
bInterpreted using 2014 IAS classification and 2014 Stanford HIVDB genotypic resistance interpretation algorithm
cThe 2014 IAS classification did not include maraviroc
The prevalence of the K103N mutation, associated with resistance to NNRTIs, was high for all three analysis groups (TDR: 7.1%; ADR: 18.8%; CDR: 20.2%) (Fig. 2). Prevalence was also high for NRTI-associated mutations M41L (TDR: 3.0%; ADR: 1.0%; CDR: 7.3%) and M184V (TDR: 2.8%; ADR: 17.8%; CDR: 22.9%). Among protease-associated mutations, L90M (TDR: 1.5%; ADR: 0.0%; CDR: 5.5%) was most prevalent in the TDR and CDR groups, and N88S (TDR: 1.3%; ADR: 2.6%; CDR: 2.6%) in the ADR group. Integrase-associated mutations were detected at nine sites: primarily at F121Y (TDR: 0.6%; ADR: 0.3%; CDR: 0.8%), E92Q (TDR: 0.3%; ADR: 0.3%; CDR: 0.2%), Q148R (TDR: 0.0%; ADR: 0.6%; CDR: 0.2%), and N155H (TDR: 0.0%; ADR: 0.0%; CDR: 0.2%). On the env gene, the most common mutation was N42T (TDR: 0.3%; ADR: 0.0%; CDR: 0.6%).
Fig. 2.
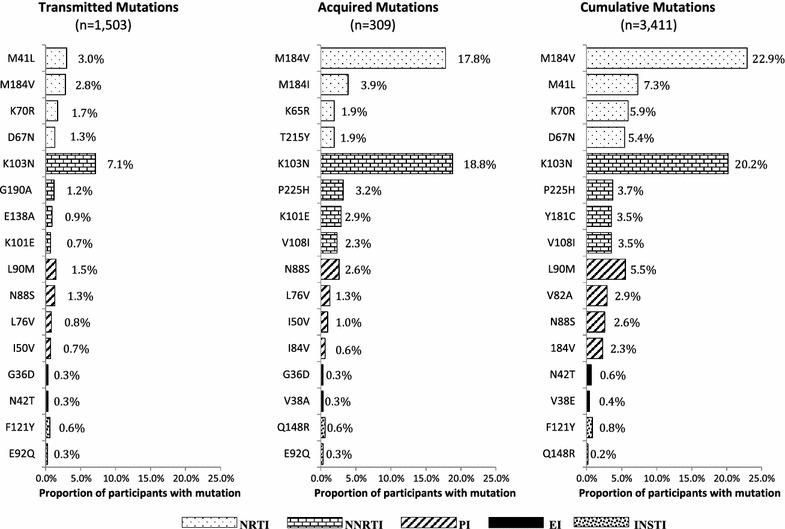
Most prevalent drug-resistant mutations for each analysis group by drug class, Washington DC, 1999–2014. NRTI, nucleoside/nucleotide analogue reverse transcriptase inhibitor (top 4); NNRTI, nonnucleoside reverse transcriptase inhibitor (top 4); PI, protease inhibitor (top 4); EI, entry/fusion inhibitor (top 2); INSTI, integrase strand transfer inhibitor (top 2)
Time trends
From 2004 to 2013, TDR was fairly stable around 20% (15.0–20.7%; p = 0.76), with a marked decrease for NRTIs (15.0 to 5.5%; p = 0.0003) and a small increase for INSTIs (0.0–1.4%; p = 0.04) (Fig. 3). Over the same time period, the proportion of newly diagnosed participants who had a genotype test within the first year of diagnosis steadily increased [5.5–65.9%; p < 0.0001 (data not shown)]. For ADR, the number of participants tested prior to 2008 was too small to permit meaningful analysis (fewer than 10 per year). The prevalence of ADR decreased from 66.7% in 2008 to 41.6% in 2013 (p = 0.003). ADR also decreased significantly for NRTIs (47.2 to 24.1%; p = 0.0004) and NNRTIs (47.2 to 26.9%; p = 0.002). Resistance to PIs rose slightly (5.6–6.3%; p = 0.25), but the difference was not significant and as noted above, no resistance was found to darunavir, which has perhaps the highest barrier to resistance. CDR prevalence to any drug class declined significantly from 70.6% in 2004 to 45.0% in 2013 (p < 0.0001). The trend was also significant for NRTIs (63.9 to 29.9%; p < 0.0001), NNRTIs (43.6 to 29.1%; p < 0.0001), PIs (32.4 to 14.8%; p < 0.0001), and to any 2 (33.6 to 17.0%; p < 0.0001) or 3 (17.9 to 6.9%; p < 0.0001) drug classes, while resistance increased for EIs (0.0–1.5%; p < 0.0001), INSTIs (0.0–1.8%; p < 0.0001), and any four drug classes (0.0–0.4%; p < 0.0001).
Fig. 3.
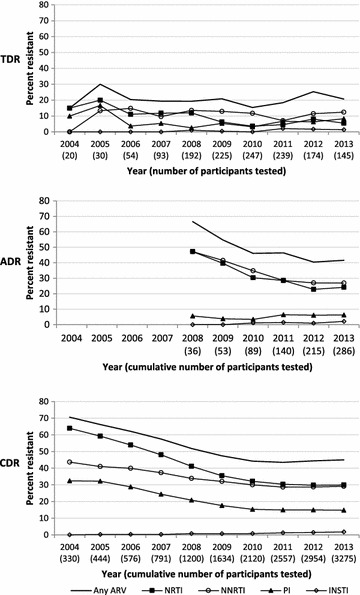
Trends in antiretroviral resistance by drug class, Washington DC, 2004–2013. TDR, transmitted drug resistance; ADR, acquired drug resistance; CDR, cumulative drug resistance. Results for 2014 are not shown due to incomplete data at the time of analysis
Logistic regression analysis
We decided a priori to include age at test, race/ethnicity, transmission risk group and years from HIV diagnosis in the multivariable regression model. Based on a statistically significant association at the α = 0.05 level in bivariate regression analysis, we added clinic type to the model. In multivariable analysis, TDR was not predicted by time between HIV diagnosis and genotype, age at genotype or race/ethnicity. Although not statistically significant, individuals infected through injection drug use (OR 1.53; 95% CI 0.79–2.97) and those receiving HIV care at community-based clinics (OR 1.27; 95% CI 0.95–1.72) were more likely to have transmitted resistance than individuals infected through male-to-male sexual contact and participants cared for at hospital-based clinics, respectively.
Discussion
To our knowledge, we are the first to report positive findings of mutations associated with transmitted resistance to INSTIs (0.9%) and EIs (1.1%). We also found evidence of resistance to these classes among participants analyzed for ADR and CDR as well as significantly increasing trends for cumulative resistance to both classes and transmitted resistance to INSTIs. Unfortunately we were not able to determine which genotypes included the INSTI and EI encoding regions, and so we report prevalence among all genotypes assessed; thus, our rates are underestimates of prevalence for these classes. The emergence of resistance to INSTIs is likely due to their increasingly widespread use in clinical practice as well as their earlier use in clinical trials. Several DC sites participated in registration trials for all three INSTIs, the first of which (raltegravir) was FDA-approved in 2007; and by the end of 2014, approximately 10% of DC Cohort participants were on INSTI-based regimens. Surprisingly, resistance to elvitegravir—as interpreted using the IAS guidelines—was higher than to raltegravir, although the latter was introduced earlier. This was mainly attributable to the presence of E92Q and T66I mutations. In the most recent US Department of Health and Human Services (DHHS) treatment guidelines, four of the five recommended regimens for ART-naïve patients were INSTI-based, while inclusion of the integrase region in routine genotype testing was still optional [31]. Given the emergence of INSTI resistance in the Washington, DC area, baseline resistance testing for integrase inhibitors should be strongly considered.
The TDR prevalence of 20.5% found in this analysis was comparable to rates reported throughout the US and in Washington, DC. We were surprised to find that 18 participants analyzed for TDR had mutations associated with resistance to three drug classes. Review of the medical records for these 18 participants is warranted to confirm that the recorded dates are accurate. Our findings of no significant association between TDR and sex, race/ethnicity, or transmission risk group support those of most [11, 14, 21, 28], but not all [10] previous studies. However, resistance appeared to be higher among injection drug users (IDUs), and although the difference was of borderline significance, it is plausible given that IDUs may have more barriers to adherence and retention in care [42].
Cumulative drug resistance may serve as a measure of a community’s burden of ARV resistance and, like community viral load, may reflect the success of treatment and adherence in that community [43]; however, these results should be interpreted with caution. First, the decrease in CDR observed between 2006 and 2010 was probably due in part to the dilution effect of increased resistance testing among newly diagnosed individuals following the 2007 DHHS recommendations [31]. Second, because patients on treatment generally have genotype testing performed when treatment fails, the prevalence of resistance in tested individuals may be higher than in the overall population of persons infected with HIV. To avoid this overestimation of resistance, others have included all treatment-experienced patients in the denominator [44] or used modeling to extend resistance estimates from tested to untested individuals [45]. By assuming that untested individuals do not have resistance, the former approach underestimates prevalence. Using this method, we found that resistance appeared to increase as the proportion of participants tested increased dramatically over the time period (results not shown). That is, the degree of underestimation decreased over time, and the resulting apparent increase in resistance was misleading. In future analysis, we hope to model resistance in the overall Cohort taking into account the rate of testing and other secular trends. Third, our measurements were based on genotype tests that do not detect minority or archived HIV strains and thus, may underestimate the true prevalence of resistance. Furthermore, some transmitted archived strains may not have been detected until after treatment was initiated, resulting in misclassification of TDR as ADR. However, since our estimates for ADR and CDR were cumulative, we maximized our ability to include archived strains within the limitations of the tests.
Other strengths of this study include the large size and representative, citywide composition of the DC Cohort, together with the availability of genotypic, demographic, and clinical data. The long-term use of INSTIs in the study population provided early evidence of resistance to this drug class, while the longitudinal data allowed us to assess acquired and cumulative resistance in a large cohort.
Conclusions
In this urban cohort of HIV-infected persons in care, almost half of participants tested had evidence of CDR, and resistance to INSTIs was increasing. If this trend continues, inclusion of the integrase-encoding region in routine genotype testing may become advisable. With new treatment guidelines recommending immediate initiation of ART for most people, innovations to promote adherence, such as co-formulations and longer-acting regimens, will be more critical than ever. Continued close surveillance of transmitted and acquired resistance will measure the success of these efforts and inform future testing and treatment guidelines.
Authors’ contributions
AMA contributed to study conception and design, analyzed and interpreted data, and drafted the manuscript. ADC and DMP contributed to study conception and design and data acquisition, and revised the manuscript. The DC Cohort Executive Board contributed to data acquisition and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Data in this manuscript were collected by the DC Cohort investigators and research staff located at: Cerner Corporation; Children’s National Medical Center Adolescent and Pediatric Clinics; Family and Medical Counseling Service; Georgetown University; George Washington Medical Faculty Associates; George Washington University Department of Epidemiology and Biostatistics; Howard University; La Clinica Del Pueblo; MetroHealth; Unity Health Care; Veterans Affairs Medical Center; Washington Hospital Center; and Whitman-Walker Health. The authors would also like to acknowledge members of the DC Cohort Data and Statistics Coordinating Center including Dr. Naji Younes, Dr. James Peterson, Ms. Maria Jaurretche and Ms. Lindsey Powers Happ, of the George Washington University Milken Institute School of Public Health as well as Dr. Jeffrey Binkley, Mr. Harlen Hays, Ms. Thilakavathy Subramanian, Ms. Dana Franklin, and Ms. Bonnie Dean of Cerner Corporation, for their work on DC Cohort data collection and processing. They would also like to acknowledge Dr. Mary Young, Dr. Carl Dieffenbach, and Dr. Deborah Goldstein for their assistance with this study as well as the research assistants at all of the participating sites and the DC Cohort Community Advisory Board. Abstract presented at ID Week 2015, San Diego, CA, October 7–11, 2015.
DC Cohort Executive Committee: Alan E. Greenberg, MD, MPH, George Washington University Milken Institute School of Public Health; Debra Benator, MD, Veterans Affairs Medical Center; Princy Kumar, MD, Georgetown University; Richard Elion, MD, Whitman-Walker Health; Maria Elena Ruiz, MD Washington Hospital Center; Angela Wood, MSW, Family and Medical Counseling Service; Lawrence D’Angelo, MD, MPH, Burgess Adolescent Clinic, Children’s National Medical Center; Natella Rakhmanina, MD, Ph.D., Special Immunology Service Pediatric Clinic, Children’s National Medical Center; Sohail Rana, MD, Pediatric Clinic, Howard University Hospital; Maya Bryant, MD, Saumil Doshi, MD, Adult Infectious Disease Clinic Howard University Hospital; Annick Hebou, MD, MetroHealth; Ricardo Fernandez, MD, La Clinica Del Pueblo; Stephen Abbott, MD, Unity Health Care; Rachel Hart, MS, Cerner Corporation; Michael Kharfen, BA, HIV/AIDS, Hepatitis, Sexually Transmitted Diseases, Tuberculosis Administration, DC Department of Health; Henry Masur, MD, National Institutes of Health.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The DC Cohort database contains numerous individual-level variables and is therefore not publicly available in keeping with the IRB approval and our informed consent process. However, a de-identified limited dataset supporting the conclusions of this article is available by request. Interested parties may contact Dr. Amanda Castel, the DC Cohort Principal Investigator, at acastel@gwu.edu to arrange access to such.
Consent for publication
All participants have consented to publication of collective data during the informed consent process.
Ethics approval and consent to participate
Written informed consent was obtained from all participants prior to enrollment in the DC Cohort. The DC Cohort study was approved by the George Washington University Institutional Review Board (IRB), which served as the IRB of record for Whitman-Walker Health, La Clinica del Pueblo, Family and Medical Counseling Service, Unity Health Care, The GW Medical Faculty Associates, MetroHealth, and Children’s National Health System (pediatric and adolescent clinics). The study was independently approved by the IRBs of Howard University Hospital (adult and pediatric clinics), MedStar Washington Hospital Center, Georgetown University, and the Veterans Affairs Medical Center.
Funding
The DC Cohort is sponsored by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (UO1 AI69503-03S2). Additionally, this publication resulted (in part) from research supported by the District of Columbia Center for AIDS Research, an NIH funded program (P30AI117970), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, FIC, NIGMS, NIDDK, and OAR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ADR
acquired drug resistance
- ART
antiretroviral therapy
- ARV
antiretroviral
- CDR
cumulative drug resistance
- DRM
drug resistance mutation
- EI
entry/fusion inhibitor
- IAS
International Antiviral Society
- IDU
injection drug user
- INSTI
integrase strand transfer inhibitor
- IRB
institutional review board
- MMS
male-to-male sex
- NNRTI
nonnucleoside reverse transcriptase inhibitor
- NRTI
nucleoside/nucleotide analogue reverse transcriptase inhibitor
- PI
protease inhibitor
- TDR
transmitted drug resistance
Contributor Information
Annette M. Aldous, Email: aaldous@gwu.edu
Amanda D. Castel, Email: acastel@gwu.edu
David M. Parenti, Phone: (202) 741-2234, Email: dparenti@mfa.gwu.edu
the DC Cohort Executive Committee:
Alan E. Greenberg, Debra Benator, Princy Kumar, Richard Elion, Maria Elena Ruiz, Angela Wood, Lawrence D’Angelo, Natella Rakhmanina, Sohail Rana, Maya Bryant, Saumil Doshi, Annick Hebou, Ricardo Fernandez, Stephen Abbott, Rachel Hart, Michael Kharfen, and Henry Masur
References
- 1.D’Aquila RT, Johnson VA, Welles SL, Japour AJ, Kuritzkes DR, DeGruttola V, et al. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. AIDS Clinical Trials Group Protocol 116B/117 Team and the Virology Committee Resistance Working Group. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 2.Havlir DV, Marschner IC, Hirsch MS, Collier AC, Tebas P, Bassett RL, et al. Maintenance antiretroviral therapies in HIV infected patients with undetectable plasma HIV RNA after triple-drug therapy. AIDS Clinical Trials Group Study 343 Team. N Engl J Med. 1998;339:1261–1268. doi: 10.1056/NEJM199810293391801. [DOI] [PubMed] [Google Scholar]
- 3.Miller V, Phillips A, Rottmann C, Staszewski S, Pauwels R, Hertogs K, et al. Dual resistance to zidovudine and lamivudine in patients treated with zidovudine–lamivudine combination therapy: association with therapy failure. J Infect Dis. 1998;177:1521–1532. doi: 10.1086/515304. [DOI] [PubMed] [Google Scholar]
- 4.Rey D, Hughes M, Pi JT, Winters M, Merigan TC, Katzenstein DA. HIV-1 reverse transcriptase codon 215 mutation in plasma RNA: immunologic and virologic responses to zidovudine. The AIDS Clinical Trials Group Study 175 Virology Team. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:203–208. doi: 10.1097/00042560-199803010-00003. [DOI] [PubMed] [Google Scholar]
- 5.Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Jones E, et al. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- 6.Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 7.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 8.Ristig MB, Arens MQ, Kennedy M, Powderly W, Tebas P. Increasing prevalence of resistance mutations in antiretroviral-naive individuals with established HIV-1 infection from 1996–2001 in St. Louis. HIV Clin Trials. 2002;3:155–160. doi: 10.1310/RUAA-TUJA-QQC3-G5FX. [DOI] [PubMed] [Google Scholar]
- 9.Holodniy M, Charlebois ED, Bangsberg DR, Zolopa AR, Schulte M, Moss AR. Prevalence of antiretroviral drug resistance in the HIV-1-infected urban indigent population in San Francisco: a representative study. Int J STD AIDS. 2004;15:543–551. doi: 10.1258/0956462041558212. [DOI] [PubMed] [Google Scholar]
- 10.Weinstock H, Respess R, Heneine W, Petropoulos CJ, Hellmann NS, Luo CC, et al. Prevalence of mutations associated with reduced antiretroviral drug susceptibility among human immunodeficiency virus type 1 seroconverters in the United States, 1993–1998. J Infect Dis. 2000;182:330–333. doi: 10.1086/315686. [DOI] [PubMed] [Google Scholar]
- 11.Grubb JR, Singhatiraj E, Mondy K, Powderly WG, Overton ET. Patterns of primary antiretroviral drug resistance in antiretroviral-naive HIV-1-infected individuals in a midwest university clinic. AIDS. 2006;20:2115–2116. doi: 10.1097/01.aids.0000247579.08449.b6. [DOI] [PubMed] [Google Scholar]
- 12.Shet A, Berry L, Mohri H, Mehandru S, Chung C, Kim A, et al. Tracking the prevalence of transmitted antiretroviral drug-resistant HIV-1: a decade of experience. J Acquir Immune Defic Syndr. 2006;41:439–446. doi: 10.1097/01.qai.0000219290.49152.6a. [DOI] [PubMed] [Google Scholar]
- 13.Hurt CB, McCoy SI, Kuruc J, Nelson JA, Kerkau M, Fiscus S, et al. Transmitted antiretroviral drug resistance among acute and recent HIV infections in North Carolina from 1998 to 2007. Antivir Ther. 2009;14:673–678. [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, US—2006. AIDS. 2010;24:1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- 15.Chan PA, Tashima K, Cartwright CP, Gillani FS, Mintz O, Zeller K, et al. Short communication: transmitted drug resistance and molecular epidemiology in antiretroviral naive HIV type 1-infected patients in Rhode Island. AIDS Res Hum Retroviruses. 2011;27:275–281. doi: 10.1089/aid.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hightow-Weidman LB, Hurt CB, Phillips G, Jones K, Magnus M, Giordano TP, et al. Transmitted HIV-1 drug resistance among young men of color who have sex with men: a multicenter cohort analysis. J Adolesc Health. 2011;48:94–99. doi: 10.1016/j.jadohealth.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Huaman MA, Aguilar J, Baxa D, Golembieski A, Brar I, Markowitz N. Late presentation and transmitted drug resistance mutations in new HIV-1 diagnoses in Detroit. Int J Infect Dis. 2011;15:e764–e768. doi: 10.1016/j.ijid.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Poon AF, Aldous JL, Mathews WC, Kitahata M, Kahn JS, Saag MS, et al. Transmitted drug resistance in the CFAR network of integrated clinical systems cohort: prevalence and effects on pre-therapy CD4 and viral load. PLoS ONE. 2011;6:e21189. doi: 10.1371/journal.pone.0021189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truong HM, Kellogg TA, McFarland W, Louie B, Klausner JD, Philip SS. Sentinel surveillance of HIV-1 transmitted drug resistance, acute infection and recent infection. PLoS ONE. 2011;6:e25281. doi: 10.1371/journal.pone.0025281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youmans E, Tripathi A, Albrecht H, Gibson JJ, Duffus WA. Transmitted antiretroviral drug resistance in individuals with newly diagnosed HIV infection: South Carolina 2005–2009. South Med J. 2011;104:95–101. doi: 10.1097/SMJ.0b013e3181fcd75b. [DOI] [PubMed] [Google Scholar]
- 21.Readhead AC, Gordon DE, Wang Z, Anderson BJ, Brousseau KS, Kouznetsova MA, et al. Transmitted antiretroviral drug resistance in New York State, 2006–2008: results from a new surveillance system. PLoS ONE. 2012;7:e40533. doi: 10.1371/journal.pone.0040533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi T, Nurutdinova D, Grubb JR, Onen NF, Shacham E, Donovan M, et al. Transmitted drug-resistant HIV type 1 remains prevalent and impacts virologic outcomes despite genotype-guided antiretroviral therapy. AIDS Res Hum Retroviruses. 2012;28:259–264. doi: 10.1089/aid.2011.0022. [DOI] [PubMed] [Google Scholar]
- 23.Yanik EL, Napravnik S, Hurt CB, Dennis A, Quinlivan EB, Sebastian J, et al. Prevalence of transmitted antiretroviral drug resistance differs between acutely and chronically HIV-infected patients. J Acquir Immune Defic Syndr. 2012;61:258–262. doi: 10.1097/QAI.0b013e3182618f05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacVeigh MS, Kosmetatos MK, McDonald JE, Reeder JL, Parrish DA, Young TP. Prevalence of drug-resistant HIV type 1 at the time of initiation of antiretroviral therapy in Portland, Oregon. AIDS Res Hum Retroviruses. 2013;29:337–342. doi: 10.1089/AID.2011.0386. [DOI] [PubMed] [Google Scholar]
- 25.Gagliardo C, Brozovich A, Birnbaum J, Radix A, Foca M, Nelson J, et al. A multicenter study of initiation of antiretroviral therapy and transmitted drug resistance in antiretroviral-naive adolescents and young adults with HIV in New York City. Clin Infect Dis. 2014;58:865–872. doi: 10.1093/cid/ciu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swierzbinski MJ, Kan VL, Parenti DM. HIV in Washington, DC: prevalence of antiretroviral resistance in treatment-naive patients, 2007–2010. J AIDS HIV Res. 2015;7:49–54. doi: 10.5897/JAHR2015.0327. [DOI] [Google Scholar]
- 27.Panichsillapakit T, Smith DM, Wertheim JO, Richman DD, Little SJ, Mehta SR. Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, California 1996–2013. J Acquir Immune Defic Syndr. 2016;71:228–236. doi: 10.1097/QAI.0000000000000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassaye SG, Grossman Z, Balamane M, Johnston-White B, Liu C, Kumar P, et al. Transmitted HIV drug resistance high and longstanding in metropolitan Washington, DC. Clin Infect Dis. 2016;63:836–843. doi: 10.1093/cid/ciw382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stekler JD, McKernan J, Milne R, Tapia KA, Mykhalchenko K, Holte S, et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007–2013. Antivir Ther. 2015;20:77–80. doi: 10.3851/IMP2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpe JM, Yang O, Petropoulos CJ, et al. Absence of integrase inhibitor resistant HIV-1 transmission in the California AIDS Healthcare Foundation Network [abstract]. ICAAC 2015, September 17–21, 2015, San Diego.
- 31.Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. https://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-treatment-guidelines/0. Accessed 7 Oct 2016.
- 32.Goldsamt LA, Clatts MC, Parker MM, Colon V, Hallack R, Messina MG. Prevalence of sexually acquired antiretroviral drug resistance in a community sample of HIV-positive men who have sex with men in New York City. AIDS Patient Care STDs. 2011;25:287–293. doi: 10.1089/apc.2011.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tilghman MW, Perez-Santiago J, Osorio G, Little SJ, Richman DD, Mathews WC, et al. Community HIV-1 drug resistance is associated with transmitted drug resistance. HIV Med. 2014;15:339–346. doi: 10.1111/hiv.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Annual epidemiology and surveillance report: surveillance data through December 2013. http://doh.dc.gov/page/annual-report-2014. Accessed 7 Oct 2016.
- 35.Boyd AC, Herzberg EM, Marshall MM, Lamparello NA, De Leon MA, Porter A, et al. Antiretroviral drug resistance among treatment-naive HIV-1-infected persons in Washington, D.C. AIDS Patient Care STDs. 2008;22:445–448. doi: 10.1089/apc.2007.0203. [DOI] [PubMed] [Google Scholar]
- 36.Gajjala J, Farhat F, Daftary M, et al. Antiretroviral drug-resistance mutations and resistance to antiretrovirals among patients with newly diagnosed HIV—experiences in inner-city hospital clinic. AIDS 2008—XVII international AIDS conference: abstract no. TUPE0037.
- 37.Castel AD, on behalf of the DC Cohort Executive Committee. Implementation of a city-wide cohort of HIV-infected persons in care in Washington DC: The DC Cohort. July 2012. Accepted for poster presentation AIDS 2012—XIX international AIDS conference.
- 38.Greenberg AE, Hays H, Castel AD, Subramanian T, Powers-Happ L, Binkley J, et al. Development of a large urban HIV/AIDS longitudinal cohort using a web-based platform to merge electronically and manually abstracted data from disparate medical record systems: technical challenges, innovative solutions. J Am Med Inform Assoc (in press). [DOI] [PMC free article] [PubMed]
- 39.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE. 2009;2009:4. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wensing AM, Calvez V, Gunthard HF, Johnson VA, Paredes R, Pillay D, et al. 2014 Update of the drug resistance mutations in HIV-1. Top Antivir Med. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 41.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liappis AP, Laake AM, Delman M. Active injection drug-abuse offsets healthcare engagement in HIV-infected patients. AIDS Behav. 2015;19:81–84. doi: 10.1007/s10461-014-0757-4. [DOI] [PubMed] [Google Scholar]
- 43.Castel AD, Befus M, Willis S, Griffin A, West T, Hader S, et al. Use of the community viral load as a population-based biomarker of HIV burden. AIDS. 2012;26:345–353. doi: 10.1097/QAD.0b013e32834de5fe. [DOI] [PubMed] [Google Scholar]
- 44.Pillay D, Green H, Matthias R, Dunn D, Phillips A, Sabin C, et al. Estimating HIV-1 drug resistance in antiretroviral-treated individuals in the United Kingdom. J Infect Dis. 2005;192:967–973. doi: 10.1086/432763. [DOI] [PubMed] [Google Scholar]
- 45.von Wyl V, Yerly S, Burgisser P, Klimkait T, Battegay M, Bernasconi E, et al. Long-term trends of HIV type 1 drug resistance prevalence among antiretroviral treatment-experienced patients in Switzerland. Clin Infect Dis Off Publ Infect Dis Soc Am. 2009;48:979–987. doi: 10.1086/597352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DC Cohort database contains numerous individual-level variables and is therefore not publicly available in keeping with the IRB approval and our informed consent process. However, a de-identified limited dataset supporting the conclusions of this article is available by request. Interested parties may contact Dr. Amanda Castel, the DC Cohort Principal Investigator, at acastel@gwu.edu to arrange access to such.


