Abstract
Novel discoveries involving the evaluation of potential therapeutics are based on newly identified molecular targets for atypical teratoid rhabdoid tumors (ATRT), which are the most common form of infantile brain tumors. Central nervous system ATRTs are rare, aggressive, and fast growing tumors of the brain and spinal cord and carry a very poor prognosis. Currently, the standard of care for ATRT patients is based on surgical resection followed by systemic chemotherapy and radiation therapy, which result in severe side effects. Since protein tyrosine kinases have proven to be actionable targets that reduce tumor growth in a number of cancers, we examined how inhibiting tyrosine kinases affected ATRT tumor growth. Here, we examine the therapeutic efficacy of the broad spectrum tyrosine kinase inhibitor Vatalanib in the treatment of ATRT. Vatalanib significantly reduced the growth of ATRT tumor cell lines, both in two-dimensional cell culture and in three-dimensional cell culture using a spheroid model. Since Vatalanib had a remarkable effect on the growth of ATRT, we decided to use a transcriptomic approach to therapy by examining new actionable targets, such as tyrosine kinases. Next generation RNA sequencing and NanoString data analysis showed a significant increase in PTK7 RNA expression levels in ATRT tumors. Inhibition of PTK7 by short interference RNA treatment significantly decreases the viability of ATRT patient-derived tumor cell lines.
Implications
These studies provide the groundwork for future preclinical in vivo studies aiming to investigate the efficacy of PTK7 inhibition on ATRT tumor growth.
Keywords: Pediatric brain tumor, Atypical teratoid rhabdoid tumors (ATRT), Protein Tyrosine kinase 7 (PTK7), Vatalanib
Introduction
Atypical teratoid rhabdoid tumors (ATRTs) are very aggressive central nervous system tumors that comprise 10% of pediatric brain tumors. ATRTs are also associated with significantly worse overall survival than other pediatric tumors, with patients succumbing to the disease between 6 months to a year from diagnosis (1). ATRT patients are quite young with 10%–20% of these patients being 3 years or younger. Surgery and radiation alleviate imminent neurological problems, but are accompanied by exceedingly morbid long-term side effects. Aggressive multiple chemotherapy approaches in addition to radiation have been attempted, yet overall survival is dismal.
A characteristic feature of ATRTs is usually the loss of one copy of the entire chromosome 22 or a deletion or translocation specifically involving chromosome band 22q11.2 (2). This aberration of chromosome 22 results in a loss of the gene SMARCB1/INI1/hSNF5 (3–5). The Smarcb1 protein is a component of the mammalian SWI/SNF complex and functions in chromatin remodeling in an ATP-dependent manner, although its role in malignancies is unclear (6). Most ATRTs are composed of rhabdoid cells juxtaposed to areas of primitive neuroepithelial tissue, which resemble a primitive neuroectodermal tumor (PNET) and have mesenchymal and/or epithelial elements (7). ATRTs are aggressive heterogeneous tumors that remain a challenge in the field of pediatric neuro-oncology. Newer therapeutic approaches are desperately needed to increase the survival of young patients. Fundamental improvements in therapeutics will help provide better care for children who harbor this deadly brain tumor. Due to the side effects and limited effectiveness of chemotherapeutics, there is a demand for target therapies based on personalized medicine. Some of the most effective drugs for cancer include tyrosine kinase inhibitors (TKI) (8). One particular TKI that may effectively treat ATRT is Vatalanib, 4-chlorophenyl-(4-pyridin-4-ylmethyl-phthalazin-1-yl) amine succinate, an orally active pan tyrosine kinase inhibitor. Vatalanib has high receptor binding affinity for all known vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR) tyrosine kinases (9,10). Vatalanib is in clinical development for patients with advanced or metastatic pancreatic adenocarcinoma and has been used to treat a number of cancers such as prostate, colorectal, kidney, melanoma, lymphoma, leukemia, non-small cells lung carcinoma (NSCLC), and colorectal adenocarcinoma (11–14). Here, we examine the therapeutic efficacy of Vatalanib in the treatment of ATRT and a potential target for treatment known as Protein Tyrosine Kinase 7 (PTK7), which we found by transcriptome analysis to be upregulated in ATRT tumors.
Also known as colon carcinoma kinase (CCK4), PTK7 is a pseudo kinase that was first identified as a protein overexpressed in colon cancer cell lines (15). Containing an extracellular domain consisting of seven immunoglobulin-like loops, a transmembrane domain, and a tyrosine kinase domain that is catalytically inactive, PTK7 is thought to be highly involved in the Wnt signaling pathway (16). While the role that overexpression of PTK7 plays in malignancies such as colon, breast, gastric, and lung cancer has been studied, no studies to date have evaluated the role of PTK7 expression in ATRT (17–23). Prior studies in this laboratory have demonstrated that the Wnt signaling pathway has a critical role in ATRT tumor growth (24), and this present study will further evaluate how components of the Wnt pathway, such as PTK7, are involved in ATRT growth.
Materials and Methods
Patient Samples and Patient-derived Cell Lines
CHLA-05-ATRT was obtained from a 2-year old male with an ATRT tumor and CHLA-06-ATRT from a 4 month-old female with an ATRT tumor, as previously described (Dr. Anat Erdreich-Epstein, 24), and will be henceforth designated in this manuscript as ATRT-05 and ATRT-06. ATRT cell lines were cultured in a neural basal medium made up of modified neurobasal 1:1 Dulbecco’s modified Eagle’s medium : F12, HEPES (15 mM), bovine fibroblast growth factor (20 ng/mL; Cell Sciences), epidermal growth factor (20 ng/mL; Invitrogen), sodium pyruvate (110 mg/L), sodium bicarbonate (1.2 g/L), and B27 (Invitrogen). Cell lines were confirmed to be ATRT by RT-PCR and western blotting to lack INI-1 / SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily B, Member 1 (SMARC2B) on both the RNA and protein level, following establishment of cell lines and periodically, every 6 months, following multiple passages. Multiple aliquots of immortalized cell lines were frozen down immediately after establishment and tested, and new frozen aliquots used for experiments discussed in this paper. Spheroids were generated by plating one thousand ATRT cells per well in a Nexcelom Ultralow attachment U bottom 96 well cell culture dish (Nexcelom Biosciences).
Transcriptome Analysis
Transcriptome analyses, including RNA sequencing and NanoString data analysis, were conducted on RNA extracted from ATRT patients and control patient samples. For the RNA sequencing, RNA was extracted from 1 tumor from an ATRT patient and 4 normal human brain samples; RNA sequencing was performed using Illumina’s HiSeq 2000 sequencing platform. Control patient samples included six brain regions, in particular, the frontal cortex, temporal cortex, and white matter, and were obtained in good condition from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland.
Probe specific NanoString analysis was performed on RNA extracted from 17 ATRT tumor tissues, 3 immortalized ATRT cell lines, and 7 patient-derived control neuronal samples as previously described (NanoString Platform, NanoString Technologies, 24). The NanoString nCounter Analysis system was used to analyze the total RNA (250ng) from each sample. Normalization and calculation of differential expression was performed using the NanoStringDiff package in R with default settings (25). Normalization was applied across individual samples in all cases except for the heatmap, where normalization was applied across the following groups: normal, frozen ATRT, ATRT-05, and ATRT-06. Of the 17 ATRT tumor tissues, 16 were deemed of sufficient quality to be included in the analysis (Supplementary Fig. S1). The log2 ratio was calculated as the log2 of the ratio of frozen ATRT tumor tissue or immortal ATRT cell lines to the patient-derived neuronal samples.
Bioinformatics Analysis
The heatmap was prepared using the R package ComplexHeatmap (26). Outliers, as defined by a log2 value above 10 or below −10, were removed from the data set for separate analysis (Supplementary Fig. S2). K-means clustering using Euclidean distances was used to define gene clusters as well as row order within clusters. The number of clusters was user defined.
Gene clusters were analyzed using the PANTHER Overrepresentation Test (release 2016-07-15) based on the gene ontology (GO) database (release 2016-09-24). The fraction of genes identified (Figure 2A) refers to the number of genes identified in the GO reference list, which references all genes in the Homo sapiens database. The annotation data set was the GO biological process complete. Expression data analysis statistics were corrected for multiple testing with the Bonferroni method.
Figure 2.
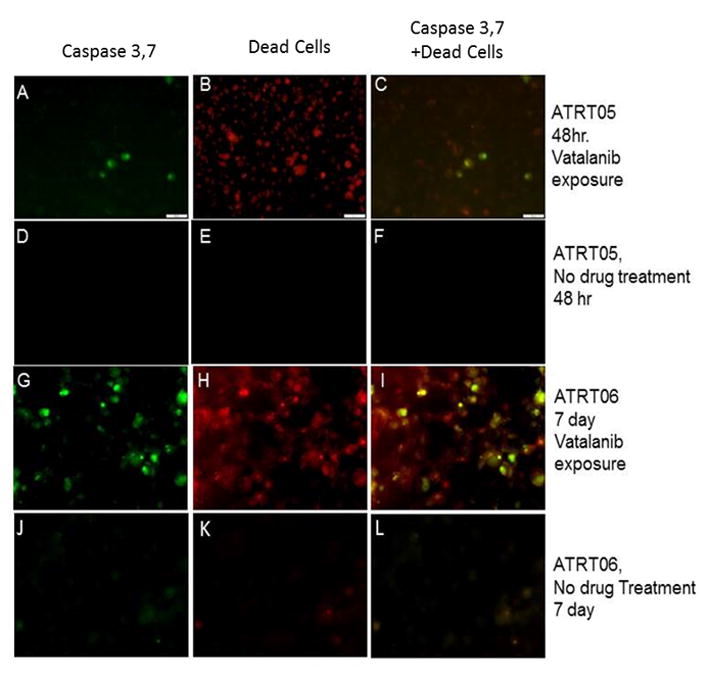
Detection of caspase 3, 7 in ATRT tumors cell lines following treatment with Vatalanib. ATRT cell lines ATRT-05 and ATRT-06 were exposed to Vatalanib (1 μM) for 48 hours (A–C) and 7 days (G–I). Caspase 3, 7 was detected in ATRT cell lines treated for 48 hours (A) or 7 days (G) using a Ready Probe to caspase 3, 7 (Molecular Probes). Dead cells were detected with NucRed® Dead 647 ReadyProbes® Reagent in ATRT cell lines treated with Vatalanib for 48 hours (B) or 7 days (H). No staining for caspase 3, 7 or dead cells were detected in cell lines not exposed to drug (D–F, J–L). 40x; Scale bar = 100 microns.
Quantitative Reverse Transcriptase PCR
Ambion® Cells-to-CT™ One Step Power SYBR green was performed on immortalized ATRT cell lines ATRT-05, ATRT-06, and control cell lines HEK-293, human neural stem cells (NSC) (MTI-Global Stem). Quantitative PCR was performed using the 7500 Real time PCR (Applied Biosystems), and the data was analyzed with 7500 System SDS software. One Step PCR was performed on lysates isolated from cells. Samples were run in triplicate, and the expression of PTK7 was normalized against the house keeping gene GAPDH. Expression levels were calculated as fold changes with ΔΔCT and a paired t test was used to evaluate significance.
Western Blotting
Cell lysates made from immortalized ATRT cell lines, HEK-293, and NSC were made using RIPA lysis buffer (Pierce) with protease inhibitors. Protein assays were performed using BCA protein assay. Equal amounts of protein were diluted in a nonreducing sample buffer (Thermoscientific) heated to 70° C and loaded on Novek Wedgewell 4–12 % TrisGlycine gels (Invitrogen). Cells were transferred to nitrocellulose using the X blot module. Ponceaus S staining was performed to verify equal amounts of protein were loaded in each lane. Blots were probed with a monoclonal antibody to PTK7 (Biorad) at 1:1000 and Goat anti-mouse IgG HRP (Thermofisher) at 1:2000 dilutions. The Supersignal West Pico Chemiluminescent substrate (Thermo Scientific) was used to detect HRP using the UVP Visionworks LS. Software on the Biospectrum 500 Imaging System (UVP).
Immunocytochemistry
For staining for PTK7 and caspase 3,7, cells were fixed with 4 % paraformaldehyde, permeabilized with 0.01% Triton-X, and stained with human anti–PTK7 Goat anti-mouse IgG Rhodamine (Santa Cruz Biotechnology) Actin 488 Ready Probes (Molecular Probes), DAPI, and Caspase 3, 7 Ready Probe (Molecular Probes). Caspase-3 immunoreactivity was also performed using a polyclonal antibody to cleaved caspase-3 (Biocare Medical).
Short Interference RNA Treatment by Reverse Transfection
HiPerFect transfection reagent, HP-validated siRNA specific for PTK7, designated here as PTK7 siRNA, and a nonspecific siRNA (AllStars Negative Control siRNA) were purchased from Qiagen. RNAi transfections were performed using siRNA constructs to human PTK7, including Hs PTK7-7, HS PTK7-14, HS PTK7-13, Hs PTK7-12, and Qiagen All Stars RNAi Controls, including an All Stars Negative control, which is highly validated for nonsilencing siRNA and is known as Flexitube siRNA unspecific All Stars, and a Cell Death control known as All Star HS cell Death (Qiagen). ATRT-06, ATRT-05, and HEK-293 cells were transfected with siRNA by HiPerfect transfection reagent. Cells were harvested during their exponential growth phase, resuspended in growth media, and divided into the following groups: nontransfected control, mock transfected with High Perfect Reagent alone, transfected with a nonspecific siRNA as a negative control, PTK7 siRNA, and a positive cell death control. Cell suspensions were transfected with 25nm siRNA in 96 well plates. Cells were cultured at 37 °C under normal growth conditions and harvested 48 hrs. post transfection for RT-PCR, western blots, cell viability assays, or fixed with 4 % paraformaldehyde for immunocytochemistry.
Viability Assays
Cell viability assays were performed using the Cell Titer Glo 2.0 (Promega). Spheroid growth was measured by measuring the diameter of the spheroids using CellSens standard 1.12 on an Olympus 1×7 microscope and spheroid volume (V) was calculated with the equation , where r =radius of the spheroid.
Apoptosis Assay
An early indicator of apoptosis is the activation of caspase-3, which was detected using a fluorogenic, no-wash indicator of activated caspase-3/7. Live cells were stained with CellEvent® Caspase-3/7 Green Ready Probes® Reagent and NucRed Dead Red 647 Ready Probe Reagent (Molecular Probes) prior to imaging. Caspase-3 immunoreactivity following PTK7 siRNA knockdown in ATRT-06 or exposure to Vatalanib was assessed using an antibody to cleave caspase-3 (Biocare Medical).
Results
1) Vatalanib is cytotoxic to ATRT growth in two- and three-dimensional cell culture
Exposure of ATRT-05 and ATRT-06 cells to Vatalanib impairs survival of ATRT tumor cells (Fig. 1). ATRT-06 cells were exposed to Vatalanib (0–150 μM) for 4 days in culture and then cell viability was measured with the Celltiter Glo (Promega). Based on 4 separate cell viability experiments, the biochemical potency for the ATRT cell lines was measured as the half maximal inhibitory concentration (IC50) and determined to be 56.03±0.001 μM. Concentrations of Vatalanib ≥ 1μM significantly reduced the viability of ATRT cell lines, according to a two-tailed heteroscedastic t test, p <0.05. Similarly, the IC50 for ATRT spheroids was 55.23 ±0.607 μM with Vatalanib exposure significantly decreasing the viability of the spheroids at concentrations ≥ 1μM Vatalanib, according to a two-tailed p test, p <0.05.
Figure 1.
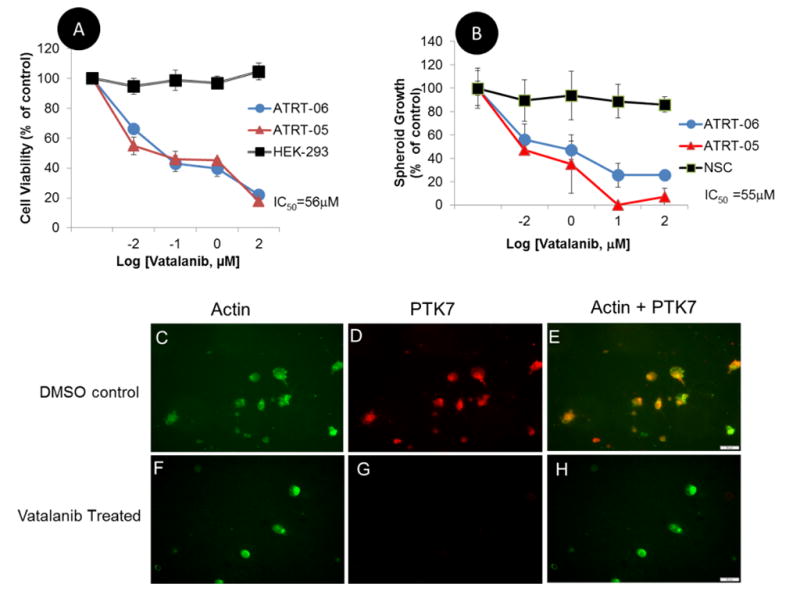
A. Pharmacologically inhibiting protein tyrosine kinases significantly reduces the viability of ATRT cell lines as compared to control HEK 293 cells. ATRT cell lines ATRT-05, ATRT-06 and control HEK 293 cells were exposed to Vatalanib for 4 days. Luminescence in each sample was normalized to a control (no drug exposure treatment) for both ATRT-05 and HEK 293. Cell viability assays were performed using Cell Titer Glo (Promega), and the Ic50 was determined to be 56 μM. Concentrations of Vatalanib ≥ 1.0 μM significantly reduced growth of ATRT cell lines according to a two-tailed heteroscedastic p test, p <0.05.
B. Similarly, Vatalanib exposure inhibited growth of spheroids in ATRT-05 and ATRT-06, but not in control spheroids generated from neural stem cells, with the IC50 for spheroids calculated at 55 μM. Concentrations of Vatalanib ≥ 1 μM significantly decreased growth of ATRT-05 and ATRT-06 spheroids, according to a two-tailed p test, p <0.05.
C–H. Immunofluorescence on ATRT-06 cells. Exposure to Vatalanib for 4 days (F–H) results in a loss of PTK7 immunoreactivity (G, H) compared to ATRT-06 exposed to a DMSO control (D, E). ATRT-06 cells were stained with anti-PTK7 (D, G) and a probe for actin (C, F) in the presence (F–H) and absence (C–E) of Vatalanib (56 μM). While actin staining was present in both Vatalanib-treated (F) and control-treated cells (C), PTK7 expression was only found in control DMSO-treated cells (D, E), and absent in Vatalanib-treated cells (G, H). Panels E and H are merged images of panels C, D and F, G, respectively. E illustrates the expression of PTK7 and actin in DMSO-treated ATRT-06, while H illustrates just actin staining in Vatalanib-treated ATRT-06 (H). Scale bar = 50 microns.
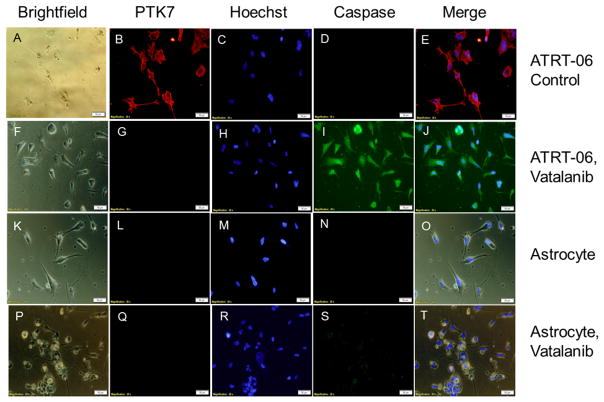
a. Exposure of ATRT cells to Vatalanib at the IC50 dose (56 μM) for four days blocked PTK7 immunoreactivity (Fig. 1G, H), which was present in DMSO-treated ATRT-06 (Fig. 1D, E), suggesting that Vatalanib inhibits PTK7 protein levels. A Stitch analysis indicates that PTK7 shares strong sequence homology to KDR and FLT4 (Supplementary Fig. 4B), two vascular endothelial growth factor receptors that have been shown to directly interact with Vatalanib (Supplementary Fig. 4A). Imaging studies indicate that exposure of ATRT cells to Vatalanib activates caspase 3/7 (Fig. 2A–C, G–I), which occurs in the early stages of apoptosis, a form of cell death characterized by cell shrinkage, blebbing of the cell membrane, and fragmentation of the cell membrane (27). Further immunocytochemical experiments were conducted to examine how Vatalanib affects caspase-3 immunoreactivity. Vatalanib exposure at the IC50 dose for ATRT cell lines (56 μM) induced expression of caspase-3 in ATRT-06 (Fig. 3I), which was absent in control ATRT-06 (Fig. 3D). Vatalanib did not induce caspase-3 immunoreactivity in normal human astrocytes (Fig. 3S) as compared to control astrocytes (Fig. 3N). Since Vatalanib had a remarkable effect on the growth of ATRT, we decided to use a transcriptomic approach to therapy by examining new actionable targets, such as tyrosine kinases.
Figure 3.
Vatalanib treatment of ATRT-06 reduced PTK7 immunoreactivity (G) as compared to control treated ATRT-06 (B). No PTK7 immunoreactivity was observed in human astrocytes (L,Q). Immunocytochemistry confirmed Vatalanib exposure at the IC50 dose for ATRT cell lines (56 μM) induced expression of caspase-3 in ATRT-06 (I), which was absent in control ATRT-06 (D). Vatalanib did not induce caspase-3 immunoreactivity in normal human astrocytes (S) as compared to control astrocytes (N).
2) Transcriptome analysis indicates increased levels of PTK7 in ATRT tumors
We performed digital multiplexed gene expression analysis (NanoString™ Technologies) using 16 ATRT tissue samples, 2 patient-derived pre-characterized primary cell lines, and 7 patient-derived control neuronal samples. Based on the results, we identified that PTK7 is significantly over-expressed in ATRT tumor tissue compared to normal brain samples, according to a heteroscedastic t test, with a p value equal to 0.0012 (Fig. 4A). The tumor cells had a lower expression of SMARCB/INI1, a tumor suppressor gene that is absent in ATRT tumors. This upregulation of PTK7 in ATRT tumor tissue is confirmed by RNA sequencing performed on an ATRT tumor and control neuronal samples (Fig 4B). Two different isoforms of PTK7 were upregulated in the ATRT tumor compared to the normal control samples.
Figure 4.
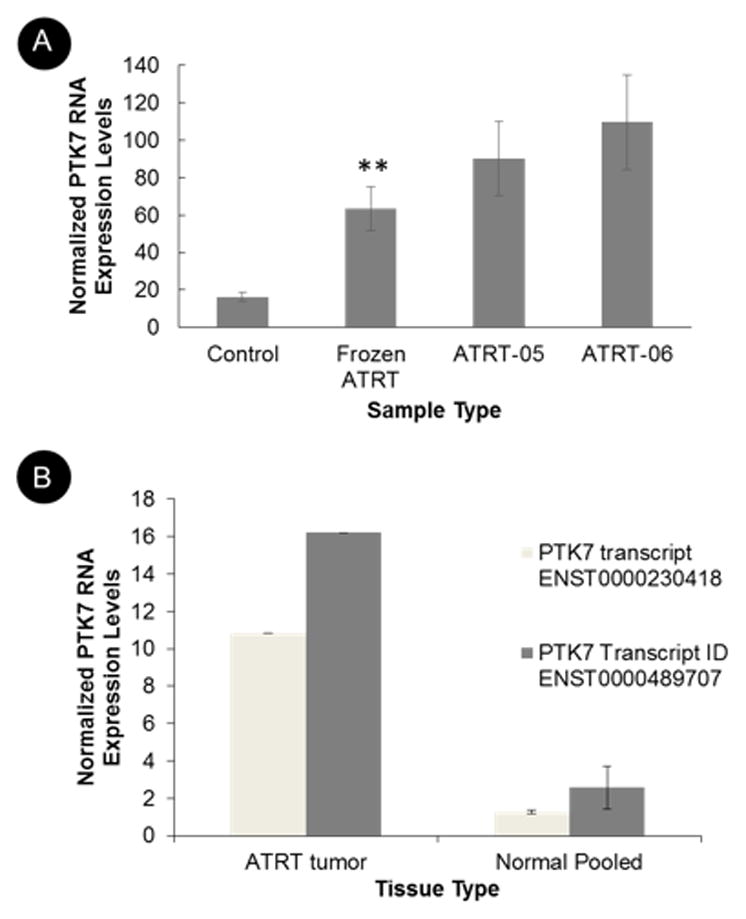
A. NanoString analysis of PTK7 expression in frozen tumors from ATRT patients (n=16), in age-matched control neuronal tissue (n=7), and in 2 ATRT cell lines, suggests increased expression of PTK7 in ATRT tumor tissue as compared to control tissue according to a two sample t test, assuming unequal variance, p value equal to 0.0012(**). B. RNA sequencing on RNA extracted from a tumor of an ATRT patient and from 4 age-matched normal control brain samples suggests increased RNA expression of PTK7 on both the gene level and in multiple transcript isoforms (B). The differential expression of PTK7 shown in this figure was calculated as logarithmic ratios of fold.
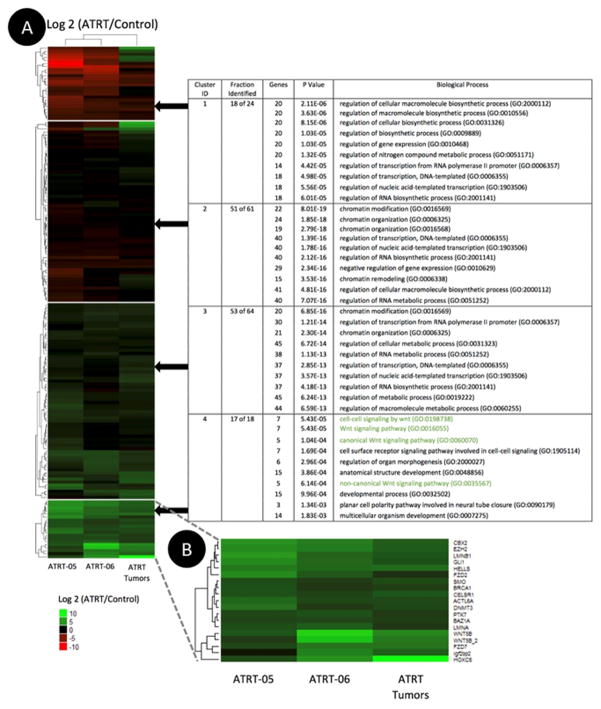
The heatmap of normalized NanoString expression data (log2 transformed) illustrates the differential expression of a number of genes in the Wnt signaling pathway, including increased expression of PTK7 (Fig. 5). Clustering of up-regulated and down-regulated genes was achieved using a k-means method. SMARCB1, which has decreased expression in ATRT tumors (28,29), appears in Cluster 1. PTK7 shows significant upregulation across all three ATRT samples and is found in the same cluster as both transcripts of WNT5B, which was previously found to be upregulated in ATRT tumor tissue (24). A gene set enrichment analysis of our four gene clusters into biological function using gene ontology (GO) shows 86 genes involved in the top ten biological processes of each cluster. Cluster 4 was identified as the cluster most related to the Wnt Pathway and contains PTK7. Low P-values (<8.0 E-19) indicate the significance of the correlation between GO terms and the genes in clusters (Fig. 5). Previous work suggests that PTK7 is involved in the non-canonical Wnt pathway (30). A differential expression analysis of the ATRT microarray data provided by Johann et al. suggests that there is no significant difference in PTK7 expression among the three newly identified epigenetic subgroups of ATRT: ATRT-TYR, ATRT-SHH, and ATRT-MYC (Supplementary Fig. S3) (31).
Figure 5.
A. Heatmap illustrating the differential expression of 167 genes and the gene ontology (GO) enrichment analysis associated with each gene cluster. Normalized NanoString expression data from 16 ATRT tumors, 2 ATRT cell lines, termed ATRT-05 and ATRT-06, and 7 neuronal samples were used to calculate the log2 ratio of frozen ATRT tumors or ATRT cell lines to normal frozen neuronal tissue. The 10 GO biological processes with the lowest p values are displayed for each gene cluster. Wnt-related biological processes–highlighted in green–appear only in the analysis for Cluster 4. Very low p values (<0.001) are associated with high significance.
This upregulation of PTK7 in ATRT tumors was further validated by a quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis (Fig. 6A), western blotting (Fig. 6B), and immunofluorescence (IHC) (Fig. 6C).
Figure 6.
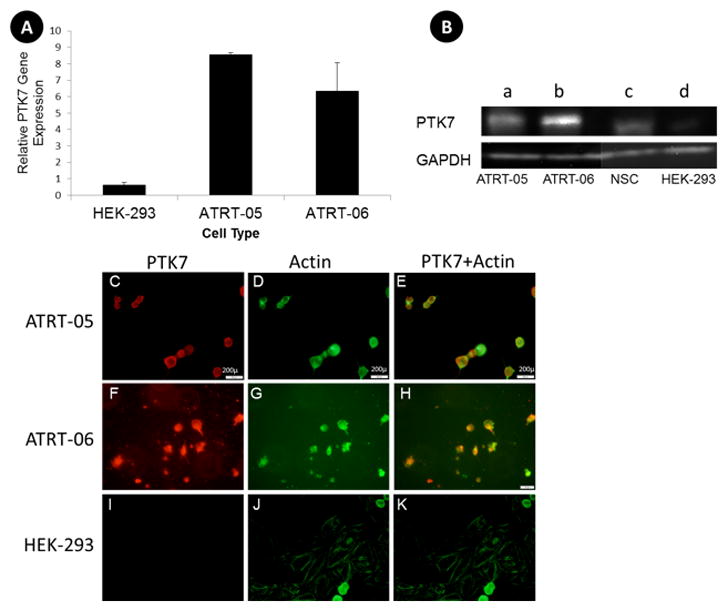
A. RT-PCR indicates increased expression of PTK7 in ATRT tumor cell lines compared to the control HEK cell line, with a p value according to two-tail t test <0.01 (**). One Step PCR was performed on lysates isolated from cells. Primers to GADPH were used to verify equal amounts of sample in each lane.
B. Western blots confirm increased expression of PTK7 in ATRT tumor cell lines (a, ATRT-05), and (b, ATRT-06) as compared to control neural stem cells (c) and HEK 293 (d), with a band corresponding to the full length PTK7 at 118 kD (top panel). Equal amounts of protein (.5μg) were loaded in each lane. HEK 293 served as a non-tumor control sample. Each lane was probed with anti-rabbit-GADPH to verify equal amounts of protein were loaded in each lane (bottom panel).
C–K. Immunocytochemistry confirms an increase in PTK7 expression in ATRT tumor cell lines that is not present in control HEK 293 lines. ATRT cell lines ATRT-05 and ATRT-06 were stained with a monoclonal antibody to PTK7 (red) and with an actin dye (green). The merged images of PTK7 and actin are depicted in E, H, and K. ATRT cell line ATRT-05 stained with PTK7 suggests expression of PTK7 in the cell membranes (C, E), whereas the same cells stained with actin reveals staining throughout the cytoplasm (D). (40x, Scale bar =200 microns). PTK7 expression was also high in ATRT-06 (F, H). No immunoreactivity for PTK7 was observed in control HEK 293 cells (I, K) exposed to anti-PTK7 or in the ATRT cell lines in the absence of primary antibody.
Increased RNA expression levels for PTK7 occurred in ATRT tumors compared to control HEK-293 (Fig. 6A), according to a two-tailed paired t test (p<0.01). Similarly, on the protein level, increased expression of PTK7 was demonstrated by western blots in ATRT-05 and ATRT-06 (Fig. 6B). Immunocytochemistry suggests increased expression of PTK7 in ATRT cells (Fig. 6C, E, F, H), which is absent in control HEK 293 cells (Fig. 6I, K) and in the absence of primary antibody. ATRT-05 stained for PTK7 suggests expression of PTK7 in the cell membrane (Fig. 6 C, E) whereas the same cells stained with actin reveals staining throughout the cytoplasm (Fig 6D). High PTK7 expression levels were observed in ATRT-06 (Fig. 6. F, H), colocalized with actin staining (Fig. 6G, H). PTK7 immunoreactivity (red) and actin staining (green) were merged in Fig. 6E, H, and K.
3) PTK7 inhibition reduces ATRT cell growth
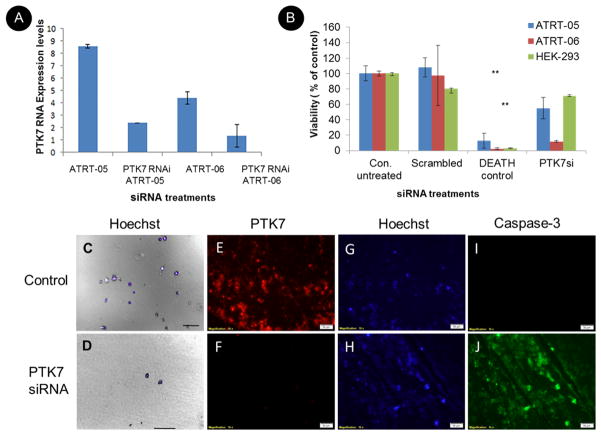
RNAi-mediated gene silencing using small interfering RNAs (siRNAs) was used to knockdown PTK7 expression. The siRNA was transiently transfected using High Perfect (Qiagen) by reverse transfection. Silencing efficacy was determined by real time PCR (Fig. 7A) and western blots. For siRNA qRT-PCR studies, a total of 1×106 cells were treated with 100 ng PTK7 siRNA (Qiagen), with 5 different PTK7 siRNAs tested. Real time PCR analysis was performed using the appropriate primers 48 hours after transfection. For controls, ATRT cell lines were transfected with an unscrambled nonspecific siRNA and as well as a positive control cell death siRNA. Statistical analysis using a paired t test indicates that PTK7 RNAi knockdown significantly reduced PTK7 RNA expression levels in ATRT cells (Fig. 7A) and significantly reduced the number of viable cells in ATRT-05 and ATRT-06, with p<0.001(**), compared to nontransfected cells (Fig. 7B). No statistical difference between the nontransfected controls and scrambled negative nonsilencing siRNA controls for ATRT 05 and ATRT-06 was observed, according to a two-tailed test. Knockdown of PTK7 via PTK7 RNAi transfection significantly reduced the viability of ATRT-05 and ATRT-06 compared to control nontransfected cells and mock transfected cells, with a two-tailed heteroscedastic t test, p<0.01. Visual confirmation of a reduced number of viable cells in ATRT-05 following knockdown of PTK7 48 hrs. following RNAi transfection (Fig. 7D) is illustrated in Fig. 7D. Few cells were observed in ATRT-05 following PTK7 knockdown (Fig. 7D) as compared to the number of healthy cells in mock transfected cells (Fig. 7C).
Figure 7. Effect of PTK7 RNAi knockdown on ATRT tumor cells in culture.
A. Real time PCR using primers to PTK7 and GAPDH indicate knockdown of PTK7 in ATRT cell lines ATRT-05 and ATRT-06. Relative gene expression of PTK7/fold change expression levels was calculated using the ΔΔCq method, using Ct values obtained with the PTK7 primers and the housekeeping gene GAPDH and plotted in the different siRNA treatments (B). Multiple cell viability assays (n=3) indicate that transfection of ATRT-05 and ATRT-06 with PTK7 RNAi significantly reduces the growth of ATRT cell lines as compared to untransfected controls (abbreviated Con. untreated), according to a two-tailed t test p<0.01 (**), while transfection of PTK7 RNAi (abbreviated PTK7siRNA) does not affect the viability of control HEK-293 cells. There is no significant difference between viability of nontransfected controls and the scrambled negative nonspecific control (abbreviated scrambled). C+D. Images of the viability of ATRT cell line ATRT-05 48 hours following RNAi transfection with PTK7 RNAi. (D) indicates the presence of very few cells following PTK7 siRNA as compared to mock transfected cells (C). Cells were stained with DAPI and visualized with DIC (10x). Scale bar= 100 microns. Transfection with PTK7 siRNA in ATRT cell lines resulted in a loss of PTK7 immunoreactivity (F) as compared to controls (E). Hoechst staining was performed to visualize nuclei in ATRT cell lines following knockdown of PTK7 (H) as compared to controls (G). Knockdown of PTK7 induced caspase-3 immunoreactivity (J) as compared to controls ().
Studies were conducted to determine how PTK7 RNAi knockdown affected caspase-3 immunoreactivity, an early indicator of apoptosis. Knockdown of PTK7 with reverse transfection inhibited PTK7 immunoreactivity in ATRT-06 cells (figure 7F) as compared to control transfection cells (Figure 7E) and induced caspase-3 expression (Fig 7j) as compared to nontransfected controls (Fig 7I). Future studies will examine more precisely the mechanisms by which PTK7 induces cell death and apoptosis.
Discussion
Treatment with the broad spectrum VEGF inhibitor Vatalanib significantly reduced the viability of ATRT cell lines, both in two- and three-dimensional aggregates of ATRT tumor cells, and inhibited expression of PTK7. Spheroids have been used for decades to simulate the in vivo tumor microenvironment. Like tumors, spheroids contain a hypoxic center with a well-oxygenated outer layer of cells, and contain both surface-exposed and deeply buried cells, as well as both proliferating and nonproliferating cells (32). In breast cancer, prostate, colon, and other cancers, spheroids have been used to model disease and identify anti-cancer therapeutics (33–37).
Imaging data suggests that the mechanism of cell death following treatment with Vatalanib may involve the caspase 3/7 activation, which is seen in early stages of apoptosis, a death pathway characterized by cell shrinkage, blebbing of the plasma membrane, and condensation and fragmentation of DNA (27). It remains for future studies to determine whether or not the induction of apoptosis by Vatalanib can be attributed to the inhibition of PTK7. KDR and FLT-1 are known to interact with Vatalanib (9) and share some structural similarity with PTK7, including in the extracellular domain. Consequently, it is possible that Vatalanib directly inhibits PTK7. Alternately, the therapeutic effect of Vatalanib may be due to inhibition of KDR. There is a biphasic relationship between PTK7 expression and KDR activity such that there is an optimal PTK7 concentration that leads to higher KDR activity (38). The therapeutic effect of Vatalanib might be due to the inhibition of KDR, which in ATRT cells may have higher activity due to increased levels of PTK7. Further studies are necessary to elucidate this mechanism, especially since defective apoptosis often occurs in cancer cells.
The effect of Vatalanib on ATRT tumor growth was examined since there have been clinical trials utilizing Vatalanib for a number of cancers, and its route of administration and its effect on multidrug resistance transporters (MDR) have also been investigated. For example, Vatalanib, in combination with a chemotherapeutic regimen, has been used in clinical trials for metastatic colorectal cancer. In addition, Vatalanib can be orally administered as compared to some drugs such as bevacizumab, which have to be administered intravenously (39). Studies have demonstrated that Vatalanib inhibits multidrug resistance transporter activity, in particular ATP-binding cassette (ABC) efflux transporter activity (40). Thus, it is hypothesized that ATRT cell lines will not develop resistance to Vatalanib. Based on clinical studies using Vatalanib in metastatic colon cancer, there is hope for true treatment with Vatalanib. Consequently, it is likely that we will pursue preclinical studies examining the effect of Vatalanib in vivo, both alone and in combination with other targeted therapeutics.
Since Vatalanib treatment significantly reduced the growth of ATRT cell lines, and its mechanism of action involves inhibition of multiple tyrosine kinases (12) we decided to take a transcriptomic approach and examine protein tyrosine kinase expression in ATRT patient tumors and cell lines. Our transcriptomic and cellular analysis indicates that PTK7 is highly expressed in ATRT tumors. Knockdown of PTK7 resulted in significantly decreased viability of ATRT cell lines, suggesting that PTK7 may be involved in tumor cell growth and proliferation. In multiple cancer types, such as colon cancer, gastric cancer, breast cancer, and acute myeloid leukemia, high PTK7 levels are correlated to poor drug response, increased metastasis, and poor patient survival rates (15,19,41–43). The role of PTK7 in different cancers is not yet known, but it is thought that PTK7 may promote tumorigenesis as expression of PTK7 in leukemia cells enhances cell migration and survival (19). In other cancers, such as glioblastomas, PTK7 is highly expressed, and knockdown of PTK7 reduces cell proliferation, interferes with tumorigenic potential, and increases apoptosis. The high expression level of PTK7 in glioblastomas predicts an unfavorable prognosis for patients (44). Furthermore, in normal human astrocytes, PTK7 enhanced anchorage-independent growth (45). It has previously been demonstrated that knockdown of PTK7 in colon cancer cells induces caspase-10-dependent apoptosis via the mitochondria pathway (46). These results suggest that PTK7 is involved in cancer cell proliferation and anchorage-independent cell growth. Despite the lack of direct kinase activity, PTK7 promotes signal transduction by interacting with other proteins. PTK7 has been shown to influence Vascular Endothelial Growth Factor (VEGF)-induced migration, invasion, and tube-formation of human umbilical vein endothelial cells (HUVECs) and angiogenesis in vivo (47). Therefore, it is thought that PTK7 at the plasma membrane plays an important role in VEGF-mediated signaling. Studies suggest that PTK7 regulates the activity of Kinase Insert Domain Receptor (KDR) and that knockdown of PTK7 inhibits VEGF-induced phosphorylation of KDR(38).
Additionally, in colorectal cancer (CRC) tissue, PTK7 was significantly up-regulated, with significant overexpression associated with reduced survival in patients. In mouse models of CRC, down-regulation of PTK7 caused reduced tumor growth, and overexpression of PTK7 in PTK7 negative cancer cells resulted in increased metastatic events (48). Studies indicate that intrahepatic cholangiocarcinoma (ICC) cells with high PTK7 overexpression had increased proliferation, DNA synthesis, migration, and invasion. Knockdown of PTK7 with small interfering RNA (siRNA) resulted in impaired DNA synthesis, migration, and invasion, and decreased expression of phosphorylated RhoA. Furthermore, knockdown of PTK7 resulted in reduced tumor size as compared to the scrambled siRNA injection in an in vivo model for ICC. ICC patients with low expression of PTK7 had better prognoses and survived longer with the disease as compared to patients who expressed high levels of PTK7, suggesting that PTK7 could potentially be used as a prognostic indicator in ICC (49).
Collectively, these studies suggest that PTK7 plays an important role in cancer cell proliferation, anchorage-independent growth, tumor growth, increased metastatic events, invasiveness of cancer cells, and poor prognosis in cancer patients. However, further studies need to be conducted to determine the precise role of PTK7 in cancer cell proliferation and growth. Thus, PTK7 may serve as an important target for future therapeutics and as a prognostic biomarker for ATRT, as well as a number of other cancers. Clearly, our recent results as well as these studies in other tumor models indicate that PTK7 is an important target for therapeutic intervention that demands further investigation in terms of its mechanisms of action and potential targets.
The RNA sequencing and NanoString analysis presented in this paper correlate well with recent findings based on whole genome DNA and RNA sequencing based on 192 ATRTs. These studies identify ATRT molecular subtypes involving regulatory networks of TYR (encoding the enzyme tyrosine), Sonic Hedgehog (SHH), and MYC (overexpression of the Myc oncogene) (31). All three of these regulatory networks are upregulated in the ATRTs described in the current study.
Conclusions
The broad spectrum tyrosine kinase inhibitor Vatalanib inhibited PTK7 immunoreactivity and significantly reduced the growth of ATRT tumor cell lines, both in two-dimensional and three-dimensional cell cultures. Since Vatalanib inhibits multiple tyrosine kinases, a transcriptomic approach examined the expression pattern of protein tyrosine kinases in ATRT patient tissue. Transcriptome profiling, including RNA sequencing and NanoString analysis, of ATRT patient tumors indicates increased expression of PTK7 RNA, which is confirmed by real time PCR in ATRT patient-derived tumor cell lines. Increased expression of PTK7 protein was observed in ATRT patient-derived tumor cell lines by immunocytochemistry and western blotting. These studies provide the groundwork and basis for future preclinical studies of the effect of Vatalanib and PTK7 inhibition on ATRT tumor growth in vivo.
Supplementary Material
Acknowledgments
Funding
Research reported in this publication was supported by the Sanford Research Imaging Core, which is supported by the National Institutes of Health COBRE grants P20 GM103620 awarded to David A. Pearce and P20 GM103548 awarded to Keith W. Miskimins. This work was partially supported by the National Science Foundation/Experimental Program to Stimulate Competitive Research (EPSCoR)
Grant IIA-1355423 awarded to Jim A. Rice, an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health P20GM103443 awarded to Barbara E. Goodman. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation.
The authors wish to thank Drs. Madhavi Chakravadhanula and Anat Erdreich-Epstein at the Barrow Neurological Institute at Phoenix Children’s Hospital for providing patient derived ATRT cell lines. The authors wish to acknowledge Kelly R. Graber for training on the microscopes.
List of abbreviations
- ATRT
Atypical teratoid rhabdoid tumors
- PTK7
Protein Tyrosine Kinase 7
- VEGFR
vascular endothelial growth factor receptor
- PDGFR
platelet-derived growth factor receptor
- GO
gene ontology
- siRNA
small interfering RNAs
- IC50
half maximal inhibitory concentration
- CRC
colorectal cancer
- ICC
intrahepatic cholangiocarcinoma
Footnotes
Declarations
The authors declare no potential conflicts of interest.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due being under further investigation and analyses by the authors of this study. However, the data sets will be available from Dr. Ratan Bhardwaj on reasonable request following completion of analysis.
References
- 1.Ginn KF, Gajjar A. Atypical teratoid rhabdoid tumor: current therapy and future directions. Front Oncol [Internet] 2012;2:114. doi: 10.3389/fonc.2012.00114. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22988546%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3439631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coccé MC, Lubieniecki F, Kordes U, Alderete D, Gallego MS. A complex karyotype in an atypical teratoid/rhabdoid tumor: Case report and review of the literature. J Neurooncol. 2011:375–80. doi: 10.1007/s11060-010-0478-0. [DOI] [PubMed] [Google Scholar]
- 3.Roberts CWM, Biegel JA. The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer Biol Ther. 2009:412–6. doi: 10.4161/cbt.8.5.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegel JA, Burk CD, Parmiter AH, Emanuel BS. Genes, Chromosom Cancer [Internet] Vol. 5. Wiley Subscription Services, Inc., A Wiley Company; 1992. Molecular analysis of a partial deletion of 22q in a central nervous system rhabdoid tumor; pp. 104–8. cited 2017 Mar 4. Available from: http://doi.wiley.com/10.1002/gcc.2870050203. [DOI] [PubMed] [Google Scholar]
- 5.Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 6.Lee MC, Park SK, Lim JS, Jung S, Kim JH, Woo YJ, et al. Atypical teratoid/rhabdoid tumor of the central nervous system: Clinico-pathological study. Neuropathology. 2002;22:252–60. doi: 10.1046/j.1440-1789.2002.00458.x. [DOI] [PubMed] [Google Scholar]
- 7.Biegel J. Molecular genetics of atypical teratoid/rhabdoid tumors. Neurosurg Focus [Internet] 2006;20:1–7. doi: 10.3171/foc.2006.20.1.12. Available from: http://thejns.org/doi/abs/10.3171/foc.2006.20.1.12. [DOI] [PubMed] [Google Scholar]
- 8.Hojjat-Farsangi M. Small-molecule inhibitors of the receptor tyrosine kinases: promising tools for targeted cancer therapies. Int J Mol Sci. 2014:13768–801. doi: 10.3390/ijms150813768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bold G, Altmann KH, Frei J, Lang M, Manley PW, Traxler P, et al. New anilinophthalazines as potent and orally well absorbed inhibitors of the VEGF receptor tyrosine kinases useful as antagonists of tumor-driven angiogenesis. J Med Chem. 2000;43:2310–23. doi: 10.1021/jm9909443. [DOI] [PubMed] [Google Scholar]
- 10.Morabito A, De Maio E, Di Maio M, Normanno N, Perrone F. Tyrosine kinase inhibitors of vascular endothelial growth factor receptors in clinical trials: current status and future directions. Oncologist [Internet] 2006;11:753–64. doi: 10.1634/theoncologist.11-7-753. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16880234. [DOI] [PubMed] [Google Scholar]
- 11.Dragovich T, Laheru D, Dayyani F, Bolejack V, Smith L, Seng J, et al. Phase II trial of vatalanib in patients with advanced or metastatic pancreatic adenocarcinoma after first-line gemcitabine therapy (PCRT O4-001) Cancer Chemother Pharmacol [Internet] 2014;74:379–87. doi: 10.1007/s00280-014-2499-4. Available from: http://dx.doi.org/10.1007/s00280-014-2499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood JM, Bold G, Buchdunger E, Cozens R, Ferrari S, Frei J, et al. PTK787/ZK 222584, a novel and potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, impairs vascular endothelial growth factor-induced responses and tumor growth after oral administration. Cancer Res [Internet] 2000;60:2178–89. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10786682. [PubMed] [Google Scholar]
- 13.Scott EN, Meinhardt G, Jacques C, Laurent D, Thomas AL. Vatalanib: the clinical development of a tyrosine kinase inhibitor of angiogenesis in solid tumours. Expert Opin Investig Drugs [Internet] 2007;16:367–79. doi: 10.1517/13543784.16.3.367. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17302531. [DOI] [PubMed] [Google Scholar]
- 14.Jones SF, Spigel DR, Yardley DA, Thompson DF, Burris HA. A phase i trial of vatalanib (PTK/ZK) in combination with bevacizumab in patients with refractory and/or advanced malignancies. Clin Adv Hematol Oncol. 2011;9:845–52. [PubMed] [Google Scholar]
- 15.Mossie K, Jallal B, Alves F, Sures I, Plowman GD, Ullrich A. Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene [Internet] 1995;11:2179–84. Available from: papers3://publication/uuid/b84ebf91-880d-47a7-86c7-c19756de1a27. [PubMed] [Google Scholar]
- 16.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 17.Speers C, Tsimelzon A, Sexton K, Herrick AM, Gutierrez C, Culhane A, et al. Identification of novel kinase targets for the treatment of estrogen receptor-negative breast cancer. Clin Cancer Res. 2009;15:6327–40. doi: 10.1158/1078-0432.CCR-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, et al. A phosphatase associated with metastasis of colorectal cancer. Science [Internet] 2001;294:1343–6. doi: 10.1126/science.1065817. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1065817%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/11675071. [DOI] [PubMed] [Google Scholar]
- 19.Prebet T, Lhoumeau AC, Arnoulet C, Aulas A, Marchetto S, Audebert S, et al. The cell polarity PTK7 receptor acts as a modulator of the chemotherapeutic response in acute myeloid leukemia and impairs clinical outcome. Blood. 2010;116:2315–23. doi: 10.1182/blood-2010-01-262352. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Zhang L-H, Wang X-H, Xing X-F, Cheng X-J, Dong B, et al. PTK7 as a novel marker for favorable gastric cancer patient survival. J Surg Oncol [Internet] 2012;106:880–6. doi: 10.1002/jso.23154. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22585737. [DOI] [PubMed] [Google Scholar]
- 21.Gorringe KL, Boussioutas A, Bowtell DDL. Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosom Cancer. 2005;42:247–59. doi: 10.1002/gcc.20136. [DOI] [PubMed] [Google Scholar]
- 22.Gobble RM, Qin LX, Brill ER, Angeles CV, Ugras S, O’Connor RB, et al. Expression profiling of liposarcoma yields a multigene predictor of patient outcome and identifies genes that contribute to liposarcomagenesis. Cancer Res. 2011;71:2697–705. doi: 10.1158/0008-5472.CAN-10-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endoh H, Tomida S, Yatabe Y, Konishi H, Osada H, Tajima K, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22:811–9. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 24.Chakravadhanula M, Hampton CN, Chodavadia P, Ozols V, Zhou L, Catchpoole D, et al. Wnt pathway in atypical teratoid rhabdoid tumors. Neuro Oncol. 2014;17:526–35. doi: 10.1093/neuonc/nou229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Horbinski C, Wu H, Liu Y, Sheng S, Liu J, et al. NanoStringDiff: a novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res [Internet] 2016:gkw677. doi: 10.1093/nar/gkw677. Available from: http://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gkw677%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/27471031. [DOI] [PMC free article] [PubMed]
- 26.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 27.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell. 2016;29:379–93. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Judkins AR, Burger PC, Hamilton RL, Kleinschmidt-DeMasters B, Perry A, Pomeroy SL, et al. INI1 protein expression distinguishes atypical teratoid/rhabdoid tumor from choroid plexus carcinoma. J Neuropathol Exp Neurol [Internet] 2005;64:391–7. doi: 10.1093/jnen/64.5.391. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15892296. [DOI] [PubMed] [Google Scholar]
- 29.Judkins AR, Mauger J, Ht A, Rorke LB, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 in pediatric CNS neoplasms. Am J Surg Pathol [Internet] 2004;28:644–50. doi: 10.1097/00000478-200405000-00013. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15105654. [DOI] [PubMed] [Google Scholar]
- 30.Hayes M, Naito M, Daulat A, Angers S, Ciruna BG. Ptk7 promotes non-canonical Wnt/PCP-mediated morphogenesis and inhibits Wnt/β-catenin-dependent cell fate decisions during vertebrate development. Development [Internet] 2013;140:1807–18. doi: 10.1242/dev.090183. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23533179. [DOI] [PubMed] [Google Scholar]
- 31.Kerr JFR, Wyllie AH, Currie AR. Apoptosis: A Basic Biological Phenomenon with Wide-ranging Implications in Tissue Kinetics. Br J Cancer [Internet] 1972;26:239–57. doi: 10.1038/bjc.1972.33. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2008650/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frieboes HB, Zheng X, Sun CH, Tromberg B, Gatenby R, Cristini V. An integrated computational/experimental model of tumor invasion. Cancer Res. 2006;66:1597–604. doi: 10.1158/0008-5472.CAN-05-3166. [DOI] [PubMed] [Google Scholar]
- 33.Calcagno AM, Salcido CD, Gillet J-P, Wu C-P, Fostel JM, Mumau MD, et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst [Internet] 2010;102:1637–52. doi: 10.1093/jnci/djq361. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2970576&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 35.Thoma CR, Stroebel S, Rösch N, Calpe B, Krek W, Kelm JM. A high-throughput-compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. J Biomol Screen [Internet] 2013;18:1330–7. doi: 10.1177/1087057113499071. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24080258. [DOI] [PubMed] [Google Scholar]
- 36.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The Use of 3-D Cultures for High-Throughput Screening: The Multicellular Spheroid Model. J Biomol Screen [Internet] SAGE Publications. 2004;9:273–85. doi: 10.1177/1087057104265040. Available from: http://dx.doi.org/10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 37.Wenzel C, Riefke B, Gründemann S, Krebs A, Christian S, Prinz F, et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323:131–43. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 38.Shin WS, Na HW, Lee ST. Biphasic effect of PTK7 on KDR activity in endothelial cells and angiogenesis. Biochim Biophys Acta - Mol Cell Res. 2015;1853:2251–60. doi: 10.1016/j.bbamcr.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Los M, Voest EE, Roodhart JM. Target practice: lessons from phase III trials with bevacizumab and vatalanib in the treatment of advanced colorectal cancer. Oncologist [Internet] 2007;12:443–50. doi: 10.1634/theoncologist.12-4-443. Available from: www.ncbi.nlm.nih.gov/pubmed/17470687. [DOI] [PubMed] [Google Scholar]
- 40.To KKW, Poon DC, Wei Y, Wang F, Lin G, Fu LW. Vatalanib sensitizes ABCB1 and ABCG2-overexpressing multidrug resistant colon cancer cells to chemotherapy under hypoxia. Biochem Pharmacol. 2015;97:27–37. doi: 10.1016/j.bcp.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 41.Kapoor S. J Surg Oncol [Internet] Vol. 107. Wiley Subscription Services, Inc., A Wiley Company; 2013. Emerging new prognostic markers of gastric tumors besides PTK7; pp. 450–450. cited 2017 Mar 4. Available from: http://doi.wiley.com/10.1002/jso.23209. [DOI] [PubMed] [Google Scholar]
- 42.Jiang G, Zhang M, Yue B, Yang M, Carter C, Al-Quran SZ, et al. PTK7: A new biomarker for immunophenotypic characterization of maturing T cells and T cell acute lymphoblastic leukemia. Leuk Res. 2012;36:1347–53. doi: 10.1016/j.leukres.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gärtner S, Gunesch A, Knyazeva T, Wolf P, Högel B, Eiermann W, et al. PTK 7 is a transforming gene and prognostic marker for breast cancer and nodal metastasis involvement. PLoS One. 2014;9:e84472. doi: 10.1371/journal.pone.0084472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Zhang C, Yuan J, Fu J, Wu M, Su J, et al. PTK7 regulates Id1 expression in CD44-high glioma cells. Neuro Oncol. 2015;17:505–15. doi: 10.1093/neuonc/nou227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Na H-W, Shin W-S, Ludwig A, Lee S-T. The cytosolic domain of PTK7, generated from sequential cleavage by ADAM17 and γ-secretase, enhances cell proliferation and migration in colon cancer cells. J Biol Chem [Internet] 2012;7:M112.348904. doi: 10.1074/jbc.M112.348904. Available from: http://www.jbc.org/cgi/content/abstract/M112.348904v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng L, Sefah K, O’Donoghue MB, Zhu G, Shangguan D, Noorali A, et al. Silencing of PTK7 in colon cancer cells: Caspase-10-dependent apoptosis via mitochondrial pathway. PLoS One. 2010:5. doi: 10.1371/journal.pone.0014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin WS, Maeng YS, Jung JW, Min JK, Kwon YG, Lee ST. Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis. Biochem Biophys Res Commun. 2008;371:793–8. doi: 10.1016/j.bbrc.2008.04.168. [DOI] [PubMed] [Google Scholar]
- 48.Lhoumeau AC, Martinez S, Boher JM, Monges G, Castellano R, Goubard A, et al. Overexpression of the promigratory and prometastatic PTK7 receptor is associated with an adverse clinical outcome in colorectal cancer. PLoS One. 2015:10. doi: 10.1371/journal.pone.0123768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin J, Ryu HS, Lee KB, Jang JJ. High expression of protein tyrosine kinase 7 significantly associates with invasiveness and poor prognosis in intrahepatic cholangiocarcinoma. PLoS One. 2014:9. doi: 10.1371/journal.pone.0090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.