Cilia cement link between squid and bacteria
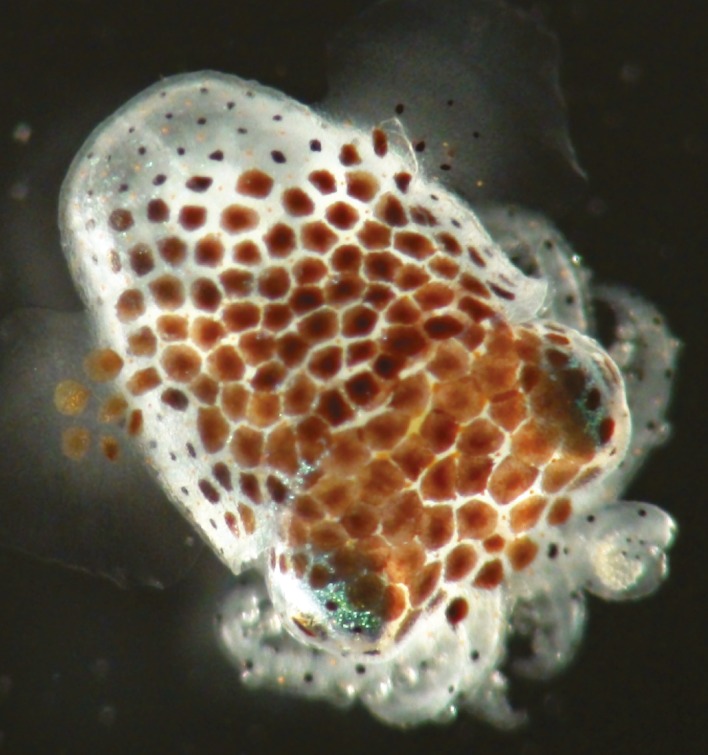
Juvenile Hawaiian bobtail squid.
Cilia found on internal mucus membranes in some animals’ bodies must not only clear toxins and pathogens but also recruit beneficial microbes through their mechanical action. Using particle tracking and video and scanning electron microscopy, Janna Nawroth et al. (pp. 9510–9516) examined how cilia on the epithelium lining the appendages of the light-emitting organ of juvenile Hawaiian bobtail squids recruit the bioluminescent bacterium Vibrio fischeri from inhaled seawater, while keeping out undesirable particulate matter. The authors report that the asymmetric beating of a group of 25-micron-long cilia along the outer surface of the appendages creates a region of counter-rotating vortices. Another group of 10-micron-long cilia thrums along in a temporally and spatially symmetric beat pattern in a sheltered zone near the light organ’s surface pores. The action of the former group of long cilia prevents the adherence of most suspended particles but funnels bacteria and similarly sized particles toward the sheltered zone. Concomitantly, the gentle mixing action of the latter group of short cilia enables diffusion of biochemical signaling molecules between the squid and the bacteria as well as bacterial colonization of the squid’s light organ. According to the authors, the findings expand the panoply of roles attributed to cilia and uncover mechanisms underlying the recruitment of symbionts. — P.N.
Novel microbes in human microbiome
Blood transports a range of biomolecules throughout the human body, including cell-free DNA, some of which can be traced back to viruses and bacteria. Using massive shotgun sequencing, Mark Kowarsky et al. (pp. 9623–9628) analyzed cell-free DNA in 1,351 human blood samples and assembled 7,190 contiguous sequences, or contigs, which were at least 1-kilobase-pair in size. When compared with known databases, more than 3,700 of the contigs, the vast majority of which contain coding sequences, exhibited little or no shared evolutionary history with other organisms. Furthermore, phylogenetic analyses suggested that many of the microorganisms represent new taxa. The authors validated the analysis with independent samples, which confirmed the existence of the novel organisms and ruled out contamination and assembly artifacts. The findings, which span a cohort of 188 patients, demonstrate that the human microbiome comprises a community of microbes that is far more diverse than previously thought, according to the authors. — T.J.
Modeling growth of prebiotic polymers into protein-like molecules
Early in Earth’s history, short-chain prebiotic polymers spontaneously grew into long chains of protein-like or nucleic acid-like molecules, but the underlying process remains unclear. To uncover a physical basis for this process, Elizaveta Guseva et al. (pp. E7460–E7468) used a computational model to study how random sequences of hydrophobic (H) and polar (P) monomers fold and bind together. The authors found that the random sequence HP chains can collapse and fold into specific compact conformations that expose hydrophobic surfaces, serving as primitive catalysts for elongating other such HP polymers. By both folding by themselves and also catalyzing the elongation of other polymers, the folding polymers, or “foldamer” catalysts, enable the growth of short chains into long chains of particular sequences. The foldamer catalysts are part of an autocatalytic set, in which any two foldamer catalyst sequences are autocatalytic to each other. The authors’ model also provides HP ensembles that can further evolve under selection. The foldamer model could help explain how random chemical processes could have resulted in long protein-like molecules, and could help devise testable hypotheses for studying the mechanisms of action of prebiotic polymers, according to the authors. — S.R.
Conservation of social behavior genes

European honeybee (Apis mellifera). Image courtesy of Wikimedia Commons/John Severns.
Sociobiology theory proposes that similarities between human and nonhuman social structures reflect common evolutionary origins, but this hypothesis remains unproven. To test the hypothesis, Hagai Shpigler et al. (pp. 9653–9658) compared behavior-associated brain gene expression between humans and honeybees. The authors exposed groups of honeybee nestmates to two different social stimuli: an unrelated bee, which elicits aggression, and a queen larva, which elicits caring behavior. The authors observed wide variation in individual bees’ responsiveness to these stimuli, including a subset of bees that consistently failed to respond to either stimulus. Next, the authors generated gene expression profiles for a high-level sensory integration center in the bees’ brains and identified genes whose expression differed significantly between unresponsive bees and other bees. Comparison of differentially expressed genes with human gene sets associated with autism spectrum disorder (ASD) revealed significant overlap. This overlap between human ASD-associated genes and genes associated with social responsiveness in honeybees suggests that genes responsible for social behavior are highly conserved across the animal kingdom. The results also illustrate how comparative genomics can be used to test the predictions of sociobiology theory, according to the authors. — B.D.
Targeted drug combinations enhance treatment efficacy in blood cancers
Precision medicine aims to tailor medical interventions to each individual’s disease. However, the molecular heterogeneity of human cancers presents challenges to finding effective drug therapies that target cancer-specific mutations. Stephen Kurtz et al. (pp. E7554–E7563) describe a strategy for sensitivity profiling to test the effectiveness of various drug combinations. Focusing on two blood cancers, acute myeloid leukemia and chronic lymphocytic leukemia, the authors created 48 drug pairs comprised of inhibitors that were either FDA-approved or in clinical development and that belonged to different drug classes targeting nonoverlapping biological pathways. Next, the authors screened the panel against 122 primary patient samples encompassing various hematologic malignancies, using a metric to quantify the relative efficacy of inhibitor combinations. The tests revealed that combinations that included a B cell lymphoma 2 inhibitor provided additional benefit to patients with myeloid malignancies, whereas lymphoid malignancies experienced gains from certain combinations with PI3K, CSF1R, or bromodomain inhibitors. The findings also demonstrate that functional ex vivo testing can be applied in clinical settings to help inform treatment decisions, according to the authors. — T.J.
Urban infrastructure, hydrology, and biodiversity
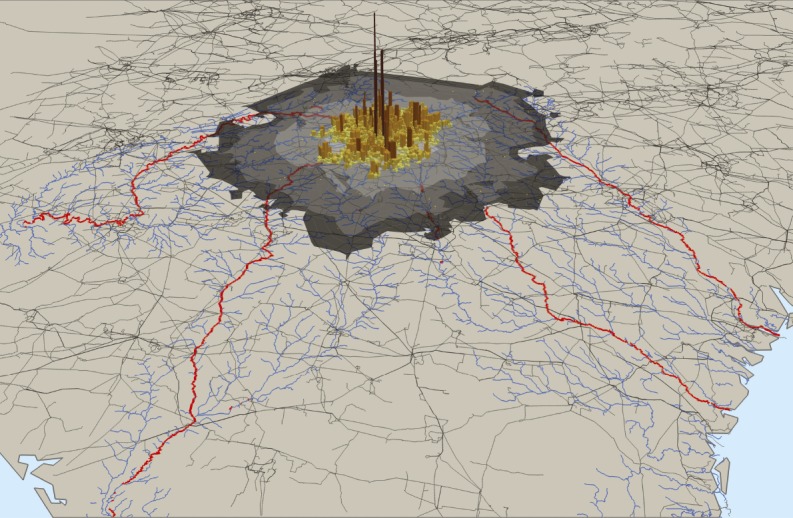
Atlanta’s electricity demand within block groups (vertical bars), energy shed (gray), and river segments altered by electricity infrastructure within Atlanta’s energy shed (red).
Cities are hubs of not only sociopolitical power and economic production, but also land transformation and resource consumption. Hard urban infrastructure systems, such as land transformation, water supplies, and electricity production, can extend well beyond a city’s boundaries and affect distant aquatic ecosystems. Ryan McManamay et al. (pp. 9581–9586) developed a data-driven model to determine the impact of hard urban infrastructure systems on the hydrology and biodiversity of rivers in the United States. The authors found that urban land transformation and electricity production have affected more than 1,200 fish, mussel, or crayfish species nationwide and contributed to 260 local extinctions. The impacts of individual cities were not associated with population, per capita energy demand, or energy efficiency, but instead reflected cities’ individual infrastructure decisions. For example, Atlanta, whose hard infrastructure network extends across multiple major river basins, affects nearly 12,500 km of rivers and streams and more than 500 indigenous species, whereas Las Vegas, a comparably sized city, affects fewer than 1,000 km of rivers and streams and 19 indigenous species. According to the authors, cities could substantially improve environmental quality in rivers and streams well beyond their boundaries through infrastructure policies. — B.D.


