Significance
Temperature dramatically affects lifespan in both vertebrates and invertebrates. Recent studies have challenged the notion that cold promotes longevity solely through thermodynamic effects. We support this idea by describing a genetic manipulation influencing the longevity response to temperature in Drosophila. Low temperatures activate 4E-BP [eukaryotic translation initiation factor 4E (eIF4E)-binding protein], triggering changes in metabolism, protein translation, and lifespan. In conjunction with recent studies, the findings support the idea that nutrition and temperature affect lifespan via partially overlapping mechanisms and highlight the 4E-BP/eIF4E pathway as a pharmacological target to treat aging-related pathologies and maximize longevity.
Keywords: aging, cold, dietary restriction, Drosophila, temperature
Abstract
Changes in body temperature can profoundly affect survival. The dramatic longevity-enhancing effect of cold has long been known in organisms ranging from invertebrates to mammals, yet the underlying mechanisms have only recently begun to be uncovered. In the nematode Caenorhabditis elegans, this process is regulated by a thermosensitive membrane TRP channel and the DAF-16/FOXO transcription factor, but in more complex organisms the underpinnings of cold-induced longevity remain largely mysterious. We report that, in Drosophila melanogaster, variation in ambient temperature triggers metabolic changes in protein translation, mitochondrial protein synthesis, and posttranslational regulation of the translation repressor, 4E-BP (eukaryotic translation initiation factor 4E-binding protein). We show that 4E-BP determines Drosophila lifespan in the context of temperature changes, revealing a genetic mechanism for cold-induced longevity in this model organism. Our results suggest that the 4E-BP pathway, chiefly thought of as a nutrient sensor, may represent a master metabolic switch responding to diverse environmental factors.
Studies on the biological underpinnings of aging have uncovered numerous genetic and environmental factors regulating animal lifespan. Longevity-promoting genes include components of the insulin-like signaling pathway, the histone deacetylase Sir2, the GTPase Ras, TRP membrane channels, and transcription factors, among many others (1, 2). Environmental manipulations extending life include changes in nutrition (often referred to as caloric or dietary restriction), sexual/reproductive history, and ambient temperature (3–5).
Of the factors known to impact aging, temperature is arguably the most promising. Lifespan extension by cold is evolutionarily conserved—documented in poikilotherms, such as worms and flies, and homeotherms, including mammals (4, 6–11)—and more robust than well-studied interventions such as dietary restriction (12, 13). However, the underlying mechanisms remain incompletely understood (10, 14, 15). In Caenorhabditis elegans, the effect of cold on survival involves the thermosensitive membrane channel TRPA-1 acting upstream of the DAF-16/FOXO transcription factor (14, 15). This seminal finding countered the notion that longevity is the passive result of general thermodynamic changes, and favored instead the view that genetic pathways actively control lifespan in response to ambient temperature. However, these results have to date not been extended or replicated in other model organisms. Explanatory theories for the effect of cold on longevity include a reduction in reactive oxygen species generated by mitochondrial uncoupling proteins, suppression of autoimmune response, and changes in neuroendocrine factors (6, 16, 17), but these views remain largely speculative given the lack of further mechanistic insight into lifespan extension by cold in model organisms.
We report that, in Drosophila, changes in temperature trigger a metabolic program impacting protein translation, mitochondrial protein synthesis, and the posttranslational regulation of the translation repressor, 4E-BP. Manipulation of 4E-BP levels affects temperature-induced mortality, shedding light on the genetic underpinnings of this process in Drosophila. Our findings support a view of the 4E-BP pathway as a master regulator of translation under stressful conditions (18), with dramatic consequences for metabolism and organismal longevity.
Results
Metabolic Effects of Temperature.
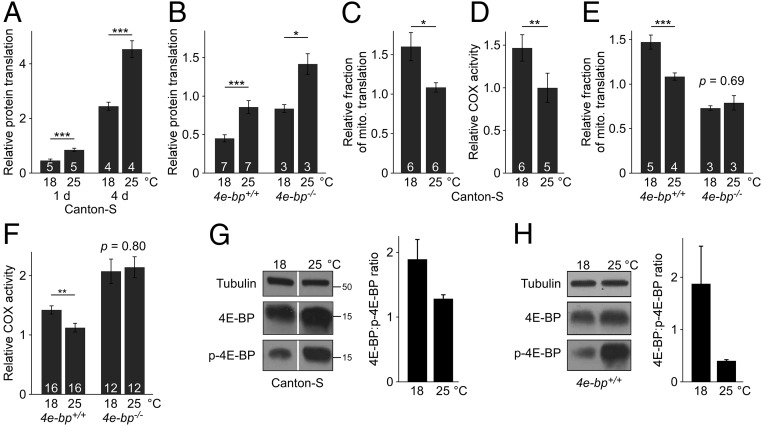
Temperature can affect protein synthesis (19–22). We tested if changes to ambient temperature modulate protein translation in Drosophila. Flies were given food supplemented with 35S-labeled methionine and the radioactive tracer was quantified thereafter in the isolated protein fraction. The ratio of labeled to unlabeled methionine in the food was kept constant, so the amount of radiolabel incorporated into nascent protein depends only on the rate of protein synthesis and not on feeding rate (23). Lowering temperature from 25 to 18 °C reduced protein translation by 50% (Fig. 1A). The radioactive tracer was not limiting since only ∼15% of the ingested label was incorporated into nascent protein. Thus, cold inhibits Drosophila protein translation in vivo.
Fig. 1.
Effect of temperature on protein translation and mitochondrial function. (A and B) Cold inhibits protein translation. Flies kept at the indicated temperature were fed medium supplemented with [35S]methionine and de novo protein synthesis was quantified by measuring radioisotope levels in the isolated protein fraction relative to total protein content. Canton-S flies were fed labeled food for 1 or 4 d (A), while the 4e-bp deletion mutant (−/−) and revertant control (+/+) were maintained on labeled food for 1 d (B). (C and D) Cold increases the proportion of mitochondrial protein translation (C) and COX activity (D) in Canton-S flies. [35S]methionine incorporation into mitochondria (mito.) was normalized to de novo protein synthesis. (E and F) 4e-bp deletion abolishes the effect of cold on mitochondrial protein translation (E) and COX activity (F). (G and H) Cold results in a higher proportion of nonphosphorylated 4E-BP (Right, average of two independent replicates) in Canton-S (G) and 4e-bp+/+ (H) flies. Each of the three blots (Left) are from a single exposure, and protein ladder molecular masses (in kilodaltons) are shown in G. 4E-BP, 4E-BP nonphosphorylated at Thr46; p-4E-BP, 4E-BP phosphorylated at Thr37 and/or Thr46 (see Methods for details). (A–F) Averages ± SEM are shown in arbitrary units; n = number of vials, superimposed on each bar. *P < 0.05; **P < 0.01; ***P < 0.001. Males were used in all studies.
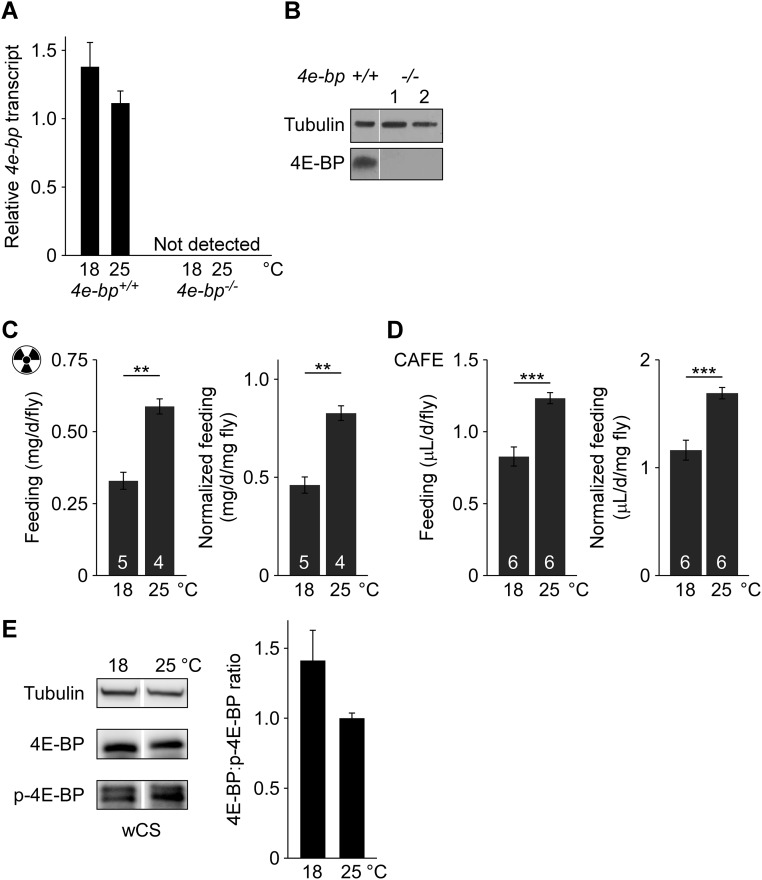
The eukaryotic translation initiation factor 4E (eIF4E) plays a crucial role in the assembly of the translation initiation complex. The eIF4E-binding protein (4E-BP) regulates translation rate by recruiting eIF4E, thereby blocking translation initiation (24). We asked if 4E-BP regulates translation in response to cold. Deletion of 4e-bp (25) stimulated translation rate at both 25 and 18 °C but did not abolish the effect of cold on protein synthesis (Fig. 1B). The deletion mutant shows no detectable 4e-bp transcript or protein and is thus a de facto null (Fig. S1 A and B).
Fig. S1.
Effect of temperature on 4e-bp expression and food consumption. (A) Relative 4e-bp transcript levels in 4e-bp+/+ (revertant control) and 4e-bp−/− (deletion null) males, normalized to total input RNA. The 4e-bp–null mutant has undetectable transcript in all conditions. n = 6 vials of 25–30 flies for each temperature. (B) The 4E-BP protein is undetectable by Western blot in the null mutant. Flies were maintained at 25 °C and two independently processed 4e-bp−/− protein samples are shown. (C and D) Cold reduces feeding. Canton-S male food intake over 24 h was determined by radioisotope-labeling of the medium (C) or CAFE assay (D). Consumption normalized to fly body mass is also shown (Right). n = number of vials or CAFE chambers, superimposed on each bar. (E) Cold results in a higher proportion of nonphosphorylated 4E-BP in a Cantonized white (wCS) line. 4E-BP, 4E-BP nonphosphorylated at Thr46; p-4E-BP, 4E-BP phosphorylated at Thr37 and/or Thr46 (see Methods for details). The two bands for phosphorylated 4E-BP resolved in these blots likely represent different phosphorylation states of the molecule. Males were used in all studies. Averages ± SEM are shown. **P < 0.01; ***P < 0.001.
The synthesis of proteins targeted to mitochondria can be regulated independently from the overall translation of bulk cytoplasmic protein (23). To assess the translation rate of mitochondria-targeted proteins, mitochondria were isolated from [35S]methionine-fed animals and 35S-labeled protein was quantified. Flies exposed to cold showed a 50% increase in the proportion of newly synthesized protein present in mitochondria (Fig. 1C). Cold also enhanced the activity of the mitochondrial protein cytochrome c oxidase (COX) (Fig. 1D), consistent with a previous study (26). Hence, cold inhibits translation in general, but the synthesis of proteins destined to the mitochondria is selectively preserved, resulting in a cellular enrichment of these molecules. Although cold inhibited feeding (Fig. S1 C and D), the metabolic effects of cold cannot be explained by a global, indiscriminate inhibition of protein synthesis due to lower amino acid availability, since the relative synthesis of mitochondria-tagged proteins was increased. Rather, our results suggest that lowering ambient temperature shifts cellular resources toward mitochondrial metabolism.
The binding protein 4E-BP influences mitochondrial activity and biogenesis in mammalian systems (27, 28). Therefore, we asked if it mediates the observed effect of cold on mitochondrial metabolism. Strikingly, the effect of temperature on mitochondrial protein translation and activity was abolished in the absence of 4E-BP (Fig. 1 E and F). These results demonstrate that 4E-BP regulates mitochondrial protein synthesis and function in response to temperature. To elucidate this process further, we sought to determine if 4E-BP is regulated by temperature. The activity of 4E-BP is highly dependent on its phosphorylation state; the active hypophosphorylated form binds to eIF4E and inhibits translation (29). 4E-BP can be phosphorylated at multiple sites but phosphorylation of Thr37/Thr46 by the mammalian target of rapamycin (mTOR) acts as a priming event required for further phosphorylation (30). Cold exposure increased the proportion of nonphosphorylated protein, largely at the expense of the primed fraction (Fig. 1 G and H and Fig. S1E). Changes in absolute levels of nonphosphorylated 4E-BP were not statistically significant or reproducible across genotypes. Taken collectively, our observations suggest that reduced ambient temperature induces a physiological state comprising posttranslational modification of 4E-BP—resulting in a lower proportion of the phosphorylated isoform primed for inactivation—and a switch from global protein translation toward mitochondrial metabolism and efficiency.
4E-BP Modulates the Effect of Temperature on Longevity.
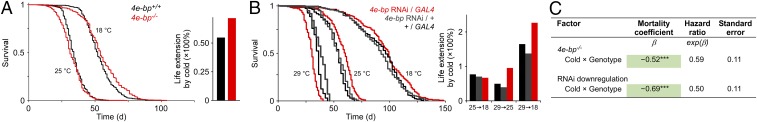
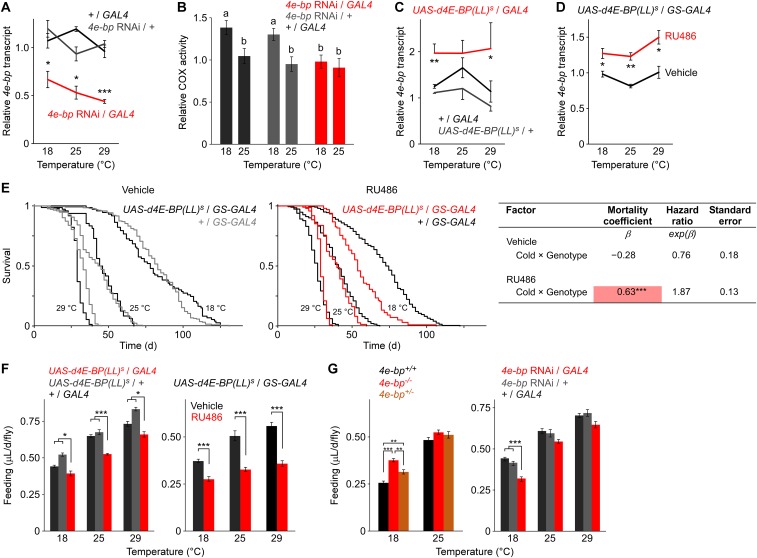
Since 4E-BP mediates some of the metabolic effects of temperature, we asked if it also modulates the effect on lifespan. As expected, temperature reduction strongly prolonged life (Fig. 2A). Interestingly, this effect was enhanced in the absence of 4E-BP. Cox proportional hazards analysis confirmed a significant interaction between genotype and temperature in determining survival, demonstrating that cold elicited a greater reduction in mortality in the deletion mutant than in the control (Fig. 2C). To further validate our findings, we down-regulated 4e-bp by RNAi. Ubiquitous 4e-bp knockdown using the daughterless-GAL4 promoter reduced 4e-bp transcript levels (Fig. S2A) and also enhanced the effect of cold on survival (Fig. 2 B and C), although a more dramatic temperature challenge (from 18 to 29 °C) was required to reveal the phenotype, possibly due to RNAi affecting 4E-BP levels more subtly than the null. Knockown of 4e-bp also blocked the effect of temperature on mitochondrial activity (Fig. S2B).
Fig. 2.
4E-BP loss of function enhances lifespan extension by cold. (A) Survival of 4e-bp null (4e-bp−/−) and control (4e-bp+/+) flies. Mean life extension by cold is enhanced in the null (Right). n = 554–601 flies. (B) Survival of a 4e-bp hypomorphic allele using RNAi and the ubiquitously expressed daughterless-GAL4/UAS-Dcr2 driver. Controls are hemizygous for each transgene. n = 156–363 flies. (C) Cox proportional hazards analysis of survival data. The negative coefficient for the interaction term indicates that the loss of function manipulations gain a greater benefit from cold than their respective controls. Genotype refers to the experimental cohort, 4e-bp−/− or 4e-bp RNAi/GAL4. Males were used in all studies. ***P < 0.001. Wald χ2 = 682.9, P < 10−6 (4e-bp−/−); χ2 = 263.5, P < 10−6 (RNAi).
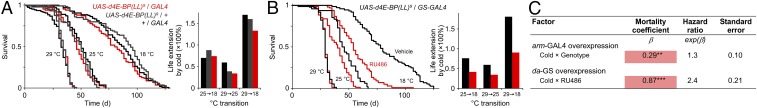
Fig. S2.
Characterization of 4e-bp transgenic manipulations. (A and B) Relative 4e-bp transcript levels measured by qRT-PCR (A) and COX activity (B) in 4e-bp RNAi lines. RNAi lines harbored a UAS-controlled RNAi construct targeting 4e-bp and the ubiquitously expressed daughterless-GAL4 driver. Controls are hemizygous for each element. In B, labeled bars without a common letter differ, P < 0.05. *P < 0.05; ***P < 0.001. (C and D) Relative 4e-bp transcript levels measured by qRT-PCR in overexpression lines. Overexpression lines harbored a UAS-controlled activated form of 4e-bp [d4E-BP(LL)] driven by either the ubiquitously expressed constitutive armadillo-GAL4 driver (C) or the inducible daughterless-GS GAL4 driver (D). Controls for the constitutive driver are hemizygous for each element. For the GS driver, addition of RU486 to the diet results in overexpression of the UAS-controlled d4E-BP(LL) transgene. GS controls were given vehicle only. Results were normalized to RpL32 transcript levels. n = 2–6 vials of 20–30 flies for each genotype and temperature (A, C, and D). Comparisons between experimental and controls are shown for each temperature (Student’s t test: *P < 0.05; **P < 0.01). (E) Survival at three temperatures of the GS driver harboring the d4E-BP(LL) transgene or a control, fed vehicle (Left) or RU486 (Center). Cox proportional hazards analysis (Right) shows no significant interaction between temperature and genotype when animals were fed vehicle. Expression of activated 4E-BP with RU486, however, results in a significant interaction. The positive mortality coefficient suggests that inducing gene expression results in attenuated lifespan extension under cold. n = 48–198 flies per condition. ***P < 0.001. Wald χ2 = 252.5, P < 10−6 (vehicle); χ2 = 199, P < 10−6 (RU486). (F) Effect of 4e-bp overexpression on food intake. Feeding over 24 h was measured using radioisotope-labeling of the medium for d4E-BP(LL) overexpression driven by a constitutive GAL4 (Left) or inducible GS (Right), and their respective controls, as described in C and D. For the constitutive driver, n = 6 vials of ∼15 flies for each condition. For the GS driver, n = 10 vials of ∼10 flies for each condition. *P < 0.05; ***P < 0.001. (G) Effect of 4e-bp loss of function on food intake. Feeding over 24 h was measured using radioisotope-labeling of the medium for the 4e-bp–null mutant (4e-bp−/−) and its control (4e-bp+/+) (Left) or for 4e-bp RNAi and its controls (Right), as described in A. Feeding rate in a heterozygous mutant at 18 °C is intermediate of that of the null and control lines, demonstrating that moderate 4e-bp down-regulation does not explain the differences in total consumption between the null and RNAi manipulation. n = 10–15 vials of 20–25 flies for each condition. Only statistically significant differences between the experimental group (red) and controls are shown (**P < 0.01; ***P < 0.001, one-way ANOVA with Tukey–Kramer post hoc test for multiple comparisons or Student’s t test). All data are shown as averages ± SEM. Males were used in all studies.
We next tested the effect of increased 4E-BP activity on survival. Overexpression of activated 4E-BP [d4E-BP(LL) (31)] increased 4e-bp transcript levels (Fig. S2C) and inhibited the effect of cold on mortality (Fig. 3 A and C). To rule out possible confounds from differences in genetic background, we used the drug-inducible GeneSwitch (GS) system, where experimental and control animals have the same genotype and gene expression is switched on by ingestion of the drug, RU486 (32, 33). Expression of d4E-BP(LL) using the ubiquitous inducible daughterless-GS driver increased 4e-bp transcript levels (Fig. S2D) and also countered lifespan extension by cold (Fig. 3 B and C). The effect of the drug required the presence of both transgenes (34) (Fig. S2E). Drug-inducible d4E-BP(LL) expression had a stronger phenotypic effect than the constitutive driver (Fig. 3C), possibly due to better control of genetic background or differential expression in specific tissues critical for the lifespan response. Regardless, our findings show that the translation repressor 4E-BP modulates survival in the context of a temperature change. Overexpression of 4E-BP inhibited feeding at all temperatures (Fig. S2F), while 4E-BP down-regulation had a significant effect only at 18 °C, and in opposite directions in the two genetic manipulations (Fig. S2G), suggesting that the effect of 4E-BP on lifespan is not explained by changes in feeding behavior.
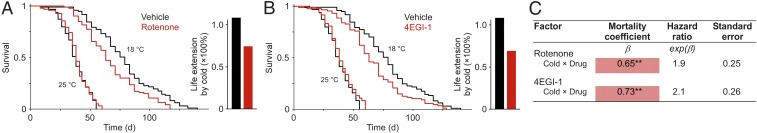
Fig. 3.
Overexpression of activated 4E-BP diminishes lifespan extension by cold. (A and B) Survival at three temperatures of animals bearing the ubiquitous armadillo-GAL4 (A) or the ubiquitous, drug-inducible daughterless-GS GAL4 (B) driver expressing an active form of 4E-BP [d4E-BP(LL)]. GS is activated upon ingestion of the drug, RU486. Controls are hemizygous for each transgene (A) or fed vehicle instead of RU486 (B). Mean life extension by cold is inhibited with 4e-bp overexpression (Right). n = 100–208 flies (A); 48–98 flies (B). (C) Cox proportional hazards analysis of survival data. The positive coefficient for the interaction term indicates that 4E-BP activity attenuates the effect of cold. Genotype refers to the experimental cohorts. Males were used in all studies. **P < 0.01; ***P < 0.001. Wald χ2 = 135.1, P < 10−6 (A); χ2 = 92.4, P < 10−6 (B).
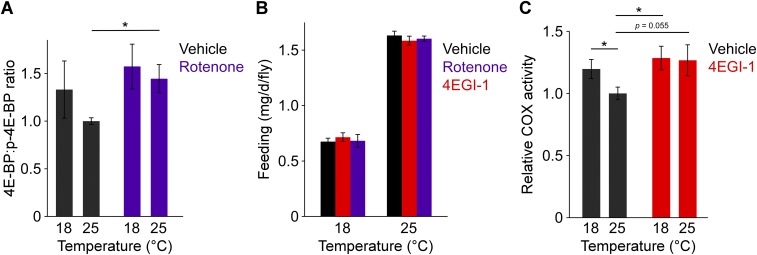
We next sought to confirm our findings pharmacologically. The protein kinase mTOR is involved in cell growth and differentiation and acts as one of the main regulators of 4E-BP function (35). mTOR mediates the phosphorylation of 4E-BP, shifting it to its inactive form, which dissociates from eIF4E, releasing the break on translation initiation (18). The naturally occurring lipophilic compound, rotenone, inhibits mTOR activity, rendering 4E-BP hypophosphorylated, i.e., active (36), an effect we also observed (Fig. S3A). One would therefore expect rotenone treatment to have an effect resembling 4E-BP overexpression (Fig. 3). Indeed, animals exposed to a nontoxic concentration of rotenone at 25 °C (37) showed blunted lifespan extension with cold (Fig. 4A). Hazard ratio analysis confirmed that rotenone treatment interacted with temperature to determine survival (Fig. 4C). Importantly, rotenone supplementation did not affect total food consumption, suggesting that the drug does not affect food palatability (Fig. S3B).
Fig. S3.
Characterization of pharmacological manipulations. (A) Rotenone treatment mimics the effect of cold by increasing the proportion of nonphosphorylated 4E-BP at 25 °C. n = 4–10 vials of flies per condition. (B) Dietary supplementation of rotenone or 4EGI-1 does not affect food intake. Feeding over 24 h was measured using radioisotope-labeling of the medium. n = 10 vials of flies per condition. (C) 4EGI-1 treatment mimics the effect of cold by increasing COX activity. n = 17–21 group of flies per condition. Canton-S males were used in all studies. *P < 0.05.
Fig. 4.
Effect of pharmacological manipulation of 4E-BP/eIF4E pathway on lifespan extension by cold. (A and B) Survival at 25 and 18 °C of Canton-S males fed rotenone (A) or 4EGI-1 (B). Controls were fed vehicle. Mean life extension by cold is inhibited with either drug (Right). n = 93–97 flies (A); 83–97 flies (B). (C) Cox proportional hazards analysis of survival data. The positive coefficient for the interaction term indicates that both drugs attenuate the effect of cold on survival. **P < 0.01. Wald χ2 = 112.4, P < 10−6 (A); χ2 = 96.7, P < 10−6 (B).
Since rotenone is also thought to modulate the activity of the electron transport chain downstream of the 4E-BP pathway—albeit at higher concentrations (38)—we sought to validate our observations with an independent compound. The small molecule 4EGI-1 promotes 4E-BP activity, stabilizing the binding of 4E-BP to eIF4E and inducing the dissociation of the translation initiation complex (39, 40). The effect of 4EGI-1 treatment on survival was identical to that of rotenone; 4EGI-1 had no effect at 25 °C but attenuated lifespan extension by cold (Fig. 4 B and C). The drug 4EGI-1 also mimicked the effect of cold on COX activity (Fig. S3C). Dietary 4EGI-1 supplementation did not affect food intake (Fig. S3B). Our cumulative data strongly support the idea that the 4E-BP/eIF4E pathway modulates longevity in response to ambient temperature.
Cold and Protein Restriction Similarly Impact Mortality Rate.
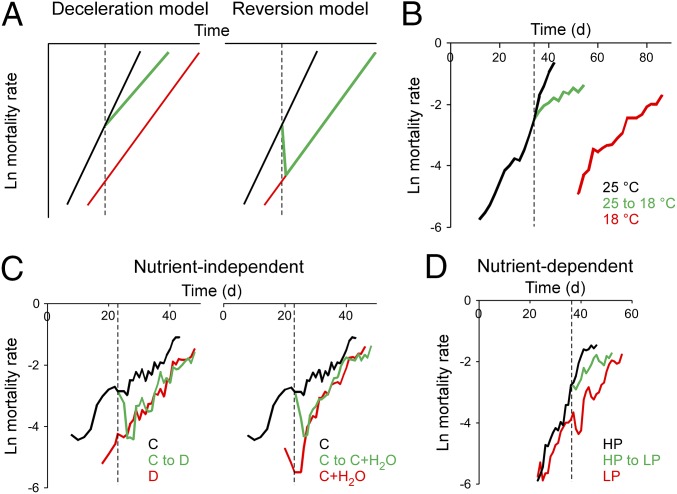
Our results show that temperature regulates 4E-BP activity and mitochondrial efficiency, with 4E-BP in turn modulating survival. Reduced protein intake has been associated with similar effects (23, 41), a surprising parallel since temperature and diet are believed to affect lifespan through different mechanisms (12). The controversy centers around the effect of these manipulations on mortality rate, defined as the probability of death at a given time. Mortality rate increases steadily with age; interventions that counter this trend can do it in two basic ways: by slowing the rate of increase or by reversing mortality rate altogether (Fig. 5A). Temperature has long been known to be a powerful decelerator of mortality rate (12, 13, 42) (Fig. 5B). Surprisingly, diet was proposed to act as a mortality reverser in flies, suggesting that the two manipulations have contrasting demographic mechanisms (12). However, these findings have been called into question, as they may be confounded by factors other than nutrient intake, such as dehydration stress, an effect compounded in whole-food dilution paradigms (43). We tested the effect of hydration on mortality rate with two independent protocols. Using a whole-food dilution paradigm, we indeed observed a mortality reversal (Fig. 5C) reminiscent of the reported effect (12). However, merely providing the animals with ad libitum water access was sufficient to recapitulate the mortality reversal (Fig. 5C and Fig. S4A), suggesting that the observed mortality trend is explained by differential hydration between the two populations rather than nutrient intake. Moreover, when the experiment was repeated on a dietary paradigm varying protein intake without affecting hydration, which more closely resembles mammalian models of dietary manipulation (43–46), diet acted as a clear mortality decelerator, not a reverser (Fig. 5D and Fig. S4 B and C and Table S1). Our observations contradict the belief that temperature and nutrition have different demographic effects and instead reveal that both environmental factors have a similar impact on mortality: they slow the rate of aging without reversing it.
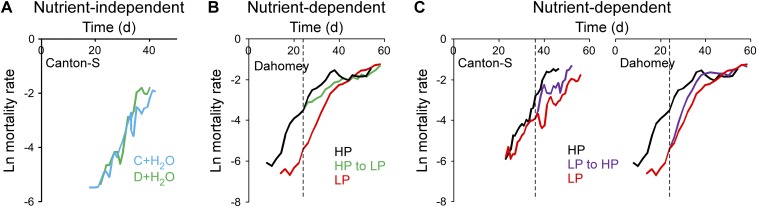
Fig. 5.
Effect of temperature and diet on mortality rate. (A) Treatments causing lifespan extension can impact mortality via two mechanisms (12), an attenuation of its rate of increase (the “deceleration” model, Left) or an acute reduction (the “reversion” model, Right). Black line, untreated; red line, chronically treated; green line, acutely treated; dashed line, start of acute treatment. (B) The effect of temperature on mortality rate is consistent with the deceleration model. A midlife switch in temperature results in a deceleration of mortality rate increase. Following the switch, all three experimental groups have significantly different mortality trajectories (Table S1). n = 783 flies, 25 °C; 192, 18 °C; 954 for the switch. (C) Whole-food dilution maintains a constant carbohydrate:protein ratio and can impact lifespan in a nutrient-independent manner (43). Under these conditions, an acute switch from concentrated (C) to diluted (D) food induces an immediate reversion of mortality to that of animals on lifelong diluted medium (Left), as reported previously (12). Water supplementation to the concentrated diet has a similar effect (Right). The mortality trajectory of switched animals is indistinguishable from that of flies on the lifelong treatment (Table S1). n = 658 flies, C; 482, D; 598, C to D switch; 500, C + H2O; 621, C to C + H2O switch. (D) In a paradigm varying carbohydrate:protein ratio, which affects lifespan in a nutrient-dependent manner (43, 44), an acute switch from HP to LP diet results in an intermediate trajectory consistent with the deceleration model and similar in nature to—albeit milder than—the effect of temperature (B). All postswitch trajectories are significantly different (Table S1). Lifelong LP results in lower age-specific mortality rate and a midlife switch induces a subsequent deceleration of mortality increase without a complete and permanent reversion. n = 247 flies, HP; 247, LP; 498 for the switch. All animals are Canton-S males. Dashed lines indicate day of switch to specified treatment (typically when mortality reached ∼25%). See Methods for food compositions.
Fig. S4.
Effect of diet on mortality rate. (A) In a dietary-restriction paradigm using whole-food dilution and therefore a constant carbohydrate:protein ratio (C:P), dilution has no effect on mortality rate in the presence of ad libitum water (P = 0.11, hazard ratio = 0.79 for C + H2O vs. DR + H2O). Canton-S males were fed control (C) or diluted (D) diet and provided with water access throughout life. n = 603 flies, C + H2O; 212, D + H2O. (B and C) In a dietary paradigm varying C:P—known to affect lifespan in a nutrient-dependent manner—an acute dietary switch from either HP to LP or vice versa results in an intermediate trajectory consistent with the deceleration model and similar to the effect of temperature. (B) Dahomey males, n = 496 flies, HP; 478, LP; 486, HP to LP switch. (C) Canton-S males, n = 247 flies for each chronic treatment; 498 for the switch. Dahomey males, n = 496 flies, HP; 478, LP; 491, switch. Dashed lines indicate day of switch to specified diet (typically when 25–35% mortality had been recorded in the HP diet group). See Methods for food compositions and also Table S1.
Table S1.
Hazard ratios for mortality rate trials
| Paradigm | Experiment ID (figure no.) | Populations compared | Hazard ratio* | P value |
| Constant C:P | Fig. S4A | C + H2O | 0.79 | 0.11 |
| D + H2O | ||||
| C | 0.35 | <0.000001 | ||
| D | ||||
| Fig. 5C | C | 0.33 | <0.000001 | |
| C → D | ||||
| D | 1.03 | 0.75 | ||
| C → D | ||||
| C | 0.32 | <0.000001 | ||
| C + H2O | ||||
| Fig. 5C | C | 0.36 | <0.000001 | |
| C → C + H2O | ||||
| C + H2O | 1.02 | 0.84 | ||
| C → C + H2O | ||||
| Δ C:P | HP | 0.29 | <0.000001 | |
| LP | ||||
| HP | 0.43 | <0.000001 | ||
| HP → LP | ||||
| Fig. 5D and Fig. S4C | LP | 2.77 | <0.000001 | |
| HP → LP | ||||
| HP | 0.52 | <0.000001 | ||
| LP → HP | ||||
| LP | 2.25 | <0.000001 | ||
| LP → HP | ||||
| HP | 0.37 | <0.000001 | ||
| LP | ||||
| HP | 0.57 | <0.000001 | ||
| HP → LP | ||||
| Fig. S4 B and C | LP | 1.33 | 0.00026 | |
| HP → LP | ||||
| HP | 0.61 | <0.000001 | ||
| LP → HP | ||||
| LP | 1.89 | <0.000001 | ||
| LP → HP | ||||
| Δ T | Fig. 5B | 25 °C | 0.26 | <0.000001 |
| 25 → 18 °C | ||||
| 18 °C | 42.8 | <0.000001 | ||
| 25 → 18 °C |
Cox proportional hazards regression analysis. For data from Fig. S4A, lifelong mortalities were compared. For all other experiments, only mortality data following the treatment switch date were compared. When the hazard ratio is close to 1, the manipulation has little effect on survival. Lower or higher ratios indicate greater or poorer survival, respectively, of the second population in each comparison. P < 0.001 are in bold.
Discussion
In C. elegans, the nutrient-sensing DAF-16/FOXO pathway has been implicated in lifespan regulation by temperature (15, 47). We report a genetic mechanism for cold-mediated longevity in Drosophila. The translational repressor 4E-BP, chiefly thought of as a nutrient sensor, is posttranslationally modified in response to cold and regulates mitochondrial metabolism and longevity. Nutrient-sensing pathways may thus play an evolutionarily conserved role in modulating lifespan in response to temperature, with 4E-BP acting as a key metabolic regulator in response to environmental triggers. Such a process would allow organisms to control metabolic investment to match favorable environmental conditions and therefore maximize evolutionary fitness. The temperatures used in our study reasonably reflect the range of natural conditions; even the largest shift used (29–18 °C = 11 °C) is not unlikely to occur during the life of a fly in certain habitats. Thus, our findings are likely to bear ecological relevance.
Some intergenotype variation was observed in response to 4e-bp manipulation. Downregulation of 4e-bp by RNAi impacted mortality at all temperatures, while overexpression and pharmacological interventions showed temperature specificity, with no effect on survival in control conditions (25 °C). Similarly, COX activity was subject to interstrain variability; 4e-bp deletion elicited a temperature-independent, supraphysiological stimulation of COX that was not seen with any of the other manipulations. Regardless, across all trials, 4E-BP function showed a consistent, cross-genotype effect on the response to cold, both at the biochemical level and for organismal longevity.
The binding protein 4E-BP interacts with the nutrient-sensing insulin/IGF pathway (48) and has been implicated in lifespan extension by dietary restriction (23), although some controversy remains (41, 49). Our results showing that 4E-BP regulates survival in response to ambient temperature therefore suggest a link between temperature and nutrition in the context of lifespan determination. 4E-BP may represent a multipronged metabolic sensor governing metabolism and lifespan in response to multiple environmental factors. In an accompanying paper, Cintron-Colon et al. (50) show that the insulin-like growth factor 1 receptor (IGF-1R) mediates the integration of nutrient and temperature signaling in the mammalian CNS, adding another intriguing link between diet and temperature, two factors classically thought of as unrelated. We also report that temperature and diet have similar effects on mortality rate. Despite these striking parallels between the two environmental manipulations, key differences exist. Temperature affects survival more dramatically than dietary manipulations (12, 13). Additionally, we show that 4E-BP activity reduces survival under cold, whereas it is commonly linked with life extension in the context of diet and nutrient sensing (23, 48). This disparity is puzzling, since both environmental factors trigger similar 4E-BP–dependent metabolic changes—such as enhanced mitochondrial function—which are thought to play a causal role in lifespan regulation (23). Different environmental factors might shift metabolism toward or past a longevity optimum, perhaps explaining how similar molecular mechanisms can be associated with contrasting effects on lifespan under different conditions.
Although one might expect ambient temperature to play a negligible role in homeothermic animals, recent evidence compellingly contradicts this notion. Even relatively mild temperature shifts, well within the range of the manipulations used in our study, strongly impact human health and disease (51). Temperature variation is the basis of therapeutic hypothermia, an induced drop in core body temperature routinely used as part of the therapeutic approach against heart attack and stroke (52, 53). In the acute phase of therapeutic hypothermia, a reduction of as little as 1 °C in core body temperature can dramatically affect patient prognosis (54). Although the beneficial effects of therapeutic hypothermia have been repeatedly documented and its practice is becoming more widespread, its mechanistic basis is incompletely understood. One of the processes thought to play an important role is inflammation. The inflammatory response is induced in the acute posttraumatic phase of stroke and other ischemic brain injuries (55) and underlies much of the pathogenesis seen in these conditions; cooling has a strong antiinflammatory effect (56, 57). Protein translation, and 4E-BP in particular, is known to regulate the inflammatory response (25, 58, 59), and drugs that influence 4E-BP function can affect the outcome of ischemic events (60, 61). Together with these observations, our finding that temperature regulates 4E-BP function raises the possibility that 4E-BP mediates the antiinflammatory, beneficial effects of therapeutic hypothermia.
Temperature robustly impacts longevity in both poikilotherms and homeotherms (4, 6–11). Elucidation of the underlying mechanisms will greatly contribute to the understanding of aging and age-related disease across species. The 4E-BP/eIF4E pathway may also mediate the effects of temperature in mammals, raising the exciting prospect of pharmacological interventions.
Methods
Fly Strains.
Flies were obtained from the Bloomington Drosophila Stock Center, unless otherwise noted. The 4e-bp−/− null and control (4e-bp+/+) flies are Thor2 and Thor1rv1, respectively (25). For some studies, daughterless-GAL4 was combined with UAS-Dcr2 for increased RNAi efficiency (62). The 4e-bp RNAi (#100739 from the Vienna Drosophila RNAi Center) is predicted to have no off-targets. For overexpression, a transgenic fly line [d4E-BP(LL)s] encoding an active form of 4E-BP under UAS control was used (31). For constitutive GAL4 experiments, genotypes were in a heterozygous background of w1118 (P0 = female) and “Cantonized” w1118 (P0 = male), achieved by outcrossing parental lines for at least 10 generations.
Fly Husbandry, Media Preparation, and Lifespan.
Fly food was prepared as described previously (43, 44). The concentrated (C) diet contained 10% yeast extract and 10% sucrose, while the diluted (D) diet contained 2.5% yeast extract and 2.5% sucrose, both in 1% agar (all wt/vol). The high protein (HP) diet contained 5% yeast extract, 5% sucrose, 8.6% cornmeal, and 0.5% agar, while the low protein (LP) diet differed only in the amount of yeast extract (0.5%). All diets contained 0.4% (vol/vol) propionic acid and 0.06% (vol/vol) phosphoric acid. Ad libitum water was supplied as 1% agar (wt/vol), as described previously (43). For lifespan, behavior, and biochemical studies, flies (2–4 d posteclosion) were randomly allocated to experimental diets and temperatures at a density of 25–30 flies per vial or ∼100 flies per bottle for demography switch experiments. Flies were transferred to fresh food every 2–3 d. Fly enclosures were placed randomly in incubators (humidity-controlled with 12/12-h light/dark cycles) and positions were rotated after each transfer to minimize the effects of microenvironment. To minimize interexperimental variation, survival data from independent trials conducted in three different laboratories were pooled for 4e-bp−/−-null studies. For biochemical and behavioral assays, flies (10–12 d old) were typically harvested or tested after 7 d on the indicated diet and temperature. HP medium was used in all experiments unless otherwise noted. For GS studies, food contained 200 μM RU486 (from a 50× stock) or the equivalent amount of a vehicle control (80% ethanol), prepared as described previously (63). For drug studies, 50 μL of 10 μM rotenone, 50 μM 4EGI-1, or vehicle (1% DMSO) were added on top of fly food and allowed to dry.
Food Intake Assays.
Food consumption was assessed using radioisotope labeling of the medium or the capillary feeder (CAFE) assay, as described previously (64). For radiolabeling, [α-32P]dCTP was used with five flies per vial, unless otherwise noted. For the CAFE assay, two capillaries of liquid food, identical to the HP diet but without cornmeal or agar, were used with four flies per chamber.
Protein Translation Analysis.
[35S]methionine incorporation was determined essentially as described previously (23). Briefly, flies were maintained at the indicated temperature on medium supplemented with 2.5 μCi [35S]methionine per milliliter of food. For each sample, five flies were homogenized in 100 μL 1% SDS and boiled for 5 min. Homogenates were cleared by centrifugation and supernatant was precipitated with 1 mL of 10% TCA. Samples were iced for 20 min and the supernatant was discarded after centrifugation at 4 °C. Pellets were washed twice with ice-cold 95% ethanol, air-dried, and resuspended in 50–100 μL 1% SDS for liquid scintillation and protein assay. Total protein content was measured using the Thermo Scientific Pierce 660 nm Protein Assay with Ionic Detergent Compatibility Reagent. Only background scintillation counts were detected in protein isolated from flies fed unlabeled food and spiked with pure [35S]methionine before homogenization, confirming that the protein precipitation procedure eliminated all unincorporated amino acids. To calculate the fraction of ingested label that was incorporated into nascent protein, total [35S]methionine accumulation within fly bodies was assessed from animals that underwent the feeding procedure.
[35S]methionine incorporation was also determined from isolated mitochondria (65). Briefly, labeled flies (five per sample) were gently crushed in a Dounce homogenizer [10 strokes with 0.4 mL of MIB buffer: 5 mM Tris, pH 7.4, 250 mM sucrose, 2 mM EGTA, and 0.3% (wt/vol) BSA]. After passing the homogenate through a 1-mL syringe packed with gauze, samples were centrifuged at 4 °C at 9,000 × g for 3 min. Pelleted mitochondria were washed with MIB and resuspended in 1% SDS for liquid scintillation.
COX Activity.
COX activity was measured essentially as described previously (23). Briefly, groups of 10 flies were homogenized in 400 μL PBST and centrifuged at 4 °C. Isolated supernatant (50 μL) was added to 950 μL assay buffer (10 mM Tris-Cl, pH 7.8, 2.5 mM MgCl2, and 10 μM cytochrome C from a freshly made stock of 0.22 mM cytochrome C reduced with 0.1 M DTT) in a cuvette and absorbance (550 nm) was monitored on a BioMate 5 spectrophotometer (Thermo Electron Corporation) for 2–3 min with 10- to 20-s intervals. Four samples, randomly selected, were processed and monitored at a time. COX activity was normalized to total protein content.
qRT-PCR.
Total RNA was extracted from whole-fly homogenates (15–30 flies per sample) using TRIzol (Life Technologies). RT-qPCR was performed using QScript XLT 1-step RT-qPCR ToughMix (Quanta Biosciences) on a CFX96 Touch Real-Time PCR Detection system (Bio-Rad) with a commercially designed primer and probe assay (TaqMan Gene Expression Assays, Life Technologies). Alternatively, first-strand cDNA was synthesized with random nonamer primers before qPCR using iQ SYBR Green Supermix (Bio-Rad), as described previously (66), and predefined primer pairs from FlyPrimerBank (67). Relative gene expression was calculated using the ΔΔCt method (68) and normalized to total input RNA.
Western Blotting.
Soluble protein (typically 25 μg) from clarified homogenates (20–30 flies per sample) were run on SDS/PAGE under reducing conditions, transferred to PVDF membrane, and blocked with 5% BSA in TBST before blotting with primary antibodies against phospho-4E-BP1 (Thr37/46, 236B4; Cell Signaling Technology, recognizes phosphorylation of Thr37 and/or Thr46), nonphospho–4E-BP1 (Thr46, 87D12, Cell Signaling Technology, recognizes nonphosphorylated Thr46), or β-tubulin (E7, Developmental Studies Hybridoma Bank, University of Iowa). Blots were then incubated with the appropriate HRP-conjugated secondary antibody, HRP goat anti-rabbit IgG, or HRP goat anti-mouse IgG, before detection using ECL Prime (GE Life Sciences). All antibodies were used at 1:1,000 dilution, except for the β-tubulin antibody (1:4,000).
Statistics.
Survival statistics, Kaplan–Meier plots, and Cox proportional hazards analysis (using the robust estimator and temperature as a categorical variable) were obtained using OASIS (69). Mortality results for demography switch experiments were calculated from raw survival data and smoothed using 3-d moving averages. Behavioral and biochemical results were compared by Student’s t test or ANOVA with Tukey–Kramer post hoc tests.
Acknowledgments
This work was supported by NIH Grants R01AG045036 and R21DK092735 (to W.W.J.), R01GM113894 (to B.C.), and the Glenn Foundation for Medical Research/American Federation for Aging Research. W.W.J. is a New Scholar in Aging of The Ellison Medical Foundation. We also acknowledge the Vienna Drosophila Resource Center and Bloomington stock centers for fly lines.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618994114/-/DCSupplemental.
References
- 1.Xiao R, Liu J, Xu XZ. Thermosensation and longevity. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2015;201:857–867. doi: 10.1007/s00359-015-1021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrella E, Longo VD. Insulin/IGF-I and related signaling pathways regulate aging in nondividing cells: From yeast to the mammalian brain. Sci World J. 2010;10:161–177. doi: 10.1100/tsw.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman T, Miyatake T, Smith HK, Partridge L. Interactions of mating, egg production and death rates in females of the Mediterranean fruit fly, Ceratitis capitata. Proc Biol Sci. 1998;265:1879–1894. doi: 10.1098/rspb.1998.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti B. Considerations on temperature, longevity and aging. Cell Mol Life Sci. 2008;65:1626–1630. doi: 10.1007/s00018-008-7536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solon-Biet SM, et al. Macronutrients and caloric intake in health and longevity. J Endocrinol. 2015;226:R17–R28. doi: 10.1530/JOE-15-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo AE, Flouris AD. Caloric restriction and longevity: Effects of reduced body temperature. Ageing Res Rev. 2011;10:153–162. doi: 10.1016/j.arr.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Conti B, et al. Transgenic mice with a reduced core body temperature have an increased life span. Science. 2006;314:825–828. doi: 10.1126/science.1132191. [DOI] [PubMed] [Google Scholar]
- 8.Klass MR. Aging in the nematode Caenorhabditis elegans: Major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 9.Lamb MJ. Temperature and lifespan in Drosophila. Nature. 1968;220:808–809. doi: 10.1038/220808a0. [DOI] [PubMed] [Google Scholar]
- 10.Loeb J, Northrop JH. Is there a temperature coefficient for the duration of life? Proc Natl Acad Sci USA. 1916;2:456–457. doi: 10.1073/pnas.2.8.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb J, Northrup JH. On the influence of food and temperature upon the duration of life. J Biol Chem. 1917;32:103–121. [Google Scholar]
- 12.Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- 13.Yen K, Mobbs CV. Dietary restriction and cold temperature both acutely reduce senescence in C. elegans. Open Longev Sci. 2007;1:8–13. [Google Scholar]
- 14.Conti B, Hansen M. A cool way to live long. Cell. 2013;152:671–672. doi: 10.1016/j.cell.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 15.Xiao R, et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. 2013;152:806–817. doi: 10.1016/j.cell.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil G, Cummings E, de Magalhães JP. Being cool: How body temperature influences ageing and longevity. Biogerontology. 2015;16:383–397. doi: 10.1007/s10522-015-9571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rikke BA, Johnson TE. Lower body temperature as a potential mechanism of life extension in homeotherms. Exp Gerontol. 2004;39:927–930. doi: 10.1016/j.exger.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afonyushkin T, Moll I, Bläsi U, Kaberdin VR. Temperature-dependent stability and translation of Escherichia coli ompA mRNA. Biochem Biophys Res Commun. 2003;311:604–609. doi: 10.1016/j.bbrc.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 20.Das AB, Prosser CL. Biochemical changes in tissues of goldfish acclimated to high and low temperatures. I. Protein synthesis. Comp Biochem Physiol. 1967;21:449–467. doi: 10.1016/0010-406x(67)90445-8. [DOI] [PubMed] [Google Scholar]
- 21.Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathers EM, Houlihan DF, McCarthy ID, Burren LJ. Rates of growth and protein synthesis correlated with nucleic acid content in fry of rainbow trout, Oncorhynchus mykiss: Effects of age and temperature. J Fish Biol. 1993;43:245–263. [Google Scholar]
- 23.Zid BM, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 25.Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci USA. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmbeck MA, Rand DM. Dietary fatty acids and temperature modulate mitochondrial function and longevity in Drosophila. J Gerontol A Biol Sci Med Sci. 2015;70:1343–1354. doi: 10.1093/gerona/glv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita M, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Tsai S, et al. Muscle-specific 4E-BP1 signaling activation improves metabolic parameters during aging and obesity. J Clin Invest. 2015;125:2952–2964. doi: 10.1172/JCI77361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonenberg N, Gingras AC. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 30.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miron M, et al. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- 32.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada R, et al. Mifepristone reduces food palatability and affects Drosophila feeding and lifespan. J Gerontol A Biol Sci Med Sci. 2016;72:173–180. doi: 10.1093/gerona/glw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q, et al. Rotenone induction of hydrogen peroxide inhibits mTOR-mediated S6K1 and 4E-BP1/eIF4E pathways, leading to neuronal apoptosis. Toxicol Sci. 2015;143:81–96. doi: 10.1093/toxsci/kfu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scialò F, et al. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coulom H, Birman S. Chronic exposure to rotenone models sporadic Parkinson’s disease in Drosophila melanogaster. J Neurosci. 2004;24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 40.Sekiyama N, et al. Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc Natl Acad Sci USA. 2015;112:E4036–E4045. doi: 10.1073/pnas.1512118112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatar M. The plate half-full: Status of research on the mechanisms of dietary restriction in Drosophila melanogaster. Exp Gerontol. 2011;46:363–368. doi: 10.1016/j.exger.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen K, Mobbs CV. Evidence for only two independent pathways for decreasing senescence in Caenorhabditis elegans. Age (Dordr) 2010;32:39–49. doi: 10.1007/s11357-009-9110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ja WW, et al. Water- and nutrient-dependent effects of dietary restriction on Drosophila lifespan. Proc Natl Acad Sci USA. 2009;106:18633–18637. doi: 10.1073/pnas.0908016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruce KD, et al. High carbohydrate-low protein consumption maximizes Drosophila lifespan. Exp Gerontol. 2013;48:1129–1135. doi: 10.1016/j.exger.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KP, et al. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci USA. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solon-Biet SM, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horikawa M, Sural S, Hsu AL, Antebi A. Co-chaperone p23 regulates C. elegans lifespan in response to temperature. PLoS Genet. 2015;11:e1005023. doi: 10.1371/journal.pgen.1005023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai H, Post S, Kang P, Tatar M. Drosophila longevity assurance conferred by reduced insulin receptor substrate chico partially requires d4eBP. PLoS One. 2015;10:e0134415. doi: 10.1371/journal.pone.0134415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Partridge L, Alic N, Bjedov I, Piper MD. Ageing in Drosophila: The role of the insulin/Igf and TOR signalling network. Exp Gerontol. 2010;46:376–381. doi: 10.1016/j.exger.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cintron-Colon R, et al. Insulin-like growth factor 1 receptor regulates hypothermia during calorie restriction. Proc Natl Acad Sci USA. 2017;114:9731–9736. doi: 10.1073/pnas.1617876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee P, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 2014;63:3686–3698. doi: 10.2337/db14-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi HA, Badjatia N, Mayer SA. Hypothermia for acute brain injury–Mechanisms and practical aspects. Nat Rev Neurol. 2012;8:214–222. doi: 10.1038/nrneurol.2012.21. [DOI] [PubMed] [Google Scholar]
- 53.Perman SM, Goyal M, Neumar RW, Topjian AA, Gaieski DF. Clinical applications of targeted temperature management. Chest. 2014;145:386–393. doi: 10.1378/chest.12-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bro-Jeppesen J, et al. The inflammatory response after out-of-hospital cardiac arrest is not modified by targeted temperature management at 33 °C or 36 °C. Resuscitation. 2014;85:1480–1487. doi: 10.1016/j.resuscitation.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Jin R, Yang G, Li G. Inflammatory mechanisms in ischemic stroke: Role of inflammatory cells. J Leukoc Biol. 2010;87:779–789. doi: 10.1189/jlb.1109766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng H, Han HS, Cheng D, Sun GH, Yenari MA. Mild hypothermia inhibits inflammation after experimental stroke and brain inflammation. Stroke. 2003;34:2495–2501. doi: 10.1161/01.STR.0000091269.67384.E7. [DOI] [PubMed] [Google Scholar]
- 57.Gundersen Y, Vaagenes P, Pharo A, Valø ET, Opstad PK. Moderate hypothermia blunts the inflammatory response and reduces organ injury after acute haemorrhage. Acta Anaesthesiol Scand. 2001;45:994–1001. doi: 10.1034/j.1399-6576.2001.450812.x. [DOI] [PubMed] [Google Scholar]
- 58.Mazumder B, Li X, Barik S. Translation control: A multifaceted regulator of inflammatory response. J Immunol. 2010;184:3311–3319. doi: 10.4049/jimmunol.0903778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colina R, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 60.Chauhan A, Sharma U, Jagannathan NR, Reeta KH, Gupta YK. Rapamycin protects against middle cerebral artery occlusion induced focal cerebral ischemia in rats. Behav Brain Res. 2011;225:603–609. doi: 10.1016/j.bbr.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 61.Xie R, et al. Alpha-lipoic acid pre- and post-treatments provide protection against in vitro ischemia-reperfusion injury in cerebral endothelial cells via Akt/mTOR signaling. Brain Res. 2012;1482:81–90. doi: 10.1016/j.brainres.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 63.Ren C, Finkel SE, Tower J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2009;44:228–235. doi: 10.1016/j.exger.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deshpande SA, et al. Quantifying Drosophila food intake: Comparative analysis of current methodology. Nat Methods. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker DW, et al. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc Natl Acad Sci USA. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ja WW, Carvalho GB, Madrigal M, Roberts RW, Benzer S. The Drosophila G protein-coupled receptor, Methuselah, exhibits a promiscuous response to peptides. Protein Sci. 2009;18:2203–2208. doi: 10.1002/pro.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y, et al. FlyPrimerBank: An online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 2013;3:1607–1616. doi: 10.1534/g3.113.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Yang JS, et al. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS One. 2011;6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]