Significance
d-3-Phosphoglycerate dehydrogenase (SerA) is a key enzyme in l-serine biosynthesis. It couples the dehydrogenation of d-3-phosphoglycerate to 3-phosphohydroxypyruvate and the reduction of 2-ketoglutarate to d-2-hydroxyglutarate (d-2-HG). This provides an example of how nonenergetically favorable and energetically favorable reactions are linked together to allow metabolic processes to proceed. d-2-HG is often considered as an abnormal metabolite produced by several enzymes with “promiscuous” activities. Our findings offer insights into how an enzymatic reaction that was considered promiscuous or accidental plays a key role in metabolism. We have identified a bacterial d-2-hydroxyglutarate dehydrogenase (D2HGDH), which converts d-2-HG produced during l-serine biosynthesis back to 2-ketoglutarate. d-2-HG is a normal metabolite that is simultaneously produced and catabolized without accumulation in bacterial metabolism.
Keywords: d-2-Hydroxyglutarate, 2-ketoglutarate, d-3-phosphoglycerate, l-serine biosynthesis, bacterial metabolism
Abstract
l-Serine biosynthesis, a crucial metabolic process in most domains of life, is initiated by d-3-phosphoglycerate (d-3-PG) dehydrogenation, a thermodynamically unfavorable reaction catalyzed by d-3-PG dehydrogenase (SerA). d-2-Hydroxyglutarate (d-2-HG) is traditionally viewed as an abnormal metabolite associated with cancer and neurometabolic disorders. Here, we reveal that bacterial anabolism and catabolism of d-2-HG are involved in l-serine biosynthesis in Pseudomonas stutzeri A1501 and Pseudomonas aeruginosa PAO1. SerA catalyzes the stereospecific reduction of 2-ketoglutarate (2-KG) to d-2-HG, responsible for the major production of d-2-HG in vivo. SerA combines the energetically favorable reaction of d-2-HG production to overcome the thermodynamic barrier of d-3-PG dehydrogenation. We identified a bacterial d-2-HG dehydrogenase (D2HGDH), a flavin adenine dinucleotide (FAD)-dependent enzyme, that converts d-2-HG back to 2-KG. Electron transfer flavoprotein (ETF) and ETF-ubiquinone oxidoreductase (ETFQO) are also essential in d-2-HG metabolism through their capacity to transfer electrons from D2HGDH. Furthermore, while the mutant with D2HGDH deletion displayed decreased growth, the defect was rescued by adding l-serine, suggesting that the D2HGDH is functionally tied to l-serine synthesis. Substantial flux flows through d-2-HG, being produced by SerA and removed by D2HGDH, ETF, and ETFQO, maintaining d-2-HG homeostasis. Overall, our results uncover that d-2-HG–mediated coupling between SerA and D2HGDH drives bacterial l-serine synthesis.
As a critical intermediate in tricarboxylic acid cycle (TCA cycle), 2-ketoglutarate (2-KG) can be converted into succinyl-CoA (1), succinate (2), l-glutamate (3), or 2-hydroxyglutarate (2-HG) (4). 2-HG exists in two stereoisomeric conformations: l-2-hydroxyglutarate (l-2-HG) (5, 6) and d-2-hydroxyglutarate (d-2-HG) (7, 8) (SI Appendix, Fig. S1). Mutations in isocitrate dehydrogenase (IDH), which result in strong 2-KG–reducing activity and accumulation of excess d-2-HG, have been detected in numerous types of cancer. d-2-HG is a key effector of IDH mutations that promote oncogenesis (7–10). Another severe disease involving d-2-HG is inborn d-2-hydroxyglutaric aciduria (d-2-HGA), with increased d-2-HG levels in body fluids as the biochemical hallmark (4). Thus, d-2-HG has been viewed as an abnormal metabolite (4, 11, 12).
d-2-HG has been detected in various organisms, including Homo sapiens (4, 13), Arabidopsis thaliana (14), and Saccharomyces cerevisiae (15). In animals, plants, and yeasts, d-2-HG can be converted into 2-KG by d-2-HG dehydrogenase (D2HGDH) (13–15). The role of d-2-HG catabolism has been extensively studied, particularly in patients with d-2-HGA (4). 2-HG has also been found in bacteria such as Pseudomonas (16) and Escherichia coli (17). However, even though D2HGDH activities have been reported in crude preparations of Pseudomonas putida (18) and Rhodospirillum rubrum (19), bacterial D2HGDH has never been characterized.
Certain enzymes exhibit, beyond their classical functions, 2-KG–reducing activity for d-2-HG production. These enzymes include hydroxyacid-oxoacid transhydrogenase (HOT) in H. sapiens (20, 21), 4-phospho-d-erythronate dehydrogenase (PdxB) in E. coli (22), UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase (WbpB) in Pseudomonas aeruginosa (23), 2-hydroxyacid dehydrogenase (McyI) in Microcystis aeruginosa (24), and d-3-phosphoglycerate dehydrogenase (SerA, also named PHGDH) in H. sapiens (25) and S. cerevisiae (15). SerA in E. coli is capable of producing 2-HG from 2-KG; however, the enantiomer of its product (2-HG) needs to be verified (26, 27).
SerA catalyzes the initial reaction for l-serine biosynthesis, a critical metabolic pathway that is essential for the survival of most organisms (26–29). Approximately 15% of the carbon flux generated from glycolysis of glucose passes through l-serine biosynthesis in E. coli (27). However, the starting point of l-serine biosynthesis, d-3-phosphoglycerate (d-3-PG) dehydrogenation catalyzed by SerA, is quite thermodynamically unfavorable (27, 30). The feasible but puzzling reaction of d-2-HG production contrasts with the unfavorable but critical physiological reaction of d-3-PG dehydrogenation. This intriguing phenomenon prompts us to study the poorly understood roles of d-2-HG production by SerA in l-serine biosynthesis.
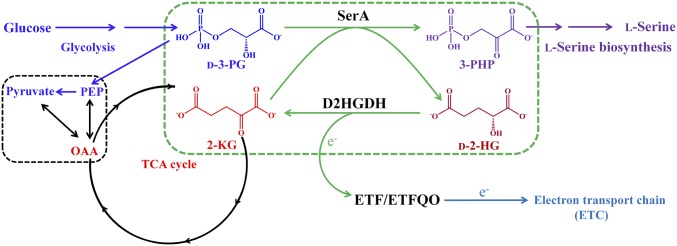
In this study, we discovered a robust coupling between SerA and D2HGDH, which constructs the interconversion between d-2-HG and 2-KG, and drives bacterial l-serine synthesis in Pseudomonas (Fig. 1). SerA combines the reaction of d-2-HG production from 2-KG to overcome the thermodynamic barrier of d-3-PG dehydrogenation. We have identified a bacterial D2HGDH, which converts d-2-HG to 2-KG, and is functionally tied to l-serine biosynthesis. The coupling between SerA and D2HGDH also establishes a linkage among the intermediates of glycolysis (d-3-PG), TCA cycle (2-KG), and l-serine biosynthesis (Fig. 1).
Fig. 1.
The coupling between SerA and D2HGDH makes the robust interconversion between d-2-HG and 2-KG (green dotted box) to overcome the thermodynamic barrier of d-3-PG dehydrogenation and facilitate the biosynthetic process of l-serine (purple pathway). The coupling establishes a linkage among the intermediates of glycolysis (d-3-PG), TCA cycle (2-KG), and l-serine biosynthesis. The black dotted box indicates the already known straightforward connections between the intermediates of glycolysis (PEP and pyruvate) and TCA cycle (OAA). OAA, oxaloacetate; PEP, phosphoenolpyruvate; 3-PHP, 3-phosphohydroxypyruvate.
Results
D2HGDH Is Involved in d-2-HG and Glucose Utilization.
All representative Pseudomonas species, including Pseudomonas stutzeri A1501, P. aeruginosa PAO1 and P. putida KT2440, possess homologous proteins that share more than 30% identity with the D2HGDH from H. sapiens (SI Appendix, Table S1). This study focuses on the bacterial metabolism of d-2-HG, using Pseudomonas species as the model system. Given their known genomic information and tractable genetics, P. stutzeri A1501 (31, 32) and P. aeruginosa PAO1 (33) were selected for further studies.
The gene d2hgdh (Gene ID: 5094894) that encodes the predicted D2HGDH in P. stutzeri A1501 was deleted and then complemented. The resulting strains were named as A1501-Δd2hgdh and A1501-Δd2hgdh-d2hgdh+, respectively. Compared with the wild-type (WT) strain and A1501-Δd2hgdh-d2hgdh+, A1501-Δd2hgdh is barely able to use d-2-HG (SI Appendix, Fig. S2 A and B). Disruption of d2hgdh to impair d-2-HG utilization also caused a relatively slower growth of P. stutzeri A1501 in the medium containing glucose (SI Appendix, Fig. S3 A and B). The maximal biomass of A1501-Δd2hgdh was lower than that of A1501-WT (SI Appendix, Fig. S3 A and B), implying that d-2-HG metabolism might be involved in glucose utilization.
D2HGDH Contributes to d-2-HG Conversion to 2-KG.
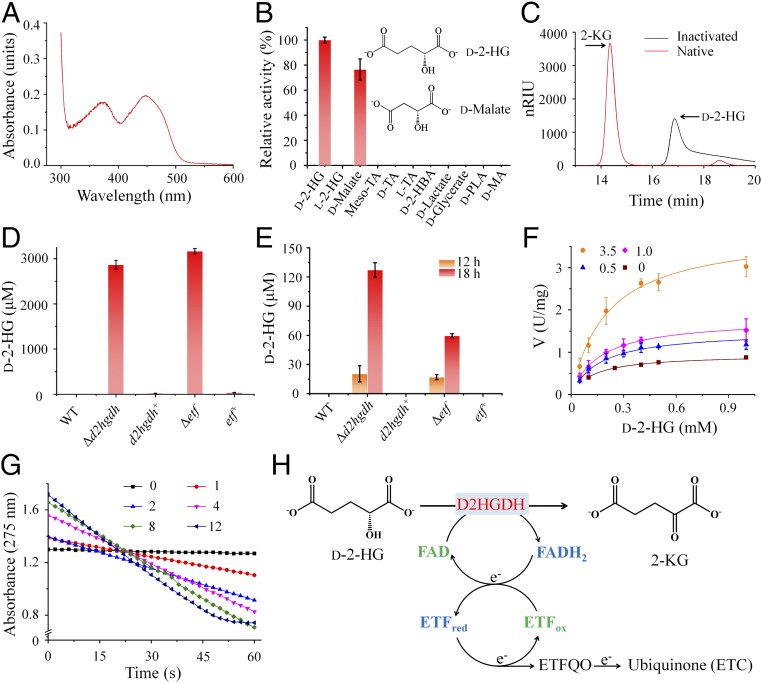
The predicted D2HGDH in P. stutzeri A1501 was expressed and purified as a His-tagged protein (SI Appendix, Fig. S4A). The purified protein was identified as a homodimer (SI Appendix, Figs. S4A and S5A) and bound one flavin adenine dinucleotide (FAD) per subunit (Fig. 2A and SI Appendix, Fig. S5B). Substrate screening revealed that only the d-isomers of 2-HG and malate could function as substrates (Fig. 2B). HPLC analysis showed that d-2-HG was converted into 2-KG by the enzyme in the presence of artificial electron acceptors (Fig. 2C), confirming the identification of a bacterial D2HGDH from P. stutzeri A1501. Other enzymatic properties of the D2HGDH were also investigated in this study (Table 1 and SI Appendix, Fig. S5 C–F).
Fig. 2.
D2HGDH plus ETF and ETFQO convert d-2-HG to 2-KG. (A) Full-wave scanning of the purified D2HGDH. (B) Substrate screening (d-2-HG in reaction mixture, 1 mM; other chemicals, 5 mM) for the purified D2HGDH (0.2 μM). d-2-HBA, d-2-hydroxybutyrate; d-MA, d-mandelate; d-PLA, d-phenyllactate; TA, tartrate. (C) HPLC analysis of the product of D2HGDH-catalyzed d-2-HG oxidation. The reaction mixtures containing d-2-HG (5 mM), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 8 mM), PMS (0.8 mM), and native or heat-inactivated D2HGDH (11.4 μM) in 50 mM Tris⋅HCl (pH 7.4) were incubated at 37 °C for 40 min. (D) The maximal concentrations of extracellular d-2-HG. (E) The intracellular concentrations of d-2-HG. The strains in D and E were grown in medium with glucose as the sole carbon source. (F) The effect of ETF (0, 0.5, 1.0, and 3.0 μM) on the kinetics of D2HGDH (0.14 μM) for d-2-HG was studied in the presence of a series of d-2-HG concentrations (0–1 mM) and DCIP (200 μM). The data were fitted to a hyperbolic curve. (G) The effect of ETFQO on the D2HGDH-catalyzed CoQ1 reduction. The reaction mixtures containing 50 mM Hepes (K+) (pH 7.4), dodecamaltoside (0.2 mM), D2HGDH (1 μM), d-2-HG (1 mM), ETF (4 μM), CoQ1 (53.2 μM), and ETFQO (0–12 μM) were monitored at 275 nm at room temperature. (H) Schematic diagram for the process of d-2-HG conversion to 2-KG. ETFox, oxidized ETF; ETFred, reduced ETF. Values are the average ± SD (n = 3).
Table 1.
Kinetic parameters of D2HGDH for d-2-HG, ETF and cytochrome c
| Substrate | Km, mM | Vmax, U·mg−1 | Kcat, min−1 | Kcat/Km, min−1·mM−1 |
| d-2-HG | 0.17 ± 0.02 | 4.56 ± 0.60 | 473.83 ± 62.87 | 2,723.55 ± 91.15 |
| ETF | (7.71 ± 1.83) × 10−3 | 7.50 ± 1.10 | 779.72 ± 114.85 | (1.03 ± 0.11) × 105 |
| Cytochrome c | (210.11 ± 18.40) × 10−3 | 0.84 ± 0.07 | 87.38 ± 6.98 | 416.08 ± 5.76 |
d-2-HG was barely detectable in the P. stutzeri A1501 growth medium during glucose utilization. For A1501-Δd2hgdh, the gradual accumulation of extracellular d-2-HG (SI Appendix, Fig. S3B) reached an unexpectedly high concentration (0.42 g·L−1, ≈2.8 mM) from 3.5 g·L−1 of glucose (Fig. 2D), implying continuous and robust d-2-HG production. By contrast, A1501-Δd2hgdh-d2hgdh+ regained the ability to catabolize d-2-HG (SI Appendix, Fig. S3C), and therefore, prevented the accumulation of d-2-HG (Fig. 2D). Accumulation of intracellular d-2-HG was observed in A1501-Δd2hgdh (Fig. 2E), further confirming the function of D2HGDH in d-2-HG catabolism. Although complementation of A1501-Δd2hgdh with the gene d2hgdh eliminated the d-2-HG accumulation, the delayed growth kinetics was not reversed (SI Appendix, Fig. S3 A and C). This may be due to the inhibitory effect of added antibiotic (gentamicin) and inducer (isopropyl β-d-thiogalactoside, IPTG), which are required for the expression of d2hgdh.
Electron Transfer Flavoprotein and Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase also Participate in d-2-HG Conversion to 2-KG.
D2HGDH is not a membrane protein and cannot directly transfer electrons to the electron transport chain (ETC). Both cytochrome c and electron transfer flavoprotein (ETF) have been shown to function as soluble electron carriers between flavoprotein dehydrogenases and electron transport chains (34). The Km and Vmax values of D2HGDH for cytochrome c were 210.11 ± 18.40 μM and 0.84 ± 0.07 U·mg−1, respectively (Table 1), implying that cytochrome c might not be an efficient electron acceptor (because of the high Km value). The ETF from P. stutzeri A1501 was expressed and purified (SI Appendix, Fig. S4B). When the purified ETF was added, increased activities of D2HGDH toward dichloroindophenol (DCIP) reduction were detected (SI Appendix, Fig. S4C). As shown in Fig. 2F, the addition of ETF strongly favored d-2-HG oxidation. As the ETF concentration increased, the Vmax values also increased continuously. When ETF was considered as the substrate for D2HGDH, the Km and Vmax values were measured as 7.71 ± 1.83 μM and 7.50 ± 1.10 U·mg−1, respectively (Table 1).
ETF uses ETF-ubiquinone oxidoreductase (ETFQO) as its essential partner to transfer electrons from flavoprotein dehydrogenases to the ubiquinone pool in ETC (34). Coenzyme Q1 (CoQ1) reduction was observed in the presence of d-2-HG, D2HGDH, ETF, and ETFQO (expressed and purified as shown in SI Appendix, Fig. S4D) (Fig. 2G). The rate of d-2-HG oxidation increased upon the addition of ETFQO (Fig. 2G), further demonstrating the role of ETFQO in d-2-HG oxidation and the accompanied ETF-mediated electron transfer to CoQ1.
An in-frame deletion of etf abolished growth of the mutant in the media containing d-2-HG, and complementation with the plasmid containing etf restored growth of the mutant (SI Appendix, Fig. S2 A and B). The strains were also tested for growth on the medium containing glucose as the sole carbon source. Intracellular and extracellular d-2-HG concentrations were also determined. The results (Fig. 2 D and E and SI Appendix, Fig. S3 D and E) were similar to those obtained with the d2hgdh deletion mutant and complemented strain (Fig. 2 D and E and SI Appendix, Fig. S3 B and C). These results indicated that D2HGDH, ETF, ETFQO, and ETC efficiently maintain d-2-HG homeostasis through the conversion of d-2-HG to 2-KG (Fig. 2H).
SerA Contributes to 2-KG Conversion to d-2-HG.
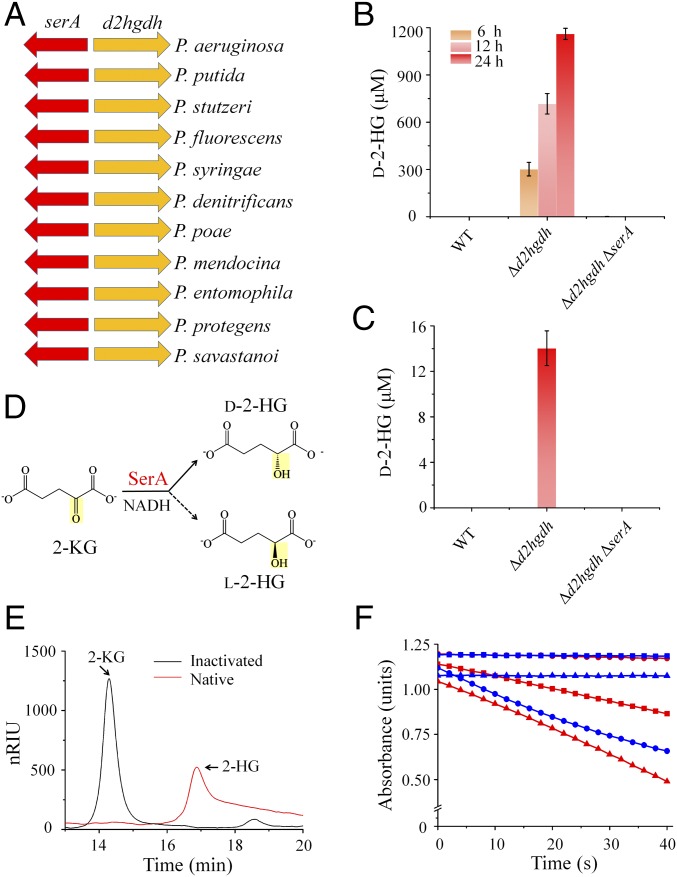
The d2hgdh gene is adjacent to the d-3-phosphoglycerate dehydrogenase (SerA, also named PHGDH)-encoding gene serA in all representative species of Pseudomonas, including P. stutzeri, P. aeruginosa, P. putida, P. fluorescens, and so on (Fig. 3A). SerA was disrupted in A1501-Δd2hgdh to generate the double-mutant A1501-Δd2hgdhΔserA. As expected, the strain exhibited l-serine auxotrophy (SI Appendix, Fig. S2 C and D), and the addition of external l-serine triggered some growth (SI Appendix, Fig. S3F). A1501-Δd2hgdhΔserA could grow relatively well (SI Appendix, Fig. S3G) in LB medium, a rich medium containing the amino acids required for bacterial growth (including l-serine). Compared with A1501-Δd2hgdh, the double-mutant A1501-Δd2hgdhΔserA in LB medium showed no accumulation of extracellular and intracellular d-2-HG (Fig. 3 B and C), indicating that SerA catalyzes d-2-HG production in vivo.
Fig. 3.
SerA contributes to 2-KG conversion to d-2-HG. (A) SerA-encoding gene serA is adjacent to the predicated D2HGDH encoding gene d2hgdh. (B) The concentrations of extracellular d-2-HG during growth. (C) The intracellular concentrations of d-2-HG (sampled at 12 h). The strains in B and C were grown in LB medium. Values are the average ± SD (n = 3). (D) The possible configurations of 2-HG produced by SerA-catalyzed 2-KG reduction. (E) HPLC analysis of the product of SerA-catalyzed 2-KG conversion in presence of NADH. The mixtures containing 2-KG (5 mM), NADH (5 mM), native SerA, or heat-inactivated SerA (5.6 μM) in 50 mM Tris (pH 7.4), were incubated at 30 °C for 3 h. (F) The configuration of the SerA-produced 2-HG was determined using an enzymatic method. The reactions using authentic d-2-HG (squares), l-2-HG (circles), or SerA-produced 2-HG (triangles) as substrates were catalyzed by the D2HGDH purified in this study (red lines) or l-2-hydroxyglutarate oxidoreductase (LGRO, blue lines), respectively. The purification and characterization of LGRO are presented in SI Appendix, Fig. S4 F and G.
SerA from P. stutzeri A1501 was expressed and purified as a His-tagged protein (SI Appendix, Fig. S4E). The purified SerA was found to reduce 2-KG to 2-HG in the presence of NADH (Fig. 3 D and E), and the results from enzymatic chirality assays clearly showed that the reaction product is d-2-HG solely (Fig. 3F and SI Appendix, Fig. S4 F and G); this indicates that SerA can reduce 2-KG with good stereoselectivity to produce d-2-HG (Fig. 3D).
2-KG Is Converted to d-2-HG During the Coupled Reaction of SerA and SerC.
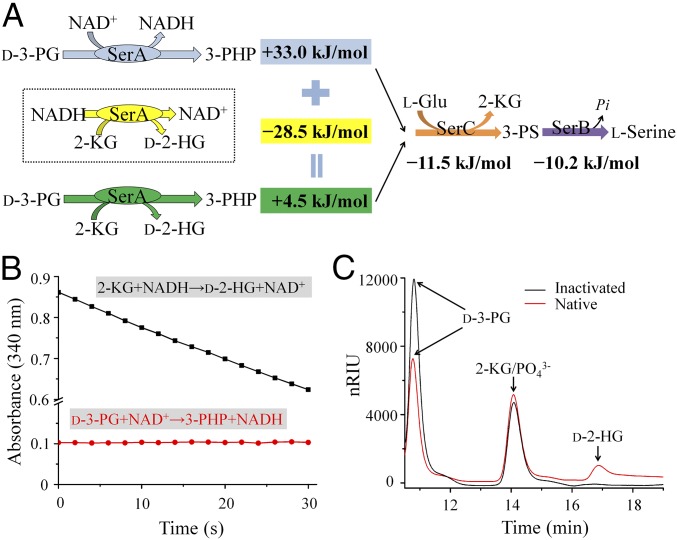
SerA catalyzes the dehydrogenation of d-3-PG to 3-phosphohydroxypyruvate (3-PHP), but it also possesses the puzzling ability to reduce 2-KG to d-2-HG (Fig. 4A). The rate of SerA-catalyzed 2-KG reduction was considerably higher than that of SerA-catalyzed d-3-PG dehydrogenation, which was below the detection limit (Fig. 4B). SerA-catalyzed d-3-PG dehydrogenation has a high ΔG°′obs value (+33.0 kJ·mol−1) (30). According to the conventional view, d-3-PG dehydrogenation is driven through coupling with the reactions catalyzed by l-phosphoserine aminotransferase (SerC) (ΔG°′obs = −11.5 kJ·mol−1) (30) and l-phosphoserine phosphatase (SerB) (ΔG°′obs = −10.2 kJ·mol−1) (35) in l-serine synthesis. The ΔG°′obs of the coupled reactions from d-3-PG to l-phosphoserine and from d-3-PG to l-serine are +21.5 kJ·mol−1 and +11.2 kJ·mol−1, respectively, indicating that these reactions are not thermodynamically favorable. However, d-3-PG was efficiently transformed by SerA and SerC (expressed and purified as shown in SI Appendix, Fig. S4H) in the presence of d-3-PG, NAD+, and l-glutamate (Fig. 4C), which are the substrates for SerA and SerC (Fig. 4A). Importantly, d-2-HG was also detected in the reaction mixture (Fig. 4C). This result was unexpected but is theoretically possible: The reaction catalyzed by SerC would produce 2-KG as the end product, which could then be reduced by SerA. The ΔG°′obs value of the 2-KG reduction to d-2-HG is approximately −28.5 kJ·mol−1 (36). The total ΔG°′obs of SerA-catalyzed d-3-PG dehydrogenation, SerC-catalyzed reaction, and SerA-catalyzed 2-KG reduction would be −7.0 kJ·mol−1 (Fig. 4A), suggesting that d-3-PG dehydrogenation catalyzed by SerC and SerA is feasible.
Fig. 4.
2-KG conversion to d-2-HG couples and drives d-3-PG dehydrogenation. (A) Thermodynamic analysis of the reactions in the l-serine biosynthesis pathway indicates that d-2-HG production serves SerA to facilitate d-3-PG dehydrogenation and l-serine biosynthesis. The ΔG°′obs values were calculated using the following formula: ΔG°′obs = −RTlnKobs. The Kobs value is the observed equilibrium constant of the reaction performed at 38 °C, pH 7.0 in previous studies. 3-PS, 3-phosphoserine. (B) The activities of 2-KG reduction reaction (2-KG, 5 mM; NADH, 0.2 mM) and d-3-PG dehydrogenation reaction (d-3-PG, 10 mM; NAD+, 1 mM) catalyzed by SerA (0.28 μM). (C) HPLC analysis of the products of the reaction coupled by SerA and SerC (heat-inactivated enzymes as a control). The mixtures containing NAD+ (4 mM), l-glutamate (10 mM), d-3-PG (20 mM), SerC (1.2 mg·mL−1), and SerA (1.03 mg·mL−1) in 50 mM Tris (pH 7.4) were reacted at 30 °C for 3 h.
SerA Couples d-3-PG Dehydrogenation with Conversion of 2-KG to d-2-HG.
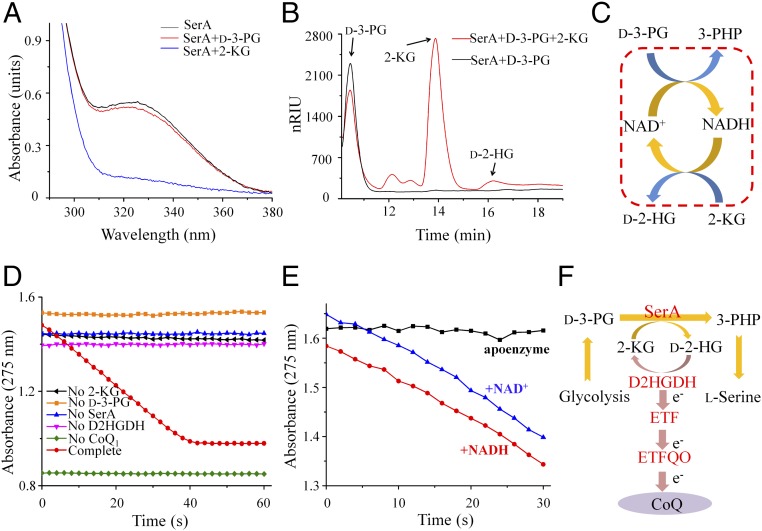
Purified SerA (as-isolated SerA) exhibited a prominent spectral signal at 325 nm (Fig. 5A), suggesting the presence of tightly bound NADH. The ratio of NADH to subunit of as-isolated SerA is ≈0.4–0.5 (SI Appendix, Fig. S4I). This was further supported by the determined high affinity of the apoprotein (SI Appendix, Fig. S4I) of SerA for NADH (equilibrium dissociation constant: KD ≈ 0.1 μM) using isothermal titration calorimetry (SI Appendix, Table S2). Additional NAD+ binding to SerA in this state (as-isolated or preincubated with NADH) was not preferred (SI Appendix, Table S2). Thus, reoxidation of the bound NADH to regenerate NAD+ timely is a prerequisite for NAD+-dependent d-3-PG dehydrogenation. When 2-KG was added to the solution containing as-isolated SerA, the absorbance at 325 nm decreased dramatically, indicating oxidation of the tightly bound NADH to NAD+ by 2-KG (Fig. 5A).
Fig. 5.
The reactions of d-2-HG production and d-3-PG dehydrogenation are coupled by redox transitions of NAD+ and NADH. (A) Full-wave scanning of SerA. Black line, as-isolated SerA; red line, as-isolated SerA treated with 5 mM d-3-PG; blue line, as-isolated SerA treated with 1.25 mM 2-KG. (B) HPLC analysis of the coupled reaction of d-3-PG dehydrogenation and 2-KG reduction catalyzed by SerA. The reaction mixtures containing as-isolated SerA (8.3 μM), d-3-PG (5 mM), and 2-KG (10 mM) as the substrates were reacted at 30 °C for 3 h. (C) Schematic diagram for the redox state transitions of NADH/NAD+ in the coupling and the stoichiometry of SerA-catalyzed coupled reaction. According to SI Appendix, Fig. S7, d-3-PG and 2-KG decrease 1.06 ± 0.04 mM and 1.04 ± 0.04 mM, respectively, while d-2-HG increases 1.01 ± 0.03 mM. Since the d-3-PG can only be converted into 3-PHP under the reaction conditions, the result can demonstrate the equimolar conversion of d-3-PG and 2-KG to 3-PHP and d-2-HG. (D) Enzymatic assays of the coupled reaction using an enzyme system including the essential elements for the HG-KG interconversion. The reaction mixtures containing 50 mM Hepes (K+) (pH 7.4), dodecamaltoside (0.2 mM), D2HGDH (1 μM), ETF (4 μM), CoQ1 (53.2 μM), 2-KG (5 mM), d-3-PG (5 mM), ETFQO (1.32 μM), and as-isolated SerA (1.66 μM) were monitored by CoQ1 reduction at 275 nm. (E) Effects of coenzyme on the activities of the coupled reaction. Apoenzyme of SerA, 0.19 μM; NAD+, 12.5 μM; NADH, 12.5 μM. (F) Schematic diagram for the multiple enzyme system assembled by SerA, D2HGDH, ETF, and ETFQO.
Therefore, we hypothesized that 2-KG conversion to d-2-HG is not a meaningless side reaction, but a reaction that couples and drives d-3-PG dehydrogenation through oxidizing NADH that binds to SerA and reducing the total ΔG°′obs of the SerA-catalyzed reaction. The sum of ΔG°′obs values of these two reactions is +4.5 kJ·mol−1, which is a value considerably lower than that of a single d-3-PG dehydrogenation reaction (Fig. 4A). Then, we analyzed SerA-catalyzed reactions containing d-3-PG, 2-KG, and NAD+. d-3-PG was very poorly oxidized in the absence of 2-KG, but when 2-KG was added, the dehydrogenation of d-3-PG was enhanced and the residual d-3-PG in the reaction mixture was diminished (SI Appendix, Fig. S6A), confirming that the coupling with 2-KG reduction is needed for the SerA-catalyzed d-3-PG dehydrogenation.
d-3-PG and 2-KG were also incubated with as-isolated SerA but without external NAD+ or NADH. As shown in Fig. 5B, the amount of the oxidized d-3-PG (estimated from the peak height) far exceeds that of the NADH contained in the enzyme (≈17 μM), suggesting that as-isolated SerA can oxidize d-3-PG with multiple turnovers by recycling of its bound coenzyme through reducing 2-KG into d-2-HG. The coupled reactions can also be simplified into a single reaction (Fig. 5C), which employs d-3-PG and 2-KG as the substrates and 3-PHP and d-2-HG as the products. The stoichiometry analysis of the SerA-catalyzed coupled reaction showed the equimolar conversion of d-3-PG and 2-KG to 3-PHP and d-2-HG (SI Appendix, Fig. S7). Due to the SerA-catalyzed d-3-PG dehydrogenation and the coupled d-2-HG production, efficient CoQ1 reduction was observed in the presence of d-3-PG, 2-KG, as-isolated SerA, D2HGDH, ETF, and ETFQO (Fig. 5D). When as-isolated SerA was replaced with apoenzyme, CoQ1 reduction disappeared. The CoQ1 reduction could be restored by adding NAD+ or NADH (Fig. 5E), suggesting the indispensable role of the coenzyme in the coupled reaction. Thus, SerA, D2HGDH, ETF, and ETFQO can be assembled into a multiple enzyme system that converts d-3-PG to 3-PHP for l-serine production and transfers the electrons produced during d-3-PG dehydrogenation to CoQ in the ETC (Fig. 5F).
l-Serine Rescues the Growth Defect of D2HGDH Knockout.
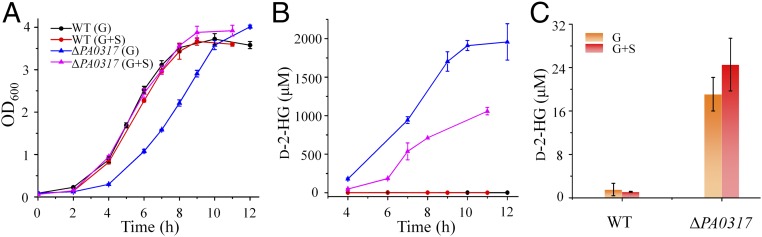
While the knockout of D2HGDH slowed the growth of P. stutzeri A1501, l-serine exhibited a strong inhibitory effect on the growth of P. stutzeri A1501 (SI Appendix, Fig. S3 H and I). Therefore, P. stutzeri A1501 is not suitable to study the capacity of l-serine to rescue the effects of D2HGDH knockout. P. aeruginosa PAO1 grows normally in the presence of a relatively high concentration of l-serine (2 mM) (Fig. 6A). The deletion of the D2HGDH-encoding gene (PA0317; Gene ID: 878323) in P. aeruginosa PAO1 (ΔPA0317) also yielded a slower growth (Fig. 6A) and accumulation of extracellular and intracellular d-2-HG (Fig. 6 B and C). Interestingly, external addition of l-serine (2 mM) was able to rescue the growth defect (Fig. 6A), suggesting that the knockout of D2HGDH decreases the flux for l-serine synthesis and that D2HGDH is functionally tied to l-serine synthesis.
Fig. 6.
l-Serine rescues the growth defect of D2HGDH knockout in P. aeruginosa PAO1. (A) The growth curves of the P. aeruginosa strains in the medium containing glucose (G) or glucose plus 2 mM l-serine (G+S). (B) The concentrations of extracellular d-2-HG during growth. (C) The concentrations of intracellular d-2-HG at 12 h. Values are the average ± SD (n = 3).
d-2-HG inhibited the SerA-catalyzed reduction of 2-KG and the inhibitory effect increased with d-2-HG concentrations (SI Appendix, Fig. S6 A and B). As 2-KG reduction is a prerequisite for d-3-PG dehydrogenation, d-2-HG would also inhibit d-3-PG dehydrogenation in the direction of l-serine synthesis. Since l-2-HG did not inhibit the activity of SerA-catalyzed 2-KG reduction (SI Appendix, Fig. S6C), the inhibitory effect is truly enantiomer-specific for d-2-HG. The real intracellular concentrations of d-2-HG (Figs. 2E and 6C) were far below extracellular ones (Figs. 2D and 6B). This means that most of the produced d-2-HG accumulated in the extracellular medium of A1501-Δd2hgdh (Figs. 2D and 6B), indicating the existence of an unknown efflux system for d-2-HG.
HG-KG Interconversion Works Robustly in Metabolism.
Coupling between SerA and D2HGDH generates an HG-KG interconversion (the interconversion between d-2-HG and 2-KG) system (SI Appendix, Fig. S8A). D2HGDH activities were detectable in cells grown in these distinct types of media and at any growth period (SI Appendix, Fig. S8B). d-2-HG was found to accumulate to high concentrations during the culture of A1501-Δd2hgdh with each carbon source investigated here (glucose, glycerol, pyruvate, succinate, 2-KG, and l-glutamate) or LB medium (SI Appendix, Table S3). These results indicated that substantial flux flows through the ongoing HG-KG interconversion during the metabolism under various conditions. Next, the effects of impaired D2HGDH function on the profiles of intracellular metabolites were investigated by GC-MS–based metabolomic analysis with glucose as the sole carbon source (SI Appendix, Fig. S8C and Dataset S1). 2-HG exhibits the most striking fold change (SI Appendix, Fig. S8C). Glycine is the metabolite that is directly related to l-serine through the action of l-serine hydroxymethyltransferase. There was no decrease in serine; however, the level of glycine decreased by fourfold (SI Appendix, Fig. S8D), indicating that the downstream pathway of l-serine biosynthesis was affected. Additionally, statistically significant changes in levels of intermediates involved in glycolysis, TCA cycle, and amino acid metabolism (SI Appendix, Fig. S8 A and C) were detected, which indicates that the dysfunction of the HG-KG interconversion influences these core metabolisms.
Phylogenetic Analysis of the Coupling Between SerA and D2HGDH.
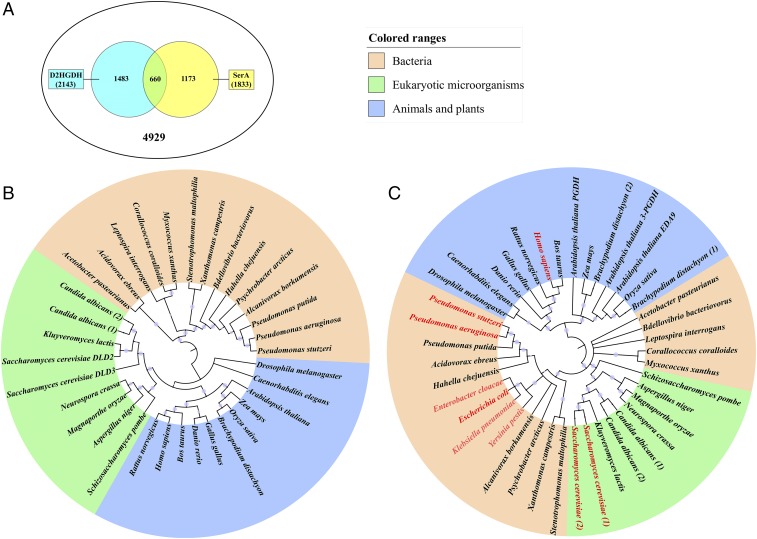
The distribution of D2HGDH and SerA in 4,929 completely sequenced bacterial genomes was studied (Fig. 7A and Datasets S2 and S3) and is illustrated in SI Appendix, Table S4. The coexistence of homologs of D2HGDH and SerA is found in 660 bacterial genomes (Fig. 7A and Dataset S4), many of which also have adjacent genomic localization of D2HGDH and SerA homologs (marked in Dataset S4). Thus, the coupling between SerA and D2HGDH is likely to be widespread in bacteria.
Fig. 7.
The type of HG-KG interconversion may exist universally across from bacteria to eukaryotes. The distribution of the homologs of D2HGDH and SerA in bacteria were analyzed (A). Phylogenetic bootstrap consensus trees of D2HGDH (B) and SerA (C) in various organisms were constructed. According to the previous reports, the organisms in which SerA can reduce 2-KG to 2-HG were marked with red color. The sequence details were shown in SI Appendix, Table S1.
Phylogenetic analysis revealed that D2HGDHs from eukaryotic microorganisms, animals, and plants (SI Appendix, Table S1) form a cluster (Fig. 7B). It is very clear that D2HGDHs found in H. sapiens (13), A. thaliana (14), and S. cerevisiae (15) catalyze the conversion of d-2-HG to 2-KG. The SerA homologs from eukaryotic microorganisms (SI Appendix, Table S1) constitute a phylogenetic group with SerA proteins from Pseudomonas (Fig. 7C), revealing the close relationship among them. SerA proteins from animals and plants (SI Appendix, Table S1) group into one separate cluster (Fig. 7C and SI Appendix, Table S1). Interestingly, SerA proteins from H. sapiens (6, 25) and S. cerevisiae (15) have been found to have the ability to produce d-2-HG from 2-KG. Combining the results obtained from these analyses, the coupling between D2HGDH and SerA may also take place in eukaryotic microorganisms, animals, and plants; however, a further detailed verification is still needed.
Discussion
According to the second law of thermodynamics, enzymatic conversions occur only in the direction with negative Gibbs free energy. There are several reported strategies to support an enzymatic conversion toward the thermodynamically unfavorable direction: the integration of thermodynamically favorable reactions catalyzed by other enzymes, the coupling with hydrolyzing reactions of high-energy phosphate bond, and redox coupling (for example, in complex II, from succinate directly to ubiquinone in ETC), and so on. The mechanism for bypassing the thermodynamic obstacle in this study may be different from the known ones. Both 2-KG reduction and d-3-PG dehydrogenation are catalyzed by SerA, and SerA combines them to make the reaction operated in the desired direction. The redox transition of NADH to NAD+ is coupled to an interesting metabolic transition of 2-KG to d-2-HG. In addition, the produced d-2-HG could be efficiently removed by D2HGDH with the assistance of ETF, ETFQO, and ETC. This study provides an example of how nonenergetically favorable and energetically favorable reactions are coupled to allow metabolic processes to proceed forward.
l-Serine is a source of one-carbon units (37) and serves as a precursor for the biosynthesis of glycine, cysteine, tryptophan, and phospholipids (27–29). SerA-involved l-serine biosynthesis pathway is the major anabolic source for l-serine (27), which could be demonstrated by the occurrence of l-serine auxotroph due to SerA knockout in this study (SI Appendix, Fig. S2C). The Gibbs free energy of the SerA-catalyzed d-3-PG dehydrogenation reaction yielding 3-PHP in real cellular environments (ΔG) has also been assessed (SI Appendix, Table S5) according to the values of ΔG°′obs (Fig. 4A) and the intracellular concentrations of related products and reactants. SerA-catalyzed d-3-PG dehydrogenation is thermodynamically unfavorable to proceed due to the high positive free energy in E. coli, MDA-MB-468, and BT-20 cells (SI Appendix, Table S5). Here, the calculated ΔG values of the reaction of d-3-PG dehydrogenation coupled with 2-KG reduction (SI Appendix, Table S5) suggest that the coupled reaction occurs easily in vivo and might be physiologically significant for l-serine biosynthesis.
The SerA-catalyzed d-3-PG dehydrogenation and 2-KG reduction follow the ordered Bi Bi mechanism (SI Appendix, Table S2). NAD+ or NADH binds first, followed by the other substrate. Following the reactions catalyzed by SerA, the products (3-PHP and d-2-HG) will fall off the enzyme before NAD+ or NADH. The presence of tightly bound NADH to as-isolated SerA indicates that NADH is not released after d-3-PG dehydrogenation. 2-KG addition would oxidize the NADH tightly bound to SerA to NAD+, and most of the NAD+ generated from NADH still bound to the enzyme (SI Appendix, Fig. S9A). However, the intracellular concentration of NAD+ in P. stutzeri A1501 was more than 100 μM, which was much higher than the KD (13 μM; SI Appendix, Table S2) and apparent Km of the apoenzyme for NAD+ (1.31 μM; SI Appendix, Fig. S9B). Therefore, the enzymatic steps for the SerA-catalyzed 2-KG reduction and d-3-PG oxidation might not include the release of NAD+ or NADH, and instead, the coenzymes could remain bound during the coupled reaction. d-3-PG and 2-KG (generated from glycolysis and TCA cycle, respectively), would be incorporated into the coupled reaction of SerA. Then, the products of SerA, d-2-HG and 3-PHP, would be continuously removed or used by cells (SI Appendix, Fig. S9C).
D2HGDH has been studied in animals, plants, and eukaryotic microorganisms (13–15). Here, we have identified a bacterial D2HGDH from Pseudomonas. Although there is a rather low level of d-2-HG under normal conditions, our results suggest substantial flux flowing through d-2-HG in physiological metabolism (SI Appendix, Table S3). In addition, there might be an efflux system for d-2-HG in Pseudomonas. If this efflux does not exist, the intracellular d-2-HG would accumulate, inhibit SerA, and cause serine auxotrophy in the absence of D2HGDH. We speculate that animal cells also have such efflux system since high levels of d-2-HG can be detected in the plasma and urine of patients with d-2-HG–related cancer and aciduria (4, 38). Despite the existence of the efflux system, the disruption of D2HGDH in Pseudomonas also led to some growth defects. Serine levels remained unchanged, but the level of glycine, the downstream metabolite of l-serine, decreased significantly (SI Appendix, Fig. S8 C and D), indicating that the process of l-serine synthesis was affected. Exogenous l-serine may replenish the cellular l-serine pool, inhibit the 2-KG reduction reaction catalyzed by SerA (SI Appendix, Fig. S6D), reduce d-2-HG production and accompanied loss of 2-KG, and as a result, rescue the growth defect of the D2HGDH mutant. Exogenous 2-KG could only replenish the 2-KG pool and, therefore, only partly improve the growth of the D2HGDH mutant (SI Appendix, Fig. S10). l-Serine biosynthesis relies on 2-KG (a substrate for SerA) and l-glutamate (a substrate for SerC that is converted to 2-KG) (Fig. 4A). Although glutamate and 2-KG were not detected directly, it is worth noting that the levels of glutamate-related amino acids (ornithine, proline, putrescine, and pyroglutamate) were dramatically reduced in A1501-Δd2hgdh (SI Appendix, Fig. S8 A and C). The impaired back conversion of d-2-HG into 2-KG and then l-glutamate may result in a short supply of these substrates for l-serine biosynthesis in the D2HGDH mutant. Thus, D2HGDH is functionally tied to l-serine biosynthesis possibly through preventing the inhibitory effect of d-2-HG on SerA and maintaining sufficient pools of substrates (2-KG and l-glutamate) for the l-serine biosynthesis.
There is no physical association between SerA and D2HGDH (SI Appendix, Fig. S11), implying that 2-KG produced from d-2-HG during l-serine biosynthesis will get mixed with the cellular 2-KG pool. P. stutzeri A1501 could use d-2-HG as the sole carbon source for growth, further supporting that 2-KG produced by D2HGDH should enter major metabolic pathways to support d-2-HG–dependent growth and other 2-KG–related metabolic processes. Notably, SerA from E. coli (also some other strains marked in light red in Fig. 7C) shows a close relationship with SerA from P. stutzeri A1501 (Fig. 7C) and was reported to have the ability to produce 2-HG. However, there was no D2HGDH homolog in E. coli, suggesting the existence of other unknown enzymes involved in the d-2-HG catabolism.
A metabolic network is a complete set of metabolites, enzymes, reactions, and pathways cross-linked with each other. Glycolysis and TCA cycle are connected by pyruvate or phosphoenolpyruvate through pyruvate carboxylase or phosphoenolpyruvate carboxykinase, respectively (the black dotted box in Fig. 1) (1, 39–41). TCA cycle and amino acid synthesis are connected by 2-KG through glutamate dehydrogenase or transaminases (1, 3). In this study, the HG-KG interconversion integrates d-3-PG (generated by glycolysis), l-serine, 2-KG (a key intermediate of the TCA cycle), and l-glutamate (SI Appendix, Fig. S8A). The HG-KG interconversion works as a bridge that closely connects glycolysis, TCA cycle, and amino acid metabolism to achieve a more sophisticated metabolic network.
Considering that d-2-HG has been viewed as an abnormal metabolite, d-2-HG conversion to 2-KG seems to be a process of metabolite repair. Here, we report that d-2-HG production is not a meaningless side reaction but serves SerA to facilitate d-3-PG dehydrogenation. For the key but toxic metabolite, both its production and elimination are essential. This is somewhat different from the reported metabolite repair mechanism, in which repair enzymes catabolize the toxic products generated from side reactions (11, 12). The findings also offer insights into the mechanism by which an enzymatic reaction previously considered promiscuous or accidental actually plays a key role in metabolism.
In eukaryotic cells, SerA-catalyzed d-2-HG production from 2-KG originates in the cytosol while D2HGDH-catalyzed d-2-HG conversion to 2-KG occurs in the mitochondrial matrix. There are active carriers that facilitate the transport of various metabolic intermediates across the mitochondrial membrane. The cytoplasmic and mitochondrial pools of different metabolites have tight connections (42, 43). A recent report indicated that deficiency in SLC25A1, which encodes the mitochondrial citrate carrier, causes d-2-HG accumulation and d-2-HGA (44), implying the presence of d-2-HG transport across mitochondrial membrane. Thus, the reactions catalyzed by cytoplasmic SerA and mitochondrial D2HGDH may be coupled through the shuttle of 2-KG and d-2-HG between the cytosol and mitochondrial matrix.
Major structures of the HG-KG interconversion modules in Pseudomonas and humans are extremely similar. D2HGDH, ETF, or ETFQO mutations can block d-2-HG elimination and lead to increased levels of d-2-HG in patients with d-2-HGA and glutaric aciduria type II (4, 45). Elevated expression of SerA results in the increased metabolic flux originating from glucose into l-serine synthesis pathway, which is essential for the growth of certain types of cancer, such as breast cancer and melanoma (28, 29). We speculate that d-2-HG may also be overproduced, accumulating in these cells, and thereby promoting cancer.
Together with SerA, other enzymes such as PdxB, HOT, and WbpB also contribute to the production of d-2-HG. These enzymes seem to contain tightly bound NADH and may also catalyze thermodynamically unfavorable reactions (20–23, 46). For example, HOT is the key enzyme in the catabolism of 4-hydroxybutyrate (GHB). Knockdown of HOT reduces the accumulation of d-2-HG in certain d-2-HG–related cancer cells (47), indicating that HOT might also participate in d-2-HG production in vivo. Thus, there may be similar HG-KG interconversion modules involving HOT and other d-2-HG–producing enzymes (SI Appendix, Fig. S12). The reaction through which 2-KG is reduced to d-2-HG is coupled to facilitate these enzymes-catalyzed unfavorable but necessary metabolic conversions (metabolite A to B). This type of HG-KG interconversion (SI Appendix, Fig. S12) represents a unique biochemical mechanism enabling reactions in thermodynamically unfavorable directions.
In summary, we have uncovered that the coupling between SerA and D2HGDH sits in the hub of l-serine biosynthesis and establishes a linkage among intermediates in glycolysis, TCA cycle, and amino acid biosynthesis. This study also illustrates that core metabolic processes might work together in a more sophisticated manner than we thought, raising the significance of digging the ancient and classic metabolic network.
Materials and Methods
Primers, plasmids, and strains used in this study are listed in SI Appendix, Table S6. The full details for bacteria culture, enzyme preparation, cofactor or coenzyme analysis, gene manipulation, metabolome analysis, isothermal titration calorimetry, and the related analytical methods are described in SI Appendix, SI Materials and Methods.
Enzyme Assays.
The enzyme assay used to assess D2HGDH activity was performed by recording the reduction of DCIP spectrophotometrically at 600 nm in a reaction mixture containing 50 mM Tris⋅HCl buffer (pH 7.4), phenazine methosulfate (PMS; 200 μM), DCIP (generally 100 μM; for the study of ETF effect on D2HGDH kinetics, 200 μM of DCIP was used), and variable concentrations of substrate incubated at 37 °C. ε value of DCIP is 22 cm−1·mM−1. The double-reciprocal plot method was used to estimate the kinetic constants of D2HGDH. One unit (U) of enzyme activity was defined as the amount that reduces 1 μmol of DCIP per min. The reduction of 2-KG by SerA was assayed by measuring the absorbance of NADH at 340 nm in 50 mM Tris⋅HCl (pH 7.4) at 37 °C. One U of enzyme activity was defined as the amount that oxidizes 1 μmol of NADH per min (26). SerA-catalyzed d-3-PG oxidation activity was measured by monitoring NADH formation (340 nm) at 37 °C.
Analytical Methods.
The reaction mixtures mentioned above were analyzed by HPLC (Agilent 1100 series; Hewlett-Packard) using a refractive index detector, as described (48). Generally, a Bio-Rad Aminex HPX-87H column was used and the mobile phase (10 mM H2SO4) was pumped at 0.4 mL·min−1 (55 °C). For quantification of d-3-PG, 2-KG, and d-2-HG, 1 mL·L−1 of formic acid (≥88%; HPLC grade; Kermel) solution was used as the mobile phase to obtain good peak shape for HPLC or liquid chromatography-tandem mass spectrometry (LC-MS; negative electrospray ionization mode). Glucose and l-glutamate concentrations were determined with a bio-analyzer (SBA-40D; Shandong Academy of Sciences).
Identification of the Conformation of the SerA-Produced 2-HG.
An enzymatic method was used to identify the configuration of the SerA-produced 2-HG. l-2-hydroxyglutarate oxidase (LGO) from E. coli showed strong stereospecificity toward l-2-HG (49). We identified the homologous l-2-hydroxyglutarate oxidoreductase (LGOR) in P. stutzeri strain SDM-LAC (50) through a BLASTX search of the whole genome (AGSX01000142.1) with the known LGO sequence (YP_490875.1). Subsequently, the putative LGOR (31% identity with LGO) was expressed and purified (SI Appendix, Fig. S4F). To examine the substrate specificity, the reaction mixtures containing 50 mM Tris, 200 μM DCIP, 200 μM PMS, 5 mM substrate, and 0.045 mg·mL−1 LGOR were assayed at 37 °C. The obtained enzyme exclusively catalyzes l-2-HG using PMS and DCIP as electron acceptors (SI Appendix, Fig. S4G). Thus, the purified D2HGDH and LGOR were used to define the configuration of the SerA-produced 2-HG. The reaction mixtures (0.8 mL) contained 200 μM PMS, 75 μM DCIP, and 16 μg of D2HGDH or 1.6 μg of LGOR in 0.1 M phosphate buffer at 37 °C. To initiate the reactions, 40 µL of 1 mM authentic d-2-HG, 1 mM authentic l-2-HG, or SerA-produced 2-HG was added, and the reactions were monitored at 600 nm using a UV/visible spectrophotometer (Ultrospec 2100 Pro; Amersham Biosciences).
Quantification of d-2-HG.
To quantify intracellular d-2-HG, three separate cell cultures were sampled (15 mL) and then rapidly transferred to prechilled centrifuge tubes. The supernatant was removed after centrifugation at 3,000 × g and −9 °C for 9 min, before washing the pellet with prechilled PBS (5 mL). Intracellular d-2-HG was extracted using the hot ethanol method (17). Ethanol (1 mL) was added to resuspend the precipitate, and the suspension was boiled for 15 min in a closed tube. After the cell debris was removed by centrifugation at 12,000 × g and 4 °C for 5 min, the supernatant was dried in a vacuum centrifuge (60 °C). The dried residue was resuspended in distilled water, and the supernatant obtained after centrifugation, as described above, was stored at −80 °C until analysis. To measure extracellular d-2-HG, aliquots of the cultures (1 mL) were sampled, and the supernatants after centrifugation were saved for further analysis.
Quantitative analysis of d-2-HG was performed as described (51). The NAD+-dependent d-2-hydroxyglutarate dehydrogenase gene from Acidaminococcus fermentans (52) was synthesized and expressed, and then the produced protein was purified. Diaphorase from Clostridium kluyveri was purchased from Sigma–Aldrich. We assumed that the cell volume of a 1-mL Pseudomonas sample at an optical density (600 nm) of 1 was 3.5 μL (53). Then, the intracellular d-2-HG concentration was estimated by dividing the amount of d-2-HG in the sample by the total cell volume.
Bioinformatics.
The phylogenetic trees of SerA and D2HGDH were constructed from the alignment of multiple proteins, followed by neighbor-joining analysis using the MEGA software program (version 6.06) with bootstrap analysis for 1,000 replications (cutoff value: 50%). The resulting trees were processed by iTOL (54). The distributions of SerA and D2HGDH in bacteria were vigorously checked by searching the sequenced bacterial genomes from GenBank (updated until April 6, 2016) with the SerA and D2HGDH in P. stutzeri A1501 as the queries. Individual genomes showing query coverages more than 90%, E values lower than e−30, and maximum identity levels higher than 30% with the query protein, were selected and further evaluated.
Supplementary Material
Acknowledgments
We thank Dr. Haijun Liu at Washington University in St. Louis, Dr. Wei Xiong at National Renewable Energy Laboratory, and Dr. Luying Xun at Washington State University for their helpful discussion and assistance in preparing the manuscript and the editor and anonymous reviewers for their critical comments that resulted in a significantly improved manuscript. The work was supported by National Natural Science Foundation of China Grants 31470199, 31470164, 31270090 and Young Scholars Program of Shandong University Grant 2015WLJH25.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619034114/-/DCSupplemental.
References
- 1.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 2.Tarhonskaya H, et al. Non-enzymatic chemistry enables 2-hydroxyglutarate-mediated activation of 2-oxoglutarate oxygenases. Nat Commun. 2014;5:3423. doi: 10.1038/ncomms4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat Chem Biol. 2011;7:894–901. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS, Jakobs C. Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis. 2012;35:571–587. doi: 10.1007/s10545-012-9462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, et al. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc Natl Acad Sci USA. 2017;114:1353–1358. doi: 10.1073/pnas.1614102114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Intlekofer AM, et al. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol. 2017;13:494–500. doi: 10.1038/nchembio.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha SK, et al. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nat Chem Biol. 2013;9:72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 12.Van Schaftingen E, et al. Metabolite proofreading, a neglected aspect of intermediary metabolism. J Inherit Metab Dis. 2013;36:427–434. doi: 10.1007/s10545-012-9571-1. [DOI] [PubMed] [Google Scholar]
- 13.Lin AP, et al. D2HGDH regulates alpha-ketoglutarate levels and dioxygenase function by modulating IDH2. Nat Commun. 2015;6:7768. doi: 10.1038/ncomms8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engqvist M, Drincovich MF, Flügge UI, Maurino VG. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J Biol Chem. 2009;284:25026–25037. doi: 10.1074/jbc.M109.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker-Kettern J, et al. Saccharomyces cerevisiae forms D-2-Hydroxyglutarate and couples its degradation to D-lactate formation via a cytosolic transhydrogenase. J Biol Chem. 2016;291:6036–6058. doi: 10.1074/jbc.M115.704494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Environ Microbiol. 2010;12:1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- 17.Winder CL, et al. Global metabolic profiling of Escherichia coli cultures: An evaluation of methods for quenching and extraction of intracellular metabolites. Anal Chem. 2008;80:2939–2948. doi: 10.1021/ac7023409. [DOI] [PubMed] [Google Scholar]
- 18.Reitz MS, Rodwell VW. Alpha-hydroxyglutarate oxidoreductase of Pseudomonas putida. J Bacteriol. 1969;100:708–714. doi: 10.1128/jb.100.2.708-714.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebisuno T, Shigesada K, Katsuki H. D-alpha-Hydroxyglutarate dehydrogenase of Rhodospirillum rubrum. J Biochem. 1975;78:1321–1329. doi: 10.1093/oxfordjournals.jbchem.a131030. [DOI] [PubMed] [Google Scholar]
- 20.Kardon T, Noël G, Vertommen D, Schaftingen EV. Identification of the gene encoding hydroxyacid-oxoacid transhydrogenase, an enzyme that metabolizes 4-hydroxybutyrate. FEBS Lett. 2006;580:2347–2350. doi: 10.1016/j.febslet.2006.02.082. [DOI] [PubMed] [Google Scholar]
- 21.Lyon RC, et al. Enzymes involved in the metabolism of gamma-hydroxybutyrate in SH-SY5Y cells: Identification of an iron-dependent alcohol dehydrogenase ADHFe1. Chem Biol Interact. 2009;178:283–287. doi: 10.1016/j.cbi.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 22.Rudolph J, Kim J, Copley SD. Multiple turnovers of the nicotino-enzyme PdxB require α-keto acids as cosubstrates. Biochemistry. 2010;49:9249–9255. doi: 10.1021/bi101291d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin A, Imperiali B. Biosynthesis of UDP-GlcNAc(3NAc)A by WbpB, WbpE, and WbpD: Enzymes in the Wbp pathway responsible for O-antigen assembly in Pseudomonas aeruginosa PAO1. Biochemistry. 2009;48:5446–5455. doi: 10.1021/bi900186u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson LA, Barrow KD, Neilan BA. Characterization of the 2-hydroxy-acid dehydrogenase McyI, encoded within the microcystin biosynthesis gene cluster of Microcystis aeruginosa PCC7806. J Biol Chem. 2007;282:4681–4692. doi: 10.1074/jbc.M606986200. [DOI] [PubMed] [Google Scholar]
- 25.Fan J, et al. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate. ACS Chem Biol. 2015;10:510–516. doi: 10.1021/cb500683c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao G, Winkler ME. A novel alpha-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol. 1996;178:232–239. doi: 10.1128/jb.178.1.232-239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant GA. Contrasting catalytic and allosteric mechanisms for phosphoglycerate dehydrogenases. Arch Biochem Biophys. 2012;519:175–185. doi: 10.1016/j.abb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrill DK, McAlexander JC, Guynn RW. Equilibrium constants under physiological conditions for the reactions of D-3-phosphoglycerate dehydrogenase and L-phosphoserine aminotransferase. Arch Biochem Biophys. 1981;212:717–729. doi: 10.1016/0003-9861(81)90416-1. [DOI] [PubMed] [Google Scholar]
- 31.Yan Y, et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA. 2008;105:7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan Y, et al. The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA. 2016;113:E4348–E4356. doi: 10.1073/pnas.1604514113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winsor GL, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toogood HS, et al. Extensive domain motion and electron transfer in the human electron transferring flavoprotein·medium chain Acyl-CoA dehydrogenase complex. J Biol Chem. 2004;279:32904–32912. doi: 10.1074/jbc.M404884200. [DOI] [PubMed] [Google Scholar]
- 35.Guynn RW, Thames H. Equilibrium constants under physiological conditions for the reactions of L-phosphoserine phosphatase and pyrophosphate: L-serine phosphotransferase. Arch Biochem Biophys. 1982;215:514–523. doi: 10.1016/0003-9861(82)90110-2. [DOI] [PubMed] [Google Scholar]
- 36.Buckel W, Miller SL. Equilibrium constants of several reactions involved in the fermentation of glutamate. Eur J Biochem. 1987;164:565–569. doi: 10.1111/j.1432-1033.1987.tb11164.x. [DOI] [PubMed] [Google Scholar]
- 37.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JH, et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci USA. 2013;110:17017–17022. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kai Y, Matsumura H, Izui K. Phosphoenolpyruvate carboxylase: Three-dimensional structure and molecular mechanisms. Arch Biochem Biophys. 2003;414:170–179. doi: 10.1016/s0003-9861(03)00170-x. [DOI] [PubMed] [Google Scholar]
- 40.Jitrapakdee S, Vidal-Puig A, Wallace JC. Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell Mol Life Sci. 2006;63:843–854. doi: 10.1007/s00018-005-5410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pircher H, et al. Identification of FAH domain-containing protein 1 (FAHD1) as oxaloacetate decarboxylase. J Biol Chem. 2015;290:6755–6762. doi: 10.1074/jbc.M114.609305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raimundo N, Baysal BE, Shadel GS. Revisiting the TCA cycle: Signaling to tumor formation. Trends Mol Med. 2011;17:641–649. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engqvist MK, Eßer C, Maier A, Lercher MJ, Maurino VG. Mitochondrial 2-hydroxyglutarate metabolism. Mitochondrion. 2014;19:275–281. doi: 10.1016/j.mito.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Nota B, et al. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined D-2- and L-2-hydroxyglutaric aciduria. Am J Hum Genet. 2013;92:627–631. doi: 10.1016/j.ajhg.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe H, et al. Identification of the D-enantiomer of 2-hydroxyglutaric acid in glutaric aciduria type II. Clin Chim Acta. 1995;238:115–124. doi: 10.1016/0009-8981(95)06074-n. [DOI] [PubMed] [Google Scholar]
- 46.Bar-Even A, Flamholz A, Noor E, Milo R. Thermodynamic constraints shape the structure of carbon fixation pathways. Biochim Biophys Acta. 2012;1817:1646–1659. doi: 10.1016/j.bbabio.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao C, et al. Efficient production of 2-oxobutyrate from 2-hydroxybutyrate by using whole cells of Pseudomonas stutzeri strain SDM. Appl Environ Microbiol. 2010;76:1679–1682. doi: 10.1128/AEM.02470-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalliri E, Mulrooney SB, Hausinger RP. Identification of Escherichia coli YgaF as an L-2-hydroxyglutarate oxidase. J Bacteriol. 2008;190:3793–3798. doi: 10.1128/JB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang T, et al. Genome sequence of Pseudomonas stutzeri SDM-LAC, a typical strain for studying the molecular mechanism of lactate utilization. J Bacteriol. 2012;194:894–895. doi: 10.1128/JB.06478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balss J, et al. Enzymatic assay for quantitative analysis of (D)-2-hydroxyglutarate. Acta Neuropathol. 2012;124:883–891. doi: 10.1007/s00401-012-1060-y. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, et al. Development of a satisfactory and general continuous assay for aminotransferases by coupling with (R)-2-hydroxyglutarate dehydrogenase. Anal Biochem. 2012;431:127–131. doi: 10.1016/j.ab.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Volkmer B, Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.