Significance
Gastrointestinal symbionts of organisms are important in the breakdown of food for the host, particularly for herbivores requiring exogenous enzymes to digest complex polysaccharides in their diet. However, their role in the digestion of algae in marine piscine herbivores remains unresolved. Here, we show that the diversity of food sources available to herbivorous surgeonfishes is directly linked with the genetic makeup of their enteric microbiota. Importantly, the genomic blueprint of dominant enteric symbionts belonging to diverse Epulopiscium clades differs according to the host diet. Thus, the acquisition of a unique enteric microbiota specialized to their diets likely shapes the nutritional ecology of piscine herbivores, in turn facilitating the coexistence of a high diversity of marine species within coral reefs.
Keywords: piscine herbivores, marine algae, carbohydrases, giant enteric symbionts, Epulopiscium
Abstract
Herbivorous surgeonfishes are an ecologically successful group of reef fish that rely on marine algae as their principal food source. Here, we elucidated the significance of giant enteric symbionts colonizing these fishes regarding their roles in the digestive processes of hosts feeding predominantly on polysiphonous red algae and brown Turbinaria algae, which contain different polysaccharide constituents. Using metagenomics, single-cell genomics, and metatranscriptomic analyses, we provide evidence of metabolic diversification of enteric microbiota involved in the degradation of algal biomass in these fishes. The enteric microbiota is also phylogenetically and functionally simple relative to the complex lignocellulose-degrading microbiota of terrestrial herbivores. Over 90% of the enzymes for deconstructing algal polysaccharides emanate from members of a single bacterial lineage, “Candidatus Epulopiscium” and related giant bacteria. These symbionts lack cellulases but encode a distinctive and lineage-specific array of mostly intracellular carbohydrases concurrent with the unique and tractable dietary resources of their hosts. Importantly, enzymes initiating the breakdown of the abundant and complex algal polysaccharides also originate from these symbionts. These are also highly transcribed and peak according to the diel lifestyle of their host, further supporting their importance and host–symbiont cospeciation. Because of their distinctive genomic blueprint, we propose the classification of these giant bacteria into three candidate genera. Collectively, our findings show that the acquisition of metabolically distinct “Epulopiscium” symbionts in hosts feeding on compositionally varied algal diets is a key niche-partitioning driver in the nutritional ecology of herbivorous surgeonfishes.
Marine herbivorous fishes are an ecologically dominant force, structuring tropical reef ecosystem functions and relationships through their grazing activities (1–3). However, they are one of the least understood guilds of herbivores regarding the mechanisms by which they digest and assimilate algal biomass, especially in contrast to plant material assimilation by terrestrial vertebrate herbivores and insects (1, 4). In terrestrial herbivores, the community of microbes inhabiting an organism’s gut—the gut microbiota—is considered to play a significant role in the organism’s ability to digest plant matter. It is thought to be a major factor leading to the evolution of vertebrate and insect-based herbivory (1–3, 5) and constitutes a potentially critical component of diet-driven speciation in mammals (1, 4, 6). Contrary to the immense literature on the nutritional ecology and plant biomass assimilation by mammals, ruminants, and arthropods (4, 7, 8), mechanistic insights into the digestive physiology of marine herbivores, especially the roles of gastrointestinal microbes of piscine herbivores in gut digestive processes, remain largely uncharacterized.
Marine algae differ vastly from the complex and refractory biomass of vascular plants primarily due to the relatively lower cellulose contents in the cell walls (only up to 10% by dry weight) and structurally variable spectrum of polysaccharides (9–11). Algal polysaccharides also show significant differences in molecular mass, monosaccharide composition, configuration of sugar units, and glycosidic linkages, as well as the presence and distribution of functional groups (e.g., carrageenan, agar, alginate, and fucoidan) (11). Marine algae also produce distinct forms of storage polysaccharides such as laminaran and floridean starch and contain comparably lower ratios of structural polysaccharides to cell contents than terrestrial plants (1–3, 12). Furthermore, most algae lack or contain only small quantities of lignin (13), which renders cellulose from plants resistant to degradation by cellulases. Given these unique properties, metabolic pathways inherent to the gut microbiota of marine herbivores supporting the utilization of algal biomass likely diverge from those canonically present in terrestrial intestinal systems assimilating the refractory lignocellulose biomass. However, these mechanisms remain poorly studied in marine systems.
Here, we illuminate the metabolic potential intrinsic to the gut microbiome of two groups of herbivorous surgeonfishes that has allowed them to use algal biomass as their principal source of carbon and energy. We specifically chose the gastrointestinal tract of the family Acanthuridae (surgeonfishes, tangs, and unicornfishes) as our model because species within this family feed on different types of algae with varied chemical constituents (1, 4, 12, 14). Secondly, their intestinal microbiota is dominated by a single group of giant bacteria, the “Epulopiscium” clade (5, 15, 16). These bacteria are morphologically and phylogenetically diverse, sometimes longer than 600 µm, and are extraordinarily polyploidic, carrying as many as 600,000 genome copies per cell (17–20). Unlike other bacteria, they also form multiple intracellular offspring in a circadian reproductive cycle that is synchronized with the feeding behavior of their hosts (21, 22). However, very little is currently known about the metabolic traits of Epulopiscium and their potential roles in host digestive process.
The antiquity of herbivory in herbivorous surgeonfishes (23) and the coevolution of Epulopiscium and related giant bacteria with their hosts (4, 7, 8, 17, 18, 20) raise the possibility that these dominant symbionts are likely essential in the breakdown of marine algae. Accordingly, we used comparative metagenomics and the single-cell genomics approach (24) to evaluate the effect of host diet on the functional microbiome of herbivorous surgeonfishes and to assess potential niche adaptations of divergent Epulopiscium symbionts colonizing distinct hosts (11, 20). Finally, we elucidated their roles in the host digestive processes by profiling community transcripts of enzymes linked to the breakdown of the main algal polysaccharides over the course of a day using RNA-sequencing (RNA-seq). Ultimately, this study not only unravels the metabolic pathways and enzymes facilitating the efficient assimilation of marine algae in a natural bioreactor, but also provides a genetic resource for enzymes facilitating the production of algal-based biofuels.
Results and Discussion
Intestinal Deconstruction of Marine Algae.
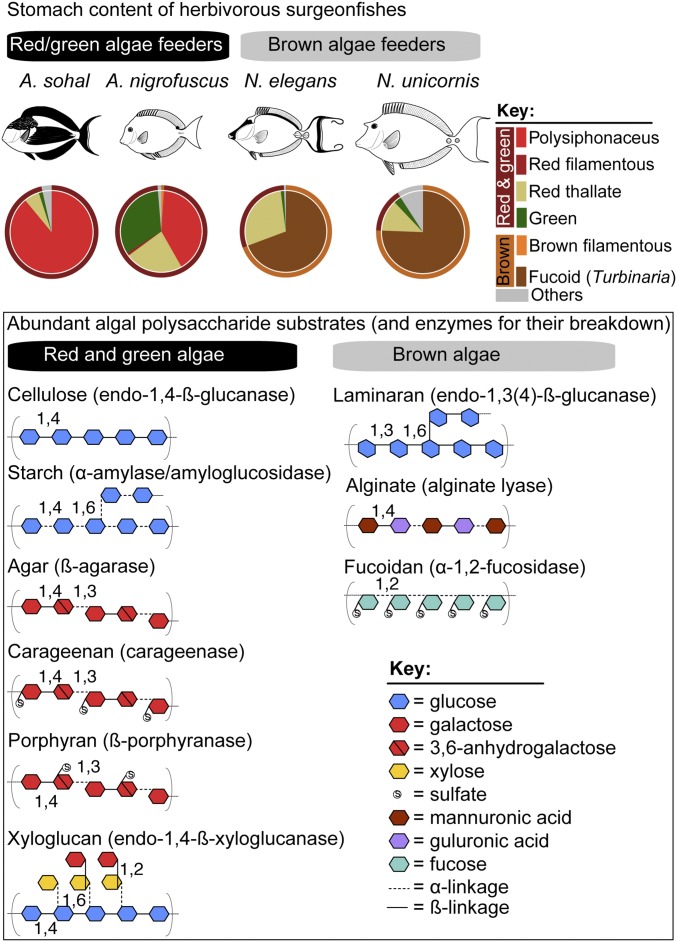
The emerging picture of the nutritional ecology of herbivorous fishes is that they consume and assimilate carbohydrate-rich diets and contain high levels of corresponding fermentation metabolites in their digestive tract (15). The surgeonfishes used in this study are reported to feed either on turfing and filamentous red and green algae [Acanthurus sohal and Acanthurus nigrofuscus (25)] or on macroscopic brown algae [Naso elegans and Naso unicornis (26, 27)]. A compilation of ingested algal species found in the stomach of these fishes from the literature (Fig. 1 and Dataset S1) shows that Acanthurus species are dominated by polysiphonous rhodophytes (18–89% of the stomach content) and several chlorophytes (Lyngbya, Cladophora, and Enteromorpha; ∼2–33% of the stomach content). In contrast, fucoid brown algae such as Turbinaria dominate in the Naso species (∼70–76% of the stomach content). Red algae, including the genus Polysiphonia, are generally characterized by high quantities of floridean starch and sulfated galactans (agar and carrageenan) in their cell walls, while most green algae usually contain starch as their food reserve and pectin as the main structural polysaccharide (28, 29). The major polysaccharides from Turbinaria and most brown algae include laminaran and fucoidan as storage glycans and alginate as the major structural polysaccharide (11, 30). Proximate chemical analyses and caloric estimates also suggest that green and red algae are nutritionally superior to brown algae, as reflected in their moderate to high levels of proteins and lipids [up to 30% (wt/wt) dry biomass], their possession of high levels of easily digestible carbohydrates (e.g., agar and starch), and their low content of noxious compounds (12, 31).
Fig. 1.
Stomach gut content of herbivorous surgeonfishes and structural information about major algal polysaccharides. (Upper) Algal constituents in the stomach of Acanthurus and Naso species compiled from literature data (Dataset S1). (Lower) Structural information about polysaccharides generally abundant in red, green, and brown seaweed. Images are not drawn to scale.
Given the variation in dietary constituents of the studied surgeonfish species (Fig. 1), we hypothesized that the structural heterogeneity of polysaccharides available to gut microbes creates functionally and ecologically discrete niches for the enteric microbiota. To this end, we constructed gut metagenomes from A. sohal, N. elegans, and N. unicornis, focusing on the midgut section (SI Appendix, Fig. S1) where the bolus of algae is usually located and the highest abundances of Epulopiscium were reported (17, 25). Initially, the lack of genomes representing diverse Epulopiscium clades in the nonredundant (NR) protein database (downloaded on 2015 June 1) hampered our ability to confidently assign metagenomic genes beyond those that were homologous to the reference draft genome of Epulopiscium species type B from Naso tonganus (32) that was present in the NR database (SI Appendix, Fig. S2A). Phylogenetic anchoring was greatly improved when we seeded the NR database with 14 Epulopiscium-like genomes generated in this study (see below)—in terms of overall protein alignment (88–95%), average amino acid identity (90–91%), and the corresponding high number of retrieved homologs (SI Appendix, Fig. S2 B–D). This was especially pivotal for the Naso species (improved by 21–60%), presumably because they harbor Epulopiscium type C and J clades that are highly divergent to the type B clade (20). Therefore, subsequent taxonomic assignments were done using the customized database, applying a BLAST score cutoff of 50 and alignment coverage of >80%. In the analyzed fishes, Epulopiscium-related sequences were found to dominate the prokaryotic coding genes (12–48% of all genes; SI Appendix, Fig. S2A), although this could be partially due to their extreme polyploidy (19). Nevertheless, the dominance of Epulopiscium is consistent with previous reports of their high abundance based on 16S rRNA gene surveys (16, 18). This is further corroborated by cell counts of Epulopiscium-like cells (57–82% of total cell counts) in DAPI-stained midgut contents of the same fishes (Dataset S2). However, the other prokaryotic cells are likely underrepresented in the cell counts due to a potential masking effect of significantly bigger (and brighter) Epulopiscium-like giant cells, which obscured DAPI signals from other smaller prokaryotes.
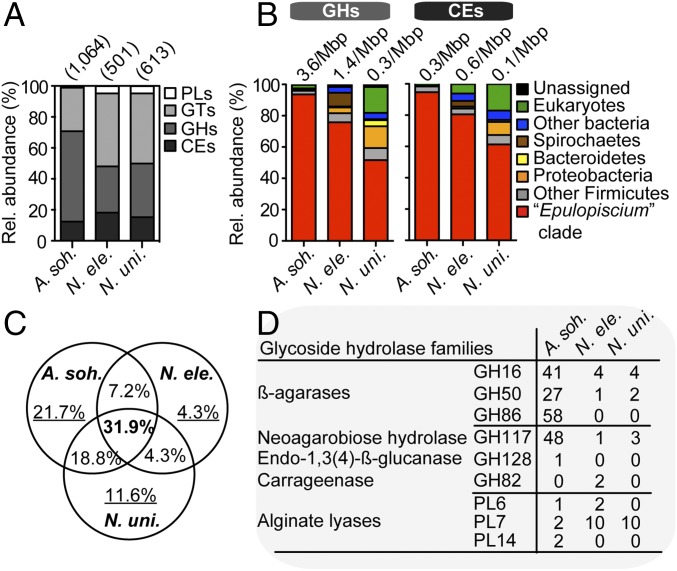
To gain further insight into the potential roles of the gut microbiota in algal biomass assimilation, we examined the distribution and putative taxonomic origin of carbohydrate active enzymes (CAZymes) among the different host species. Of the CAZymes directly involved in the digestion of polysaccharides, we observed that A. sohal had a relatively higher fraction of glycoside hydrolases (GHs) compared with the Naso species (Fig. 2A), which had proportionately more polysaccharide lyases (PLs) and carbohydrate esterases (CEs). Consequently, a much richer GH complement was found in A. sohal than in the Naso species (∼3.6 vs. 0.3–1.4 GHs/Mbp) (Fig. 2B); however, the densities of PLs (0.08 vs. 0.04–0.14 PLs/Mbp) and CEs (0.6 vs. 0.1–0.9 CEs/Mbp) were within the same range.
Fig. 2.
Distribution of CAZYmes in gut metagenomes. (A) The relative abundance of CAZyme families in the intestinal metagenomes of three surgeonfishes. Values in parentheses denote the total number of predicted CAZyme-encoding genes. (B) The putative taxonomic origin of GHs and CEs. Values above the bars denote size-scaled abundances in each metagenome. (C) The proportion of nonredundant GH families (from a total of 69) that are shared or unique to each metagenome. (D) Counts of GHs and PLs involved in the digestion of rhodophyte- and phaeophyte-specific polysaccharides in the three metagenomes.
Overall, 52–95% of CAZymes catalyzing the removal of ester substituents from glycan chains (by CEs) and the cleavage of glycosidic bonds between carbohydrates (by GHs) were predicted to originate from Epulopiscium (Fig. 2B). In the case of Naso species, other diverse microbes, including members of the phyla Proteobacteria, Spirochaetes, and Bacteroidetes, appear to be equally important. However, only 32% of the predicted GHs are shared among the three fish species (Fig. 2C), suggesting that their capacity to assimilate different algal diets require functionally distinct carbohydrases (Fig. 2D). Reflecting this is the high number of genes encoding putative β-agarases in A. sohal and alginate lyases in Naso species capable of breaking down agar (in red algae) and alginates (in brown algae), respectively. Collectively, these results support our hypothesis that the chemical variations in the host algal diet—that is, the differences between red/green and brown algae—likely drive the coevolution and intrinsic metabolic traits of the gut microbiota of herbivores surgeonfishes and, consequently, their nutritional ecology.
Phylogenomic Attributes of Epulopiscium.
The predominance and distinct profiles of Epulopiscium-like carbohydrases in the intestine of surgeonfishes feeding on contrasting algal diets warranted further investigation into the metabolic capacity of the multiple, phylogenetically divergent Epulopiscium symbionts colonizing their guts and their roles in host digestive processes. As “Candidatus (hereafter, “Ca.”) Epulopiscium fishelsoni” and related giant bacteria have not yet been cultivated, we used single-cell genomics (24) and a metagenome-resolved genome-binning approach (33) to reconstruct genomes of different Epulopiscium clades and infer their potential metabolism (see details in SI Appendix). Here, we first provide a general description of the reconstructed genomes.
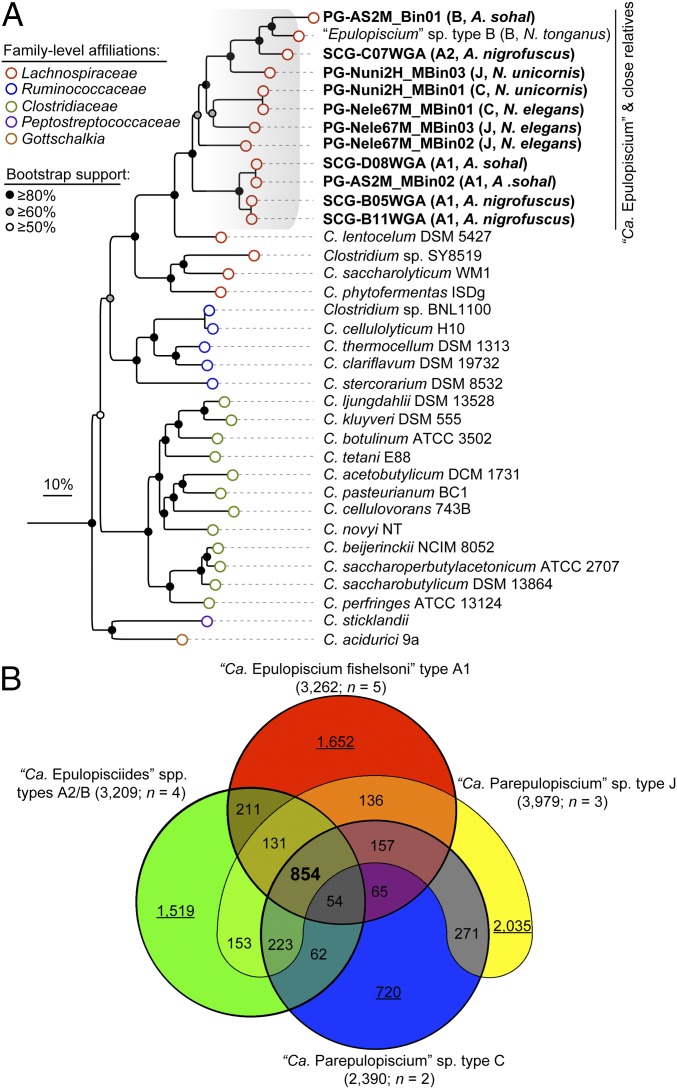
In total, seven single-cell amplified genomes (SAGs) and seven population genomes (PGs) were obtained from the midgut contents of four surgeonfishes commonly found in the Red Sea (A. sohal, A. nigrofuscus, N. elegans, and N. unicornis) (Table 1 and Dataset S3). Phylogenetic inference based on the 16S rRNA gene (SI Appendix, Fig. S3) and 16 concatenated conserved proteins (Fig. 3A and Dataset S4) classified these Epulopiscium genomes as belonging to four of the major Epulopiscium subclades placed within Clostridium cluster IV, which share common ancestry with Clostridium lentocellum (34). These draft genomes range in size from 688 kb to 3.4 Mbp, have a GC content of 29.3–34.5%, and encode 488–2,718 protein-encoding genes (Table 1). Overall, 62–82% of the coding genes were predicted to have a probable biological function. The generated genomes are estimated to have a completeness of ∼25–97% (mean of 76%) based on a profile of 202 marker genes conserved in Clostridia (Dataset S3), from which we infer that the maximum genome size of Epulopiscium is around 3.5 Mbp. These attributes together with the coding density (58–87%) imply that Epulopiscium clade members are typical Firmicutes, with features generally similar to other fully sequenced Clostridia (35).
Table 1.
Genomic attributes of Epulopiscium clade bacteria from Red Sea surgeonfishes affiliated with the genera Acanthurus and Naso
| Genome ID | Host (source) | Subclade* | Size, Mbp | Scaffolds | Completeness†, % | GC, % | Coverage‡, × | Coding, % | Protein-coding genes | |
| Counts | % with function | |||||||||
| SCG-B05WGA | A. nigrofuscus | Type A1 | 3.00 | 145 | 93.9 | 31.1 | 213 | 82 | 2,440 | 78 |
| SCG-B11WGA | A. nigrofuscus | Type A1 | 3.38 | 109 | 96.5 | 31.3 | 368 | 81 | 2,718 | 76 |
| SCG-D08WGA | A. sohal | Type A1 | 2.16 | 316 | 81.9 | 30.0 | 63 | 71 | 1,574 | 79 |
| SCG-C06WGA | A. nigrofuscus | Type A1 | 1.10 | 160 | 38.3 | 31.4 | 37 | 82 | 948 | 79 |
| PG-AS2M_MBin02 | A. sohal | Type A1 | 2.43 | 403 | 80.8 | 29.3 | 13 | 67 | 1,779 | 76 |
| SCG-C07WGA | A. nigrofuscus | Type A2 | 2.48 | 89 | 88.2 | 30.4 | 128 | 83 | 1,973 | 76 |
| SCG-B10WGA | A. nigrofuscus | Type A2 | 0.69 | 172 | 24.6 | 31.3 | 251 | 61 | 488 | 68 |
| PG-AS2M_MBin0 | A. sohal | Type B | 3.19 | 166 | 94.7 | 33.9 | 35 | 78 | 2,165 | 75 |
| SCG-B10WGA | A. nigrofuscus | Type B | 1.09 | 338 | 31.7 | 34.9 | 579 | 58 | 743 | 63 |
| PG-Nele67M_MBin01 | N. elegans | Type C | 1.88 | 242 | 79.0 | 34.1 | 69 | 78 | 1,522 | 76 |
| PG-Nele67M_MBin02 | N. elegans | Type J | 2.09 | 90 | 87.1 | 32.3 | 34 | 81 | 1,702 | 70 |
| PG-Nele67M_MBin03 | N. elegans | Type J | 2.88 | 229 | 84.9 | 34.5 | 31 | 82 | 2,475 | 62 |
| PG-Nuni2H_MBin01 | N. unicornis | Type C | 2.71 | 88 | 96.2 | 34.5 | 16 | 87 | 2,185 | 77 |
| PG-Nuni2H_MBin03 | N. unicornis | Type J | 1.85 | 221 | 84.6 | 32.5 | 10 | 85 | 1,656 | 82 |
Additional information is provided in Dataset S3. PG, population genome; SCG, single-cell genome.
Based on phylogenetic inference as shown in Fig. 3A and SI Appendix, Fig. S3.
Estimated using CheckM (64).
For population genomes, coverage is estimated based on the respective metagenomic data.
Fig. 3.
Phylogenomic relationship of Epulopiscium clade members. (A) Maximum likelihood phylogeny based on 16 conserved single-copy genes (Dataset S4). Values in parentheses show the clade affiliation and the host origin of each Epulopiscium-affiliated genome (more details are given in Table 1). Bootstrap support values ≥50% are indicated at nodes by dots. (B) Venn diagram inventorying the shared (in bold) and unique (underlined values) genetic repertoire of the four major Epulopiscium genotypes based on their pan-genomes. Values in parenthesis after the genotype name show the pan-genome size and the number of genomes in the respective genotype.
A comparison of the amino acid identity (AAI) of orthologous genes in these genomes indicates that they likely encompass organisms from distinct genera (SI Appendix, Fig. S4), assuming the operational AAI cutoff range of 66–72% for this taxonomic rank (36). Given their uncultivated status and their phylogenetically discrete clusters together with their inferred physiology (see below), we propose that these Epulopiscium-like bacteria belong to three candidate genera encompassing multiple species (SI Appendix, Fig. S4; see SI Appendix for a full description). These three genera and accommodating species are “Ca. Epulopiscium fishelsoni” type A1 (79–94% AAI; n = 5); “Ca. Epulopisciides gigas” type A2 and “Ca. Epulopisciides saccharus” type B (65–88% AAI; n = 4); and “Ca. Parepulopiscium” types C and J (64–98% AAI; n = 5). This provisional nomenclature is consistent with a recent suggestion (20). Unless otherwise mentioned, we henceforth refer to all genomes as belonging to the Epulopiscium clade when describing their overall properties and also relative to other Clostridia and use the term “genotypes” when describing the genomic inventory of a given genus in comparison with the others.
The pan-genomes of the proposed genera possess 2,390–3,370 nonredundant genes (Fig. 3B) with a relatively low fraction of conserved genes (14–55% for genomes ≥79% complete), suggesting differing metabolic capabilities. On the basis of Clusters of Orthologous Groups (COGs) functional categories, our analyses indicate that genes for energy production and conversion are overrepresented in Ca. Epulopiscium (one-way ANOVA, P < 0.01), whereas Ca. Parepulopiscium are significantly enriched with genes for replication, recombination and repair, posttranslational modifications, and protein turnover (P < 0.01) (SI Appendix, Fig. S5).
Epulopiscium Clade Members Encode Distinct Carbohydrases.
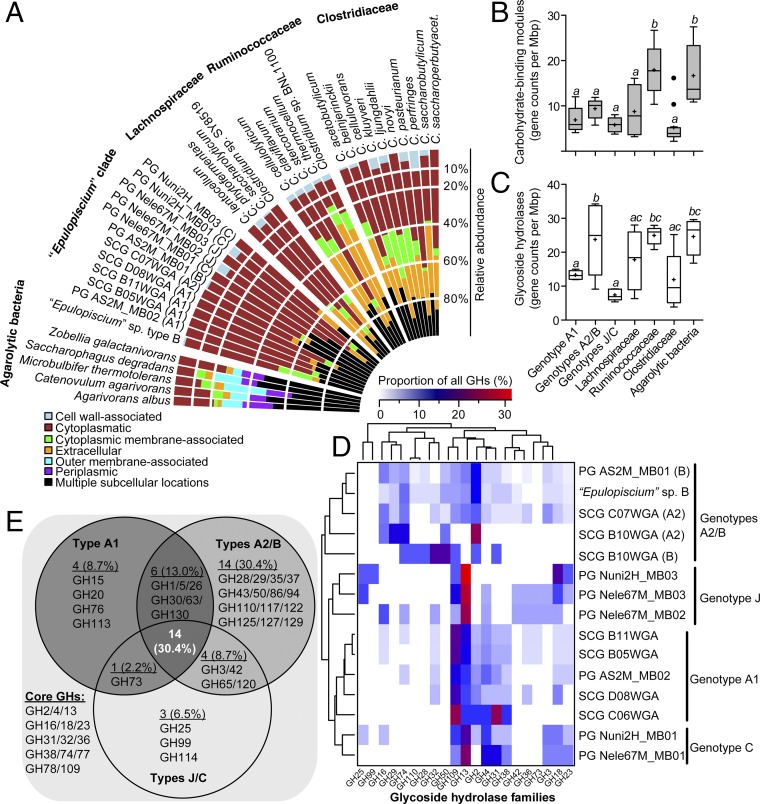
Gene-centric metagenomics analysis of intestinal carbohydrases from A. sohal and Naso species indicated the existence of distinct enzymes among the hosts, but the majority of them originate from Epulopiscium. Examination of the array of predicted GHs in Epulopiscium genomes showed that they are mostly intracellular (52–83% of all GHs) in comparison with those encoded in bacteria known to deconstruct algal or plant biomass (Fig. 4A). The latter possess many GHs secreted outside the cell (26–36%) or associated with the outer cell membrane (5–23%). The genomic blueprint of Epulopiscium also encodes significantly fewer carbohydrate-binding modules (CBMs) (approximately eight CBMs/Mbp; one-way ANOVA, P < 0.01) than that of free-living agarolytic bacteria or mammalian enteric Clostridia [i.e., Ruminococcaceae (8)] harboring ∼18 CBMs/Mbp (Fig. 4B). Furthermore, Epulopiscium clade genomes are also depleted in the enzymatic machinery enabling the degradation of cellulose; only families GH5 and GH74 (endoglucanases) and GH26 and GH28 (endohemicellulases) are detected (Datasets S5 and S6). Taken together, these findings imply that Epulopiscium clade members lack cellulosome-based systems, an adaptive trait that is consistent with the lower cellulose deposits of most algae (≤10% of their dry weight) compared with the 20–50% generally found in vascular plants (37). Interestingly, glycosyltransferases (GTs) that usually play a role in the synthesis of complex glycans are significantly depleted in genotypes A2/B (approximately five GTs/Mbp) compared with other Clostridia (∼10 GTs/Mbp; P < 0.01) (SI Appendix, Fig. S6). Moreover, polysaccharide lyases are also significantly depleted in Epulopiscium genomes compared with canonical agarolytic bacteria (P < 0.001) (SI Appendix, Fig. S6). However, this is largely driven by the lack of alginate lyases for processing alginate polysaccharides of brown algae in genotypes A1, A2, and B (see below), which dominate in hosts feeding on rhodophytes.
Fig. 4.
Distribution of CAZymes in Ca. Epulopiscium and related giant bacteria. (A) Predicted subcellular localization of GHs in Epulopiscium clade genomes in comparison with free-living agarolytic bacteria and other Clostridia. (B and C) Genome-size normalized counts of CBMs (B) and GHs (C) in different genotypes and in reference genomes. Different letters above the boxplots indicate statistically different groups as measured by one-way ANOVA (P < 0.05). The bottoms and tops of the boxes indicate the first and third interquartiles, respectively. The inside line and plus sign denote the median and mean counts, respectively. The whiskers are located at 1.5× the interquartile range above and below the box. (D) Genome-size scaled relative abundances of 21 most prevalent GHs (>1%) in Epulopiscium genomes. Dendrograms show hierarchical clustering of genotypes (Left) or GH (Top) based on the Bray–Curtis dissimilarity matrix using the average linkage method. (E) Venn diagram showing the inventory of GHs that are conserved, variable, or unique to the pan-genomes of Epulopiscium genotypes.
To gain more insight into the role of different Epulopiscium clade members that colonize distinct host species (20), we evaluated whether the divergent genotypes possessed a unique suite of GHs. Remarkably, the general trend in the distribution of carbohydrases in the Epulopiscium clade genomes (SI Appendix, Fig. S7) reflects that of the corresponding intestinal metagenomes (Fig. 1A). For instance, the least and highest GH-harboring genotypes, respectively, are the J/C types (abundant in Naso species) and the A2/B types (abundant in Acanthurus species). More specifically, the A2/B types encode significantly more GH complements (24 ± 11 GHs/Mbp) relative to the A1 (13 ± 1 GHs/Mbp) and J/C (7 ± 2 GHs/Mbp) genotypes (P < 0.05 and 0.001, respectively) (Fig. 4C) and also cluster separately from the others based on counts of GHs normalized by genome size (Fig. 4D), suggesting that this metabolic trait is taxonomically restricted. Reinforcing this is the fact that only 30% of the GHs in the pan-genomes of these three genotypes are conserved (Fig. 4E). These include mostly enzymes capable of cleaving oligosaccharides or the α-acetylgalactosaminyl moieties of glyconjugates, namely α-amylase (GH13), α-glucosidases/galactosidases (GH2, GH4, GH31, and GH36), endo-glucanase (GH74), chitinase (GH18), lysozyme (GH23), α-mannosidase (GH38), and α-N-acetlygalactosaminidase (GH109).
Importantly, the few putative GH-encoding genes in Epulopiscium genomes of mixed subcellular localization encompass those that are genotypically variable but phylogenetically closely related to enzymes that depolymerize the major polysaccharide constituents of rhodophytes (agar, porphyran, and carrageenan), chlorophytes (xyloglucan), and phaeophytes (alginate). Specifically, Ca. Epulopisciides saccharus type B and Ca. Epulopisciides gigas type A2 [abundant in Acanthurus species (20)], possess three to nine copies of β-agarases (in families 50 and 86) and β-porphyranases (in family 16). These enzymes are phylogenetically related to those catalyzing the hydrolysis of β-1,4 linkages in red seaweed glycans (SI Appendix, Fig. S8 and refs. 38–41). The multiple copies of such enzymes in these genotypes but their absence in Ca. Epulopiscium fishelsoni type A1—also colonizing Acanthurus species—and Ca. Parepulopiscium genotypes C and J from the Naso species implies niche partitioning in the symbiotic breakdown of agar polysaccharides. However, all three genotypes, as well as types C and J from the Naso species, are predicted to encode extracellular κ-carrageenases (in family 16) (SI Appendix, Fig. S8), which typically attack the β-1,4 linkage between d-galactose and 3,6-anhydro-d-galactose in κ-carrageenase (42). This implies that most of these symbionts are capable of handling this sulfated algal polysaccharide.
Additionally, Epulopiscium genotypes A2 and B from the Acanthurus species possess several putative endo-glucanases and endo-xyloglucanases (SI Appendix, Fig. S9) possibly involved in the degradation of xyloglucan, a quantitatively prominent component of chlorophycean green algae cell wall matrix (9). The deduced endoglucanase-like proteins show ∼30% amino acid sequence identity to characterized GH5 and GH74 enzymes from bacteria and eukaryotes but are phylogenetically distinguishable from these validated proteins (SI Appendix, Fig. S9). Moreover, although the predicted endo-xyloglucanases in GH74-II also appear to be secreted, they lack CBMs and many of the tryptophan residues (SI Appendix, Fig. S10) known to be essential for processive degradation by endo-processive xyloglucanases (43). These putative GH74-like xyloglucanases also lack an active site-blocking extra loop (G397–H400) (SI Appendix, Fig. S10) that is responsible for exo-activity in some GH74 xyloglucanases (44). Interestingly, endo-processive–type xyloglucanases can be altered to endo-dissociative types by mutation of these tryptophan residues (43). On the other hand, all the predicted GH5-like endo-glucanases in cluster GH5-D and GH5-E lack secretion signal motifs (SI Appendix, Fig. S9), suggesting that they possibly degrade oligosaccharides imported into the cytoplasm. Functional characterization is thus required to validate the activity of these (xylo)glucanases.
Interestingly, the genotypes A2 and B also encode the potential machinery for degrading fucan (SI Appendix, Fig. S11), which is surprising since these highly sulfated polymers are found mostly in the cell wall matrix of brown algae (10). The putative fucoidanases in these genotypes have sequence similarities of 30–40% to the enzyme from Fusarium graminearum (AFR68935), which has been suggested to play a role in plant pathogenesis (45). This enzyme shows preference toward α-1,2–linked fucosyl substrates rather than fucan containing α-1,3 (4) linkages as reported for other GH29 fucosidases. However, the predicted fucosidase-like enzymes in these genotypes lack a signal peptide characteristic of secreted proteins as well as CBMs found in some lineages of GH29 fucoidanases (SI Appendix, Fig. S11). Presumably, they play a role in the degradation of other fucosylated substrates, for instance those decorating xyloglucans (46) or mucin glycans (47). The lack of α-1,2-fucoidanase-like enzymes in Epulopiscium genomes from Naso species is intriguing, but the presence of fucosidase-like proteins (approximately seven) in the enteric metagenomes of Naso species that are homologous to other Clostridia (63% identity) suggests that other enteric microbes might be involved in the breakdown of brown seaweed fucans. However, all genotypes are predicted to encode enzymes involved in the assimilation of monomeric fucose, namely fucose isomerase, fuculose kinase, and fuculose aldolase.
All genotypes also possess the ability to assimilate mannitol, a sugar alcohol found mostly in brown algae (at up to 50% of their dry weight) (48) as a one-linked side-chain substituent of laminaran (10). Mannitol can be converted to fructose 6-phosphate by mannitol 1-phosphate dehydrogenase in a reversible reaction that assimilates it through glycolysis, which suggests that it may serve as an important fermentation substrate for Epulopiscium.
Consistent with the unique structural polysaccharides of brown algae, we found that Ca. Parepulopiscium species types C and J from the Naso species alone harbor alginate lyases, which canonically depolymerize alginates into oligomers and monomers (guluronate and mannuronate) through β-elimination reactions (49). These genotypes possess multiple copies of four alginate lyase families (PL7, 12, 15, and 17) that are phylogenetically closely affiliated to experimentally characterized enzymes, but only the PL7 family-encoded genes have a secretion signal (SI Appendix, Fig. S12). The putatively secreted PL7 alginate lyases presumably degrade alginate in brown algae endolytically into unsaturated oligo-alginates with various chain lengths (50), while the putatively cytoplasmatic oligoalginate lyases in families 15 and 17 likely degrade the produced oligosaccharides into monomers (49). Because alginate is found in high abundance in phaeophytes [varying in content from 20 to 47% of the dry cell weight (10)], and considering the paucity of endoglucanases and fucoidanases capable of hydrolyzing laminaran and fucoidan in these genotypes, it is likely to be a major nutrient source for the Naso species. As well as using alginates, these species appear also to preferentially mobilize algal peptides and proteins based on the significant enrichment of serine peptidases (P < 0.01) in their dominant symbionts (genotypes C and J) relative to the GH-rich A2/B genotypes (SI Appendix, Fig. S13 and Datasets S7 and S8). Note also that genotype A1 from the Acanthurus species is also significantly enriched with peptidases (Dataset S7) that are potentially nutritionally relevant, with the ability to nonspecifically cleave proteins into oligopeptides (e.g., families S01, S08, S09, and S33). Collectively, these results imply that the codiversification of distinct Epulopiscium clade members (20) with different hosts is partly due to their intrinsic ability to assimilate the major polysaccharide constituents of their host algal diets.
Carbohydrases from Epulopiscium Are Highly Transcribed.
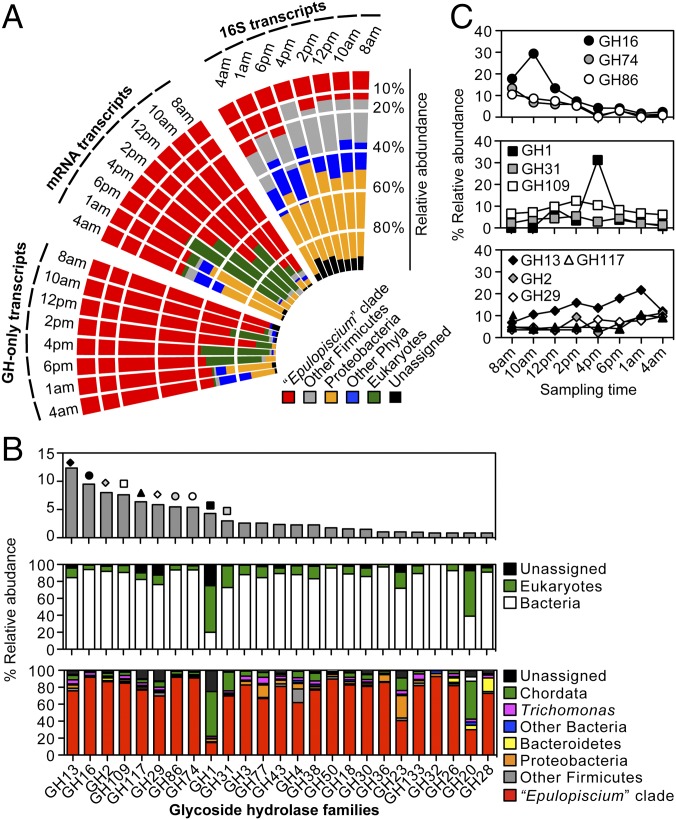
Recurring models of plant biomass digestion in wood-feeding insects and marine invertebrates implicate enzymes of both host and symbionts as mutualistic systems allowing efficient depolymerization of lignocellulose (4, 51). Because the gut microbiota is coupled tightly with the evolution of herbivory in vertebrates (5, 8), we reasoned that herbivorous surgeonfishes resemble other vertebrates in their inability to directly gain energy from ingested plant biomass (8, 52, 53). Hence, their enteric symbionts must contribute significantly to the turnover and assimilation of algal polysaccharides. To address this question, we conducted a metatranscriptomic analysis over a course of Epulopiscium’s life cycle (24 h) to evaluate the composition of active symbiotic communities and to simultaneously capture changes in host and symbiont gene expression in the midgut of A. sohal (Dataset S9). Samples were taken at eight time intervals (between 8 AM of one day and 4 AM of the next day) mirroring the active feeding and resting stages of the host (n = 3–5 replicates per time point) (Fig. 5A). Based on community 16S rRNA transcripts, the most active bacterial phyla were Proteobacteria (41 ± 10% SD) and Firmicutes (39 ± 9%), with Enterobacter (30 ± 9%) and Epulopiscium clade members (18 ± 12%) being the most dominant groups. Also, besides metazoan-associated 18S transcripts, those emanating from parabasalid flagellates were the second most abundant eukaryotes, corroborating previous microscopy-based observations (17). They increased in relative abundance at 8 AM and 10 AM (∼4–24%) (Dataset S9).
Fig. 5.
Metatranscriptomic community profile of the active enteric microbiota in A. sohal. (A) Polar histogram showing the temporal abundance of the Epulopiscium clade (relative to other microorganisms) based on transcripts of the 16S rRNA, mRNAs, and GHs. Values represent the means of three to five independent replicates per sampling point. (B) Ranked abundance of the 25 most prevalent GH transcripts (Top) as proportions of all predicted GH transcripts for all the time points and their putative taxonomic origin at the domain (Middle) and phylum (Bottom) levels. (C) The temporal relative abundance of the top 10 most prominent GH families. Symbols correspond to those depicted in B.
Complementary analyses based on nearly full-length mRNA transcripts (Dataset S10), which are likely to carry functionally relevant metabolic traits, showed that 20–99% of these transcripts are prokaryotic (Fig. 5A). The majority were bacteria (>99%), mainly Epulopiscium (61 ± 26%) and members of Proteobacteria (12 ± 17%), which starkly contrasts with the abundance based on 16S transcripts. Interestingly, the relative abundance of putative Epulopiscium mRNA transcripts fluctuates over the course of a day from 91% (8 AM) to 45% (4 AM), being replaced by transcripts from Eukaryotes (30–55% between 12 and 6 PM) and Proteobacteria (41 and 33% at 1 and 4 AM, respectively). Considering our sequencing approach—that is, sequencing host and symbiont mRNAs—the increase of eukaryotic transcripts toward midday is possibly the result of host-associated inputs as reflected by the high percentage of putative actinopterygian transcripts (88–99% of eukaryotic transcripts) (Dataset S11). The overall distributions of mRNA transcripts suggest that members of the Epulopiscium clade are the single most functionally active microorganisms in the midgut of A. sohal.
To specifically assess whether members of the Epulopiscium clade actively contribute in the turnover of algal biomass, we next examined the community profiles of GHs in the assembled metatranscriptomes as a proxy for the ability to convert algal polysaccharides to simple sugars. Concurrent with the above metagenomic analysis of A. sohal, our data illustrate that GH transcripts from the Epulopiscium clade are overall the most abundant (76 ± 22%) (Fig. 5A). These are significantly enriched between 8 AM and 2 PM (88 ± 11%; one-way ANOVA, P < 0.001) relative to those putatively originating from other bacteria (∼1%) or eukaryotes (∼8%). Moreover, an examination of the 25 most predominant GH families (with an abundance of ≥1%) further reveals that only four of these are substantially contributed by the host (i.e., Chordata) (Fig. 5B), including β-glucosidase (GH1; 53%), β-hexosaminidase (GH20; 45%), α-glucosidase (GH31; ∼22%), and lysozyme (GH23; 15%). Thus, with the additional exception of transcripts of GH families 4 (α-glucosidase), 28 (polygalacturonase), and 77 (amylomaltase), which are of diverse taxonomic origin, most transcripts of the abundant families (21 in total) are attributed to the Epulopiscium clade (with abundances of 67–92%; average of 82 ± 8%). These putatively Epulopiscium-associated GHs encode endo-β-1,4-glucanases (GH74), β-agarases (GH50 and GH86), and β-porphyranases/κ-carrageenases (GH16) that depolymerize complex polysaccharides of rhodophytes such as xyloglucan, agar, and carrageenans (28, 54). Unsurprisingly, these are also highly transcribed in the morning in contrast to transcripts of enzymes responsible for the hydrolysis of simpler oligosaccharides (families 1, 13, 31, and 117) or host-associated glycopeptides (families 29 and 109) that peak at midday, or became increasingly higher over the course of the day (Fig. 5C). This transcriptional pattern essentially shows that there is a correlation between the feeding lifestyle of A. sohal and the metabolic activities of its major symbionts, which supports the diel feeding hypothesis of marine piscine herbivorous (55)—that is, feeding activity is greater around midday when algal nutritional value is maximal. Analysis of Epulopiscium-specific 16S transcript abundances (SI Appendix, Fig. S14A) and differential expression of coding genes in the three most abundant genotypes (A1, A2, and B) (SI Appendix, Figs. S14 B and C and S15) reveal Ca. Epulopiscium saccharus type B cells as the most transcriptionally active subclade in the midgut of A. sohal. Collectively, the high transcriptional activity of Epulopiscium carbohydrases, along with their host-dependent coevolutionary patterns (20), suggests their substantial contribution in the digestion and assimilation of structural polysaccharides of marine algae in A. sohal.
Central Carbon and Energy Metabolism of the Epulopiscium Clade.
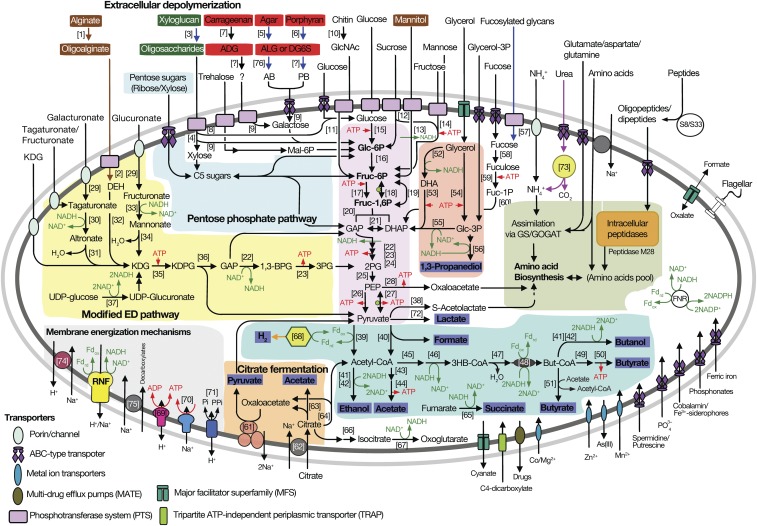
To complete the picture of how Epulopiscium transforms algal biomass into metabolites useful for the host, we reconstructed pathways of carbon and energy metabolism of these symbionts (Fig. 6 and SI Appendix, Fig. S16). For this purpose, we used all genomes with an estimated completeness above 85%, representing all symbiont subclades and host species (Table 1).
Fig. 6.
Metabolic potential of Ca. Epulopiscium and related giant bacteria. Thick arrows are color-coded to indicate pathways or genes found to be present in all genotypes (in black) or only in B and A2 (blue), C and J (brown), A1 only (orange), or in all except A1 (purple). The thin red and green arrows indicate mechanisms for energy transduction and for generating reducing equivalents, respectively. Algal polysaccharides are indicated by color-coded rectangles based on data in Fig. 1 representing phaeophytes (brown), chlorophytes (green), and rhodophytes (red), and fermentation products are highlighted with a blue rectangle. Numbers in parentheses refer to enzymes as follows: [1] alginate lyase; [2] oligoalginate lyase; [3] endo-xyloglucanase; [4] endo-glucanase; [5] β-agarase; [6] β-porphyranase; [7] κ-carrageenase; [8] β-glucosidase; [9] β-galactosidase; [10] chitinase; [11] N-acetylglucosaminidase; [12] sucrose-α-glucosidase; [13] mannitol-1-phosphate dehydrogenase; [14] fructose kinase; [15] hexose kinase; [16] phosphoglucose isomerase; [17] 6-phosphofructokinase; [18] pyrophosphate-dependent phosphofructokinase; [19] fructose-1,6-bisphosphatase; [20] fructose bisphosphate aldolase; [21] triosephosphate isomerase; [22] glyceraldehyde-3-phosphate dehydrogenase; [23] phosphoglycerate kinase; [24] phosphoglycerate kinase; [25] enolase; [26] pyruvate kinase; [27] pyruvate phosphate dikinase; [28] phosphoenolpyruvate carboxykinase; [29] uronate isomerase; [30] tagaturonate reductase; [31] altronate dehydratase; [32] 5-dehydro-4-deoxy-glucuronate reductase; [33] fructuronate dehydrogenase; [34] mannonate dehydratase; [35] 2-keto-3-deoxy-d-gluconate kinase; [36] 2-keto-3-deoxy-phosphogluconate aldolase; [37] UDP-glucose dehydrogenase; [38] acetolactate synthase; [39] pyruvate:ferredoxin oxidoreductase; [40] pyruvate formate lyase; [41] alcohol dehydrogenase; [42] aldehyde dehydrogenase; [43] phosphotransacetylase; [44] acetate kinase; [45] acetoacetyl-CoA transferase; [46] 3-hydroxybutyryl-CoA dehydrogenase; [47] 3-hydroxybutyryl-CoA dehydratase; [48] butyryl-CoA dehydrogenase; [49] phosphotransbutyrylase; [50] butyrate kinase; [51] butyrate:acetate-CoA transferase; [52] glycerol dehydrogenase; [53] dihydroxyacetone kinase; [54] glycerol kinase; [55] glycerol-3-phosphate dehydrogenase; [56] 1,3-propanediol dehydrogenase; [57] α-1,2-fucosidase; [58] fuculose isomerase; [59] fuculose kinase; [60] fuculose aldolase; [61] oxaloacetate decarboxylase; [62] citrate transporter; [63] citrate lyase; [64] citrate synthase; [65] fumarate reductase; [66] aconitase; [67] isocitrate dehydrogenase; [68] Fe-Fe hydrogenase; [69] V-ATPase; [70] F-ATPase; [71] P-PPase; [72] lactate dehydrogenase; [73] urease; [74] sodium:proton antiporter; [75] sodium-solute symporter; and [76] neoagarobiose hydrolase. Other abbreviations: AB, agarobiose; ADG, 3,6-anhydro-d-galactose; ALG, 3,6-anhydro-l-galactose; ATP, adenosine trisphosphate; But-CoA, butyryl-CoA; DEH, 5-dehydro-4-deoxy-glucuronate; DG6S, d-galactose-6-sulfate; DHAP, dihydroxyacetone phosphate; Fdox/Fdrd, ferredoxin, oxidized and reduced form; FNR, ferredoxin:NAD(P)+ oxidoreductases; Fruc-1,6P, fructose-1,6-bisphosphate; Fruc-6P, fructose-6-phosphate; GAP, glyceraladehyde-3-phosphate; Glc-3P, glycerol-3-phosphate; Glc-6P, glucose-6-phosphate; GlcNAc, N-acetylglucosamine; GOGAT, glutamine oxoglutarate aminotransferase; GS, glutamine synthetase; KDG, 2-keto-3-deoxygluconate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; Mal-6P, maltose-6-phosphate; NAD+/NADH, nicotineamide adenine dinucleotide, oxidized and reduced form; PB, porphyranobiose; PEP, phosphoenolpyruvate; Treh-6P, trehalose-6-phosphate; 1,3BPG, 1,3-bisphosphoglycerate; 2PG, 2-phosphoglycerate; 3HB-CoA, β-hydroxybutyryl-CoA; 3PG, 3-phosphoglycerate.
The metabolic reconstruction shows that sugars released from algal polysaccharides are transported into the cell and converted to pyruvate via Embden–Meyerhof–Parnas, Entner–Duodoroff, or pentose phosphate pathways. Pyruvate can be fermented to formate, acetate, butyrate, lactate, ethanol, butanol, and hydrogen. Glycerol, another compound released from the hydrolysis of storage glycans or membrane polypeptides, is fermented to 1,3-propanediol. ATP is generated by both substrate-level and electron transport phosphorylation. Electron transport phosphorylation solely involves a membrane-bound Rnf complex, which can generate an electrochemical potential (ΔμNa+) to drive ATP synthesis by a sodium-dependent F1F0-type ATP synthase (SI Appendix, Figs. S16B and S17). For glucose fermentation, the ATP yield is between 2 and 3.5 ATPs (SI Appendix, Fig. S16C).
Citrate can also be fermented, generating about 1.25 ATP per mole of citrate by substrate-level and electron transport phosphorylation. The combined action of a sodium-dependent citrate symporter and oxaloacetate decarboxylase likely generates an electrochemical potential to drive ATP synthesis (56) by electron transport phosphorylation (SI Appendix, Fig. S16D). Interestingly, the ability to use citrate is unique, as few Clostridia species have this symporter and decarboxylase (and citrate lyase) (SI Appendix, Fig. S18). In turn, the potential ability for Epulopiscium to generate ATP from this fermentation may help them reach their giant size.
In addition to the sodium-dependent ATPase, all genomes encode a H+-dependent V-type ATPase. This ATPase is rare in Clostridia (SI Appendix, Fig. S18) and in many bacteria (57). This ATPase typically operates in the direction of ATP hydrolysis (58) but can catalyze ATP synthesis in some bacteria (59). This latter possibility might be advantageous to Epulopiscium, considering the energetic challenges faced by giant single-celled prokaryotes (60). In summary, we deduce that all Epulopiscium clades are obligate anaerobic fermenters that produce energy from the fermentation of sugars. Fermentation of citrate and the possibility of generating ATP with V-type ATPase might support their energy-demanding size.
Conclusions
Fundamental insights related to the nutritional ecology of marine herbivorous fishes, particularly the roles of enteric symbionts in algal biomass assimilation and host digestive processes, have remained largely uncharacterized in comparison with terrestrial herbivores. Our investigation addressed this question in herbivorous surgeonfishes that principally feed on red and brown algae and specifically addressed the role of the enigmatic giant symbionts in the degradation and turnover of algal polysaccharides among different hosts. Based on the reconstructed metabolism of several nearly complete genomes encompassing the major Epulopiscium subclades, we elucidated the link between the host diet and the symbionts’ metabolism. Our findings illustrate that the capacity of Acanthurus species to digest and assimilate rhodophyte algae owes largely to their resident Epulopiscium genotypes, which are enriched with glycoside-hydrolyzing enzymes, in contrast to the genotypes found in Naso species that possess a greater capacity for degrading and assimilating proteins from brown algae. Given their inferred distinct physiologies, we propose that these organisms constitute at least three separate genera, whose ecological niche is defined by the host’s feeding behavior. Importantly, Epulopiscium clade members were transcriptionally the most active microbes in the midgut of A. sohal based on community metatranscriptomic analyses. Collectively, these findings provide the most comprehensive genomic and transcriptomic insights to date into the gut ecology of algivorous surgeonfishes, suggesting that a single prokaryotic lineage can contribute substantially to algal biomass assimilation, in contrast to the complexity of microbial players in gut ecosystems of terrestrial herbivores. In turn, the relative simplicity of the natural fermentative bioreactor displayed here and the underlying mechanism for assimilating different marine algal constituents illustrate fine-scale niche adaptations that help explain how key marine species partition themselves in an ecosystem as diverse as coral reefs. While the mechanisms sustaining herbivory in the marine realm remain an open question (see ref. 61), our work shows that the acquisition of a unique enteric microbiota is likely one of the mechanisms allowing piscine herbivores to evolve dietary specializations and corresponding niches, thus facilitating the coexistence of a high diversity of species within an ecosystem.
Methods Summary
Detailed description of materials and methods is provided in SI Appendix.
An adapted protocol was used for single-cell sorting, cell lysis, and DNA extraction from different Epulopiscium cells in the midgut of A. nigrofuscus and A. sohal. Genomic DNA was whole-genome amplified using the REPLI-g kit (Qiagen) and subjected to PCR analysis. The amplified DNA was sequenced using the Illumina HiSeq. 2000 platform followed by quality trimming and assembly using Trimmomatic (62) and SPAdes (63), respectively. In a second approach, PGs were obtained through a metagenome-resolved binning approach using intestinal microbiome sequences from A. sohal, N. elegans, and N. unicornis (Table 1). Genome binning was done using MetaBAT (33) complemented by the fidelity and validation workflows of CheckM (64). Automated annotations of SAGs and PGs were done using RAST (65) and PGAAP (66) followed by manual curation. CAZymes and the subcellular locations of GHs were predicted using dbCAN (67) and PSORTb (68), respectively. Temporal metatranscriptomic data were generated from the midgut of A. sohal based on Illumina’s RNA-Seq protocol to track the active gut community and evaluate gene-expression patterns of metabolically relevant genes in different Epulopiscium clades. Metatranscriptomes were assembled and processed into nearly full-length transcripts using Trinity (69) before taxonomic assignment by homology search against a customized National Center for Biotechnology (NCBI) NR protein database containing the 14 Epulopiscium genomes using DIAMOND (70) and the NCBI-based taxonomy (SI Appendix, Fig. S2). The putative taxonomic origin of 16S transcripts was deduced using MOTHUR’s implementation of the SILVA classification (71). Differential gene-expression analyses were conducted using DESeq2 (72) after mapping mRNA reads to coding genes of each genotype using bowtie2 (73). Additional details are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the King Abdullah University of Science and Technology (KAUST) Bioscience Core Lab, the Coastal and Marine Resources Core Lab, and T. Sinclair-Taylor for their technical and logistical support. We also thank John Howard Choat (James Cook University, Queensland) for his insights on surgeonfish nutrition, Andreas Brune (Max Planck Institute for Terrestrial Microbiology, Marburg) for assistance with bacterial nomenclature, and Calder J. Atta (KAUST) for the fish illustrations. This work was supported by KAUST through the Saudi Economic and Development Company Research Excellence Award Program (U.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The whole-genome sequences reported in this paper are deposited in DNA Data Bank of Japan/European Nucleoside Archive/GenBank under BioProject number PRJNA294498 [accession numbers LJDB00000000 and LJHC00000000–LJHH00000000 (for single-cell amplified genomes) and LNZM00000000–LNZR00000000, MDJM00000000, and MDJN00000000 (for population genomes)]. The assembled metagenomic sequences are deposited in the National Center for Biotechnology Information under BioProject number PRJNA338239 [accession numbers MDSO00000000 (Acanthurus sohal), MDSP00000000 (Naso elegans), and MDSQ00000000 (Naso unicornis)]; the raw metatranscriptomic sequence reads have been deposited in the Short Reads Archive (accession number SRP083815).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703070114/-/DCSupplemental.
References
- 1.Choat JH, Clements KD. Vertebrate herbivores in marine and terrestrial environments: A nutritional ecology perspective. Annu Rev Ecol Syst. 1998;29:375–403. [Google Scholar]
- 2.Floeter SR, Behrens MD, Ferreira CEL, Paddack MJ, Horn MH. Geographical gradients of marine herbivorous fishes: Patterns and processes. Mar Biol. 2005;147:1435–1447. [Google Scholar]
- 3.Clements KD, Raubenheimer D, Choat HJ. Nutritional ecology of marine herbivorous fishes: Ten years on. Funct Ecol. 2009;23:79–92. [Google Scholar]
- 4.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. Symbiosis as a general principle in eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6:a016113. doi: 10.1101/cshperspect.a016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 8.White BA, Lamed R, Bayer EA, Flint HJ. Biomass utilization by gut microbiomes. Annu Rev Microbiol. 2014;68:279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 9.Domozych DS. Algal cell walls. eLS. 2006 doi: 10.1002/9780470015902.a0000315.pub3. [DOI] [Google Scholar]
- 10.Rinaudo M. Seaweed polysaccharides. In: Kamerling JP, editor. Comprehensive Glycoscience from Chemistry to Systems Biology. Vol 2. Elsevier; London: 2007. pp. 691–735. [Google Scholar]
- 11.Stengel DB, Connan S, Popper ZA. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol Adv. 2011;29:483–501. doi: 10.1016/j.biotechadv.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery WL, Gerking SD. Marine macroalgae as foods for fishes–An evaluation of potential food quality. Environ Biol Fishes. 1980;5:143–153. [Google Scholar]
- 13.Martone PT, et al. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol. 2009;19:169–175. doi: 10.1016/j.cub.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Wei N, Quarterman J, Jin Y-S. Marine macroalgae: An untapped resource for producing fuels and chemicals. Trends Biotechnol. 2013;31:70–77. doi: 10.1016/j.tibtech.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Clements KD, Angert ER, Montgomery WL, Choat JH. Intestinal microbiota in fishes: What’s known and what’s not. Mol Ecol. 2014;23:1891–1898. doi: 10.1111/mec.12699. [DOI] [PubMed] [Google Scholar]
- 16.Miyake S, Ngugi DK, Stingl U. Diet strongly influences the gut microbiota of surgeonfishes. Mol Ecol. 2015;24:656–672. doi: 10.1111/mec.13050. [DOI] [PubMed] [Google Scholar]
- 17.Fishelson L, Montgomery WL, Myrberg AA. A unique symbiosis in the gut of tropical herbivorous surgeonfish (acanthuridae: teleostei) from the red sea. Science. 1985;229:49–51. doi: 10.1126/science.229.4708.49. [DOI] [PubMed] [Google Scholar]
- 18.Clements KD, Sutton DC, Choat JH. Occurrence and characteristics of unusual protistan symbionts from surgeonfishes (Acanthuridae) of the Great Barrier Reef, Australia. Mar Biol. 1989;102:403–412. [Google Scholar]
- 19.Mendell JE, Clements KD, Choat JH, Angert ER. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci USA. 2008;105:6730–6734. doi: 10.1073/pnas.0707522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake S, Ngugi DK, Stingl U. Phylogenetic diversity, distribution, and cophylogeny of giant bacteria (Epulopiscium) with their surgeonfish hosts in the Red Sea. Front Microbiol. 2016;7:285. doi: 10.3389/fmicb.2016.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angert ER, Clements KD. Initiation of intracellular offspring in Epulopiscium. Mol Microbiol. 2004;51:827–835. doi: 10.1046/j.1365-2958.2003.03869.x. [DOI] [PubMed] [Google Scholar]
- 22.Flint JF, Drzymalski D, Montgomery WL, Southam G, Angert ER. Nocturnal production of endospores in natural populations of epulopiscium-like surgeonfish symbionts. J Bacteriol. 2005;187:7460–7470. doi: 10.1128/JB.187.21.7460-7470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellwood DR, Goatley CHR, Brandl SJ, Bellwood O. Fifty million years of herbivory on coral reefs: Fossils, fish and functional innovations. Proc Biol Sci. 2014;281:20133046. doi: 10.1098/rspb.2013.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stepanauskas R. Single cell genomics: An individual look at microbes. Curr Opin Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery WL, Myrberg AA, Fishelson L. Feeding ecology of surgeonfishes (Acanthuridae) in the northern Red Sea, with particular reference to Acanthurus nigrofuscus (Forsskål) J Exp Mar Biol Ecol. 1989;132:179–207. [Google Scholar]
- 26.Crossman DJ, Choat JH. Nutritional ecology of nominally herbivorous fishes on coral reefs. Mar Ecol Prog Ser. 2005;296:129–142. [Google Scholar]
- 27.Choat HJ, Clements KD, Robbins WD. The trophic status of herbivorous fishes on coral reefs. Mar Biol. 2002;140:613–623. [Google Scholar]
- 28.Michel G, Nyval-Collen P, Barbeyron T, Czjzek M, Helbert W. Bioconversion of red seaweed galactans: A focus on bacterial agarases and carrageenases. Appl Microbiol Biotechnol. 2006;71:23–33. doi: 10.1007/s00253-006-0377-7. [DOI] [PubMed] [Google Scholar]
- 29.Prado HJ, Ciancia M, Matulewicz MC. Agarans from the red seaweed Polysiphonia nigrescens (Rhodomelaceae, Ceramiales) Carbohydr Res. 2008;343:711–718. doi: 10.1016/j.carres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Ermakova SP, et al. Structure, chemical and enzymatic modification, and anticancer activity of polysaccharides from the brown alga Turbinaria ornata. J Appl Phycol. 2015;28:2495–2505. [Google Scholar]
- 31.Hay ME, Fenical W. Marine plant-herbivore interactions: The ecology of chemical defense. Annu Rev Ecol Syst. 1988;19:111–145. [Google Scholar]
- 32.Miller DA, Suen G, Clements KD, Angert ER. The genomic basis for the evolution of a novel form of cellular reproduction in the bacterium Epulopiscium. BMC Genomics. 2012;13:265. doi: 10.1186/1471-2164-13-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins MD, et al. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 35.Zhou C, et al. New insights into Clostridia through comparative analyses of their 40 genomes. Bioenergy Res. 2014;7:1481–1492. [Google Scholar]
- 36.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Popper ZA, Tuohy MG. Beyond the green: Understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol. 2010;153:373–383. doi: 10.1104/pp.110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jam M, et al. The endo-beta-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: Two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem J. 2005;385:703–713. doi: 10.1042/BJ20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohta Y, et al. Cloning, expression, and characterization of a glycoside hydrolase family 86 β-agarase from a deep-sea Microbulbifer-like isolate. Appl Microbiol Biotechnol. 2004;66:266–275. doi: 10.1007/s00253-004-1757-5. [DOI] [PubMed] [Google Scholar]
- 40.Ekborg NA, et al. Genomic and proteomic analyses of the agarolytic system expressed by Saccharophagus degradans 2-40. Appl Environ Microbiol. 2006;72:3396–3405. doi: 10.1128/AEM.72.5.3396-3405.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hehemann J-H, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 42.Michel G, et al. The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of Clan-B glycoside hydrolases. Structure. 2001;9:513–525. doi: 10.1016/s0969-2126(01)00612-8. [DOI] [PubMed] [Google Scholar]
- 43.Matsuzawa T, Saito Y, Yaoi K. Key amino acid residues for the endo-processive activity of GH74 xyloglucanase. FEBS Lett. 2014;588:1731–1738. doi: 10.1016/j.febslet.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 44.Yaoi K, et al. The structural basis for the exo-mode of action in GH74 oligoxyloglucan reducing end-specific cellobiohydrolase. J Mol Biol. 2007;370:53–62. doi: 10.1016/j.jmb.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 45.Cao H, Walton JD, Brumm P, Phillips GN., Jr Structure and substrate specificity of a eukaryotic fucosidase from Fusarium graminearum. J Biol Chem. 2014;289:25624–25638. doi: 10.1074/jbc.M114.583286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrin RM, et al. Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol. 2003;132:768–778. doi: 10.1104/pp.102.016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White WL, Coveny AH, Robertson J, Clements KD. Utilisation of mannitol by temperate marine herbivorous fishes. J Exp Mar Biol Ecol. 2010;391:50–56. [Google Scholar]
- 49.Jagtap SS, Hehemann JH, Polz MF, Lee JK, Zhao H. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl Environ Microbiol. 2014;80:4207–4214. doi: 10.1128/AEM.01285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas F, et al. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem. 2013;288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davison A, Blaxter M. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol Biol Evol. 2005;22:1273–1284. doi: 10.1093/molbev/msi107. [DOI] [PubMed] [Google Scholar]
- 52.Lobel PS. Trophic biology of herbivorous reef fishes: Alimentary pH and digestive capabilities. J Fish Biol. 1981;19:365–397. [Google Scholar]
- 53.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi W-J, Chang Y-K, Hong S-K. Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol. 2012;94:917–930. doi: 10.1007/s00253-012-4023-2. [DOI] [PubMed] [Google Scholar]
- 55.Zemke-White L, Choat J, Clements K. A re-evaluation of the diel feeding hypothesis for marine herbivorous fishes. Mar Biol. 2002;141:571–579. [Google Scholar]
- 56.Schäfer G, Penefsky H. Bioenergetics: Energy Conservation and Conversion. Springer; Berlin: 2008. [PubMed] [Google Scholar]
- 57.Lolkema JS, Chaban Y, Boekema EJ. Subunit composition, structure, and distribution of bacterial V-type ATPases. J Bioenerg Biomembr. 2003;35:323–335. doi: 10.1023/a:1025776831494. [DOI] [PubMed] [Google Scholar]
- 58.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: Molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 59.Yokoyama K, et al. V-Type H+-ATPase/synthase from a thermophilic eubacterium, Thermus thermophilus. Subunit structure and operon. J Biol Chem. 2000;275:13955–13961. doi: 10.1074/jbc.275.18.13955. [DOI] [PubMed] [Google Scholar]
- 60.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 61.Steneck RS, Bellwood DR, Hay ME. Herbivory in the marine realm. Curr Biol. 2017;27:R484–R489. doi: 10.1016/j.cub.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aziz RK, et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Angiuoli SV, et al. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin Y, et al. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 71.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.