Significance
Mononuclear cells obtained from freshly isolated patient samples with various hematologic malignancies were evaluated for sensitivities to combinations of drugs that target specific cell-signaling pathways. The diagnostic, genetic/cytogenetic, and cellular features of the patient samples were correlated with effective drug combinations. For myeloid-derived tumors, such as acute myeloid leukemia, several combinations of targeted agents that include a kinase inhibitor and venetoclax, a selective inhibitor of BCL2, are effective.
Keywords: targeted therapies, drug combinations, ex vivo assay, hematologic malignancies
Abstract
Translating the genetic and epigenetic heterogeneity underlying human cancers into therapeutic strategies is an ongoing challenge. Large-scale sequencing efforts have uncovered a spectrum of mutations in many hematologic malignancies, including acute myeloid leukemia (AML), suggesting that combinations of agents will be required to treat these diseases effectively. Combinatorial approaches will also be critical for combating the emergence of genetically heterogeneous subclones, rescue signals in the microenvironment, and tumor-intrinsic feedback pathways that all contribute to disease relapse. To identify novel and effective drug combinations, we performed ex vivo sensitivity profiling of 122 primary patient samples from a variety of hematologic malignancies against a panel of 48 drug combinations. The combinations were designed as drug pairs that target nonoverlapping biological pathways and comprise drugs from different classes, preferably with Food and Drug Administration approval. A combination ratio (CR) was derived for each drug pair, and CRs were evaluated with respect to diagnostic categories as well as against genetic, cytogenetic, and cellular phenotypes of specimens from the two largest disease categories: AML and chronic lymphocytic leukemia (CLL). Nearly all tested combinations involving a BCL2 inhibitor showed additional benefit in patients with myeloid malignancies, whereas select combinations involving PI3K, CSF1R, or bromodomain inhibitors showed preferential benefit in lymphoid malignancies. Expanded analyses of patients with AML and CLL revealed specific patterns of ex vivo drug combination efficacy that were associated with select genetic, cytogenetic, and phenotypic disease subsets, warranting further evaluation. These findings highlight the heuristic value of an integrated functional genomic approach to the identification of novel treatment strategies for hematologic malignancies.
The promise of precision medicine is the ability to align medical interventions with individual patients at the time of diagnosis and to alter treatment regimens as new mutations arise and responses diminish. Although technical developments in next-generation sequencing and computational biology have accelerated progress in precision medicine, fundamental challenges remain. Whole-genome and whole-exome sequencing technologies can identify many target mutations, but these techniques are analytically intensive and may not reliably detect translocations, zygosity changes, and low-allele burden mutations with clinical significance. A further hindrance to the clinical utility of mutation status is a lack of drug therapies that selectively target cancer-associated mutations, with effective Food and Drug Administration (FDA)-approved drugs existing for only a subset of the genes currently known to underlie tumorigenesis (1). These limitations invite a complementary strategy that assesses drug sensitivities obtained with targeted agents designed to inhibit discrete cellular processes as a means of identifying phenotypic indications for specific cancers (2). Associating phenotypic responses with particular genetic alterations may then beget precision-based therapies.
For blood cancers, the success with targeted therapies developed to inhibit BCR-ABL fusions that drive chronic myelogenous leukemia has spurred efforts to identify similar therapies for other hematopoietic malignancy types. Acute myeloid leukemia (AML), which results from the enhanced proliferation and impaired differentiation of hematopoietic stem and progenitor cells, has substantial underlying heterogeneity, a feature that indicates the need for multiple targeted therapies or the use of combinations.
AML diagnosis relies on cytogenetic analysis, as recurrent chromosomal variations represent established prognostic markers, even though nearly half of patients with AML have a normal karyotype.
DNA sequencing of 200 patients with AML uncovered an average of 13 somatic mutations in each genome, of which 5 mutations were recurrent (3). Common mutations in AML that are also driver mutations represent potential therapeutic targets. Recurrent mutations in transcription factors and epigenetic regulators identified in AML suggest that aberrant transcriptional circuits are a common feature underlying leukemogenesis (3, 4). These circuits may drive oncogenic gene expression programs to alter differentiation, activate self-renewal, and generate leukemia stem cells responsible for the initiation and propagation of disease (5–7). Of the many genes commonly mutated in AML, targeted therapies in clinical trials or clinical use have been developed for only five: PML-RARA, FLT3, KIT, IDH1, and IDH2.
The standard treatment for AML is chemotherapy consisting of cytarabine and anthracyclines, which is more effective in adults younger than 60 y of age; however, the overall survival rate 5 y after diagnosis is 25% (8). The outcome in older patients, who represent the majority of patients with this disease and are unable to receive intensive chemotherapy, is poor, with a median survival of 5–10 mo (9). It is also noteworthy that a significant proportion of older patients with AML do not receive any antileukemic therapy (10). A key exception is the subset of patients with AML with acute promyelocytic leukemia, for which the use of all-trans retinoic acid therapy results in excellent and durable responses, suggesting the potential value of targeted therapies for other AML subgroups (11, 12). Recent advancements in understanding of the molecular pathogenesis of AML have resulted in a growing number of molecularly targeted drug candidates. However, several factors hinder the development of effective single-agent targeted treatments, including the intratumoral heterogeneity of hematologic malignancies, the emergence of genetically heterogeneous subclones leading to relapse, and rescue signals from the tumor microenvironment. Attempts to develop small-molecule inhibitors of the tyrosine kinase FLT3, in which activating mutations are detected in approximately 30% of adult AML cases (13, 14), illuminate the difficulty for effective single-agent targeted therapies. The short duration of response to FLT3 inhibitors is largely attributable to the rapid selection for and expansion of drug-resistant subclones (15–17). Targeted drugs may yet improve treatment outcomes. However, it may be difficult for these compounds, if used as single agents, to produce durable remissions necessary for long-term disease management or “bridging” the patient to successful bone marrow transplantation therapy, the only current potential for cure. Combinations that modulate distinct pathways may provide an opportunity for improved responses (18). For example, the combination of an MEK inhibitor (trametinib) with an RAF inhibitor (dabrafenib) is now an approved therapy for BRAF mutation-positive metastatic melanoma (19). A similarly attractive alternative strategy for AML, supported by emerging data, is the use of molecularly guided drug combinations, such as quizartinib and azacitidine, which inhibit FLT3 and DNA methyltransferase activities, respectively (20).
In the absence of a comprehensive portfolio of therapeutic drugs targeting specific mutations, we used ex vivo functional screening to identify drug sensitivities in primary samples from patients with various hematologic malignancies. Based on data accumulated from this assay to date, many instances of ex vivo sensitivity to small-molecule kinase inhibitors have been validated against known genetic targets (e.g., BCR-ABL, FLT3-ITD, RAS), and many novel drug/mutation associations have been discovered (21–24). These data suggest that a similar screening platform may identify combinations of targeted agents that are more effective than either of their respective single agents, thus defining and enabling a rational program for selecting clinically relevant combinatorial therapies. Thus, to identify new therapeutic combinations for AML and other hematologic malignancies, we assessed the sensitivity of primary patient samples to various drug combinations by using this ex vivo functional platform.
Results
Freshly isolated primary mononuclear cells from patients with various hematologic malignancies (N = 122) were cultured in the presence of a panel of 48 drug combinations, each in a fixed molar dose series. The drug combinations were designed as pairs of inhibitors that target nonoverlapping biological pathways, comprising different classes of compounds, including kinase inhibitors, bromodomain inhibitors, BH3 mimetics, and histone deacetylase (HDAC) inhibitors. To maximize the translational impact of any findings, combinations used FDA-approved drugs if possible. For comparison, cells were also tested against graded concentrations of each inhibitor alone, and sensitivity was assessed by a methanethiosulfonate (MTS)-based viability assay after 3 d. The efficacy of each combination relative to its respective single agents was quantified with combination ratio (CR) values, defined as the IC50 or area under the fitted dose–response curve for the combination divided by the lowest IC50 or area under the curve (AUC) value for either single agent. By this metric, a CR value of less than 1 indicates the drug combination is more effective than either single agent. We derived these CR values because of known limitations of applying conventional synergy calculations when one or more of the single agents is completely ineffective on particular samples (25).
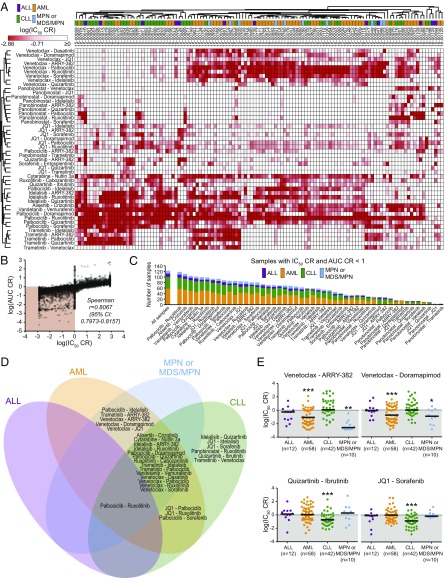
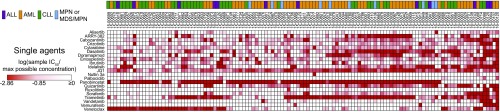
Patients were classified according to four general diagnostic groups: AML, chronic lymphocytic leukemia (CLL), acute lymphoblastic leukemia (ALL), and myeloproliferative neoplasms (MPNs) or myelodysplastic syndromes (MPNs; Table 1 and Dataset S1). Unsupervised hierarchical clustering of CR values for each drug combination revealed several distinct patterns of efficacy (Fig. 1A). Myeloid leukemia patient samples were enriched within a cluster of sensitivity to combinations pairing the BCL2 inhibitor venetoclax with select kinase inhibitors [dasatinib (multikinase), doramapimod (p38), sorafenib (multikinase), or idelalisib (PI3KCD)]. This clustering pattern segregated the majority of myeloid vs. lymphoid samples. Within each of these larger myeloid and lymphoid clusters, subsets of samples showed sensitivity to combinations involving the MEK inhibitor trametinib and a second kinase inhibitor [idelalisib, palbociclib (CDK4/6), or quizartinib (FLT3/CSF1R)]. In addition, a small cluster of patients with leukemia showed sensitivity to combinations of the HDAC inhibitor panobinostat in tandem with the JAK inhibitor ruxolitinib and/or the multikinase inhibitor sorafenib. To confirm that these clusters result from the effectiveness of the combination rather than that of a single agent, IC50 values for each single agent were mapped according to the combination cluster pattern. Importantly, apart from venetoclax, which, as a single agent, demonstrated potent and selective efficacy in lymphoid (predominantly CLL) patient samples, single-agent efficacies did not align uniquely to a combination efficacy-derived myeloid or lymphoid cluster (Fig. S1). This pattern of venetoclax sensitivity is consistent with its recent approval for treatment of patients with CLL with 17p deletions (26).
Table 1.
Summary of select patient characteristics for surveyed sample cohort
| Characteristic | All patients (n = 122) | ALL (n = 12) | AML (n = 58) | CLL (n = 42) | MPN or MDS/MPN (n = 10) |
| Sex (n evaluable) | n = 116 | n = 12 | n = 56 | n = 42 | n = 6 |
| Female | 59 | 8 | 30 | 20 | 1 |
| Male | 57 | 4 | 26 | 22 | 5 |
| Age, y (n evaluable) | n = 106 | n = 9 | n = 54 | n = 37 | n = 6 |
| Median | 62.1 | 29.7 | 53.7 | 65.9 | 60.7 |
| Range | 5.5–83.8 | 5.5–70.2 | 8.2–83.8 | 44.7–79.4 | 35.2–74.9 |
| Sample type (n evaluable) | n = 122 | n = 12 | n = 58 | n = 42 | n = 10 |
| Peripheral blood | 86 | 7 | 31 | 41 | 7 |
| Bone marrow aspirate | 34 | 5 | 25 | 1 | 3 |
| Leukapheresis | 2 | 0 | 2 | 0 | 0 |
| WBC count (n evaluable) | n = 115 | n = 9 | n = 55 | n = 42 | n = 9 |
| Median, ×1,000/µL | 42.9 | 60.0 | 34.6 | 45.8 | 92.7 |
| Range, ×1,000/µL | 0.5–309 | 10.3–169 | 0.5–240 | 5.8–309 | 26.3–240 |
In total, primary samples from 122 patients with a variety of hematologic malignancies were screened for ex vivo sensitivity to inhibitor combinations.
Fig. 1.
Differential patterns of selective efficacy of small-molecule combinations relative to single agents. (A) Unsupervised hierarchical clustering of IC50 CR values for 122 patients with leukemia across 48 tested combinations. IC50 CR values were log-transformed and row- and column-clustered using Pearson correlation pairwise average linkage method. Darker red color (lower CR values) indicates drug combinations exhibiting higher efficacy than either single agent alone. Diagnostic category annotation of each sample is also shown. (B) Correlation of IC50 CR and AUC CR effect measure values. Shaded region indicates sample/drug pairs for which the combination was more effective than either single agent (CR < 1) by both effect measures. (C) Distribution of effective drug combinations based on frequency and diagnostic category. Bar labeled “all samples” indicates diagnosis group breakdown for all 122 surveyed patient samples for comparison. (D) Overlap of significantly effective inhibitor combinations among four general diagnosis subgroups. Combinations for which median IC50 CR and AUC CR were significantly <1 within each diagnostic subgroup are shown; combinations listed in overlapping regions were significantly effective in each subgroup. (E) Select examples of combinations by diagnostic subgroup. Scatter plots of log-transformed IC50 CR values for select combinations. Black horizontal bars represent median CR. (*Median CR significantly <1 in that subgroup.)
Fig. S1.
Single-agent IC50 heat map for all 122 evaluated leukemia patient samples. IC50 values for 122 patients with leukemia across each of the 21 individually tested small-molecule inhibitors were normalized to the maximum tested concentration (1 µM for dasatinib, 10 µM for all other inhibitors) and log-transformed. Patient samples are represented in columns in the identical clustered order as identified from the corresponding combination data in Fig. 1A. Darker red color indicates increased sensitivity, and the diagnostic category annotation of each sample is shown.
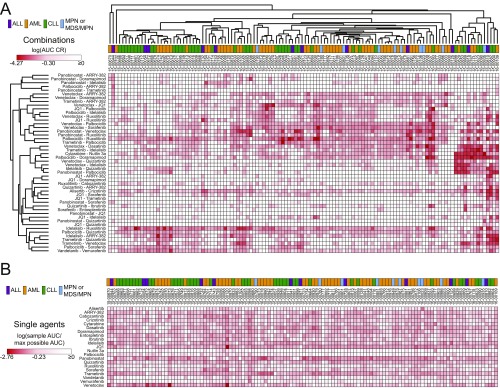
To enable comparisons among different measures of combination efficacy, AUC values were also calculated for each sample/drug pair, and IC50 and AUC values are highly correlated (Spearman ρ = 0.806; Fig. 1B). Clustering of AUC CR values yielded similar sensitivity clusters as those seen with IC50 CR results (Fig. S2). In addition, there was a broad distribution in the frequency and type of samples demonstrating combination efficacy. For instance, the palbociclib/ruxolitinib, alisertib/crizotinib, vandetanib/vemurafenib, and palbociclib/quizartinib combinations were effective, according to both CR measures, in more than 100 of 122 patient samples surveyed, encompassing all four diagnosis categories. Furthermore, the HDAC inhibitor panobinostat as a single agent showed potency across most of the patient samples tested (Fig. S3). To relate these findings with other definitions of synergy, IC50 and AUC CR effect measures for a subset of combinations and samples were compared with excess over Bliss (EOB) determinations (27). A high level of agreement was observed between the two methods (Spearman r values for IC50 CR or AUC CR vs. EOB, 0.953 and 0.928, respectively; P < 0.0001).
Fig. S2.
Clustering of AUC-based effect measures of combination and single-agent efficacy for all 122 evaluated leukemia patient samples. (A) Unsupervised hierarchical clustering of AUC CR values for 122 patients with leukemia across 48 tested combinations. AUC CR values (defined as the ratio of the combination AUC to that of the smallest AUC of either drug alone) were log-transformed and row- and column-clustered by using a Pearson correlation pairwise average linkage method. Darker red color indicates lower CR values, and diagnostic category annotation of each sample is shown. (B) Single-agent AUC heat map for all 122 evaluated leukemia patient samples. AUC values for all samples across each of the 21 individually tested small-molecule inhibitors were normalized to the maximum possible AUC value of 286.27 and log-transformed. Patient samples are represented in columns in the identical clustered order as identified from the corresponding combination data in A. Darker red color indicates increased sensitivity, and the diagnostic category annotation of each sample is shown.
Fig. S3.
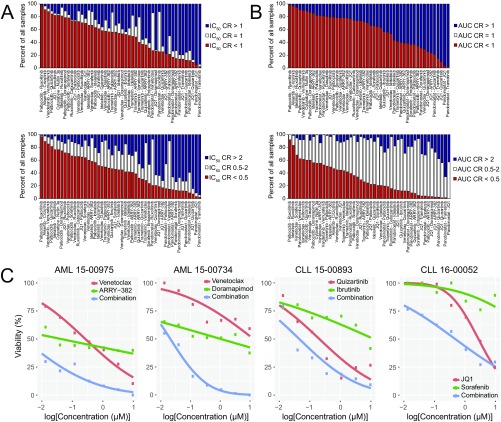
Distribution of IC50 and AUC CRs across all tested patient samples. For each combination tested, frequency of samples with CR values are shown for IC50 (A) and AUC (B) effect measures. Two different sets of cut points were examined: CR > 1/CR = 1/CR < 1 (Upper) or CR > 2/CR = 0.5–2/CR < 0.5 (Lower). Combinations are sorted according to frequency of samples with CR values <1 or <0.5, respectively. (C) Representative dose–response curves for patients with AML or CLL exhibiting CR values <1 for select combinations shown in Fig. 1E. Probit-derived curves for each single agent and the combination are shown.
To identify drug combinations that were more frequently effective within specific diagnostic categories, the median CR values for IC50 and AUC for each combination within each of the four diagnostic subgroups were compared with a CR reference value of 1 (Datasets S2 and S3). Combinations in which median IC50 CR and AUC CR were significantly <1 within a specific diagnostic subgroup were mapped to regions of a four-way Venn diagram depending on subgroup efficacy observed (Fig. 1D). Consistent with findings from clustering of CR values, several combinations exhibited median CR values significantly less than 1 selectively among patient samples with myeloid malignancies, including combinations of venetoclax and the CSF1R inhibitor ARRY-382, p38 MAPK inhibitor doramapimod, or bromodomain inhibitor JQ1 (Fig. 1E). In contrast, six combinations were selectively effective in CLL patient samples only, including quizartinib/ibrutinib and JQ1/sorafenib (Fig. 1E). Representative single-agent and combination dose–response curves for these combinations are provided in Fig. S3. Several combinations were significantly effective beyond either single agent in myeloid and lymphoid samples, with the most common broad efficacy attributable to the palbociclib/ruxolitinib combination. Although no unique effective combinations were identified for ALL patient samples, this was likely attributable to its smaller cohort size (n = 12) among the diagnostic categories surveyed. For a more complete evaluation of combination benefit, median IC50 and AUC values for each single agent were also compared across diagnosis subgroups (Dataset S4). In particular, venetoclax sensitivity was significantly different among disease types, with the lowest median IC50 and AUC values observed for CLL samples (median IC50, 51.4 nM vs. 2,105 nM for AML samples). Contrastingly, sensitivities to JQ1, quizartinib, sorafenib, and cabozantinib were also different across subgroups, with selective potency for AML samples by both CR effect measures. Notably, none of the single agents except venetoclax and quizartinib demonstrated significant selectivity for the same diagnosis subgroup alone and in combination. Furthermore, CR values across the patient cohort were not significantly associated with the general patient characteristics of sex, age (either as a categorical or continuous variable), or type of specimen analyzed (Dataset S4). Viewed comprehensively, these findings suggest patterns of disease-selective enhanced efficacy of targeted therapy combinations that highlight potential opportunities for the treatment of heterogeneous diagnostic subtypes of myeloid and lymphoid malignancies.
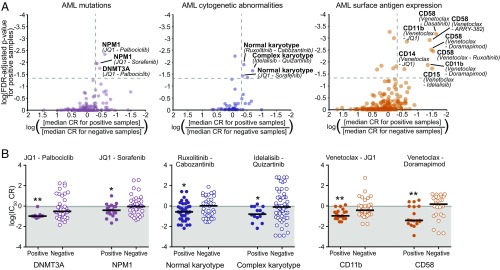
For the two largest diagnostic groups, AML and CLL, expanded panels of clinical, prognostic, mutational, cytogenetic, and surface-antigen data were compiled for comparisons to CR values for each drug combination. For AML samples (n = 58), beyond general clinical characteristics such as age, sex, and white blood cell (WBC) count, additional annotations examined included mutational profiles from a focused panel of genes commonly mutated in AML, cytogenetic features from standard chromosome analysis, and cell-surface antigen expression according to flow cytometry (Fig. 2 and Datasets S5 and S6). Notably, the most prevalent mutation in this cohort was NPM1 (33%), and approximately 50% of the cohort featured normal karyotype. Patients harboring mutations in NPM1 demonstrated significantly enhanced sensitivity to the JQ1/sorafenib combination [median IC50 CR, 0.437; false discovery rate (FDR)-adjusted P = 0.010]. Patients harboring mutations in DNMT3A exhibited significantly enhanced sensitivity to the JQ1/palbociclib combination (median IC50 CR, 0.119; FDR-adjusted P = 0.017; Fig. 3, Left). Patients with normal cytogenetics were significantly sensitive to combinations of ruxolitinib/cabozantinib and JQ1/sorafenib, whereas those harboring complex karyotypes were significantly sensitive to the combination of idelalisib/quizartinib (Fig. 3, Middle). Surface expression of several specific myeloid markers was also associated with significant sensitivity to combinations involving venetoclax, including CD11b (integrin αM) to venetoclax/JQ1 (median IC50 CR, 0.121; FDR-adjusted P = 0.001) and CD58 (LFA-3) venetoclax/doramapimod (median IC50 CR, 0.040; FDR-adjusted P = 0.003; Fig. 3, Right).
Fig. 2.
Clinical and genetic features of patients with AML surveyed. Panels of the indicated disease-specific clinical, prognostic, mutation, cytogenetic, and surface antigen features were compared among all 58 patients with AML in the study. The number of patients evaluable for each feature is given, along with the number of positive samples for that feature (when relevant for categorical variables). Gray boxes indicate unavailable information. Each patient is shown in a unique column, and samples are sorted from left to right according to frequency of genetic mutations.
Fig. 3.
Associations of selective inhibitor combination benefit with mutation, cytogenetic, and surface antigen expression features in AML. (A) Scatter plots of combination/feature pairing test significance vs. difference in median CR between subgroups. Summaries for mutation (Left), cytogenetic (Middle), and surface antigens (Right). All plotted points correspond to combination/feature pairings in which (i) median IC50 and AUC CR of the negative samples were not significantly <1 and (ii) positive and negative subgroups contained at least 15% of total evaluable samples each. Points above the horizontal dashed gray line demonstrated median IC50 CR and AUC CR values for positive subgroup that were significantly <1 (i.e., FDR-adjusted P < 0.05). Points to the right of the vertical dashed gray line represent those in which the median IC50 CR value of the positive subgroup was at least twofold lower than that of the negative subgroup. (B) Scatter plots of log-transformed IC50 CR values for select combinations by AML mutation (Left), cytogenetic (Middle), and surface antigen expression subgroups (Right). Black horizontal bars represent median CR value. (*Median CR significantly <1 in that subgroup.)
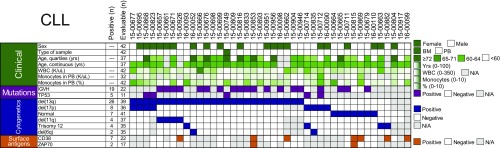
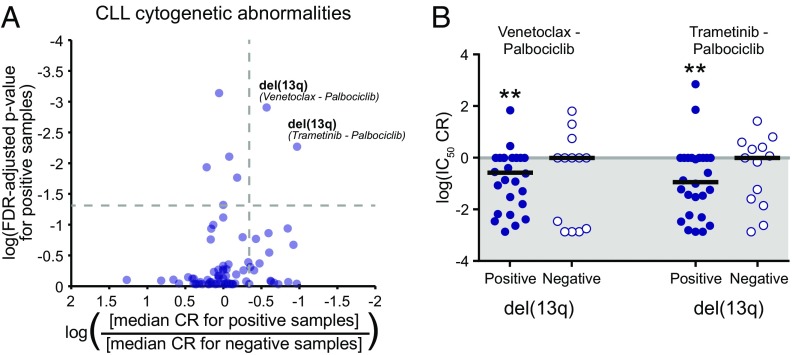
CLL samples were characterized for the mutational status of IgVH and TP53. Cytogenetic features for chromosomal deletions and trisomy, as well as prognostic cell surface antigens such as CD38 and ZAP70, were determined by standard chromosome analysis and flow cytometry, respectively (Fig. 4 and Datasets S7 and S8). Among the tested combinations, the most significant associations with respect to disease characteristics were observed in patients harboring deletion of 13q, who showed significant sensitivity to combinations of palbociclib with venetoclax (median IC50 CR, 0.040; FDR-adjusted P = 0.003) or trametinib (median IC50 CR, 0.116; P = 0.006; Fig. 5).
Fig. 4.
Clinical and genetic features of patients with CLL surveyed. Panels of the indicated disease-specific clinical, mutation, cytogenetic, and surface antigen features were compared among all 42 patients with CLL in the study. The number of patients evaluable for each feature is given, along with the number of positive samples for a given feature (when relevant for categorical variables). Gray boxes indicate unavailable information. Each patient is shown in a unique column, and samples are sorted from left to right according to frequency of cytogenetic abnormalities.
Fig. 5.
Sensitivity of CLL patient samples harboring del(13q)to combinations with the CDK4/6 inhibitor palbociclib. (A) Scatter plot of combination-feature pairing test significance vs. difference in median CR between subgroups. All plotted points correspond to combination/feature pairings in which (i) the median IC50 and AUC CR of the negative samples were not significantly <1 and (ii) the positive and negative subgroups contained at least 15% of the total evaluable samples each. Points above the horizontal dashed gray line demonstrate median IC50 CR and AUC CR values for the positive subgroup that were significantly <1 (i.e., FDR-adjusted P < 0.05). Points to the right of the vertical dashed gray line represent those for which the median IC50 CR value of the positive subgroup was at least twofold lower than that of the negative subgroup. (B) Scatter plot of log-transformed IC50 CR values for select combinations effective in del(13q)-positive CLL samples. Black horizontal bars represent the median CR value. (*Median CR is significantly <1 in that subgroup.)
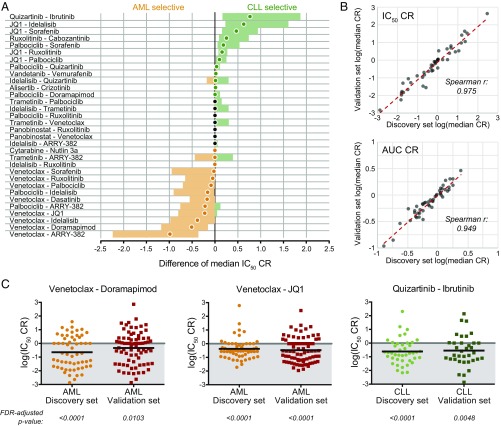
Although strict association of a particular combination with a patient feature may require a more complete understanding of the inter- and intrasubgroup heterogeneity and expanded sample size, we found statistically significant associations for select prevalent biomarkers (Table 2). Furthermore, in the broader context of the two largest diagnosis categories (AML and CLL), certain inhibitor combinations demonstrated disease-specific selective efficacy irrespective of genetic and clinical features. For example, several combinations of venetoclax with a kinase inhibitor have enhanced efficacy in AML (e.g., venetoclax/doramapimod), whereas combinations of JQ1 with multiple kinase inhibitors exhibit an enhanced efficacy preference for CLL (JQ1/sorafenib; Fig. 6). Importantly, these results, obtained by directly comparing the median CR values of the two largest disease subsets, are consistent with our earlier findings from unsupervised clustering. Thus, this approach uncovers multiple potential opportunities for application of combination therapies in distinct diagnostic, genetic/cytogenetic, and phenotypic subsets of patients with AML and CLL.
Table 2.
Selective inhibitor combination sensitivities by feature among AML and CLL patient samples surveyed
| Feature type | Diagnosis category | Combination | IC50 | AUC | ||
| Median CR | P value* | Median CR | P value* | |||
| Mutation | ||||||
| DNMT3A | AML | JQ1/palbociclib | 0.1187 | 0.0171 | 0.5693 | 0.0189 |
| NPM1 | AML | JQ1/palbociclib | 0.1138 | 0.0045 | 0.6830 | 0.0120 |
| NPM1 | AML | JQ1/sorafenib | 0.4375 | 0.0108 | 0.6952 | 0.0091 |
| Cytogenetics | ||||||
| Complex karyotype | AML | Idelalisib/quizartinib | 0.1753 | 0.0227 | 0.6872 | 0.0078 |
| Normal karyotype | AML | Ruxolitinib/cabozantinib | 0.3289 | 0.0126 | 0.8040 | 0.0015 |
| Normal karyotype | AML | JQ1/sorafenib | 0.4375 | 0.0378 | 0.8495 | 0.0197 |
| del(13q) | CLL | Trametinib/palbociclib | 0.1161 | 0.0057 | 0.4121 | <0.0001 |
| del(13q) | CLL | Venetoclax/palbociclib | 0.2673 | 0.0013 | 0.5269 | <0.0001 |
| Surface antigen | ||||||
| CD11b | AML | Venetoclax/doramapimod | 0.0441 | 0.0136 | 0.4503 | 0.0185 |
| CD11b | AML | Venetoclax/JQ1 | 0.1205 | 0.0014 | 0.2539 | 0.0070 |
| CD14 | AML | Venetoclax/JQ1 | 0.1004 | 0.0201 | 0.2539 | 0.0061 |
| CD15 | AML | Venetoclax/idelalisib | 0.0308 | 0.0189 | 0.6769 | 0.0309 |
| CD58 | AML | Venetoclax / ARRY-382 | 0.0381 | 0.0012 | 0.2796 | 0.0006 |
| CD58 | AML | Venetoclax/dasatinib | 0.0229 | 0.0007 | 0.2594 | 0.0021 |
| CD58 | AML | Venetoclax/doramapimod | 0.0402 | 0.0031 | 0.4396 | 0.0035 |
| CD58 | AML | Venetoclax/ruxolitinib | 0.0027 | 0.0036 | 0.3359 | 0.0053 |
P values are FDR-adjusted.
Fig. 6.
Validation of combination selectivity between AML and CLL. (A) The difference of median IC50 CR values (AML – CLL) was computed for each of 48 indicated combinations by using the Hodges-Lehmann method. The median difference is represented by a closed circle, and the 95% CI is shown as the colored bar. AML-selective and CLL-selective combinations are colored orange or green, respectively. (B) Spearman correlation of log-transformed median IC50 CR and AUC CR values between the discovery sample cohort (N = 122) and independent validation cohort (N = 151). (C) Validation of select effective combinations within AML or CLL diagnostic subgroups. Scatter plots of log-transformed IC50 CR values for the indicated combinations. Black horizontal bars represent median CR; FDR-adjusted P values (Wilcoxon signed-rank test of median) are shown.
To address the reproducibility of these results, we compiled an independent validation dataset (N = 151) of a similar distribution of prospectively collected primary leukemia patient samples. There is a high level of correlation for the median IC50 and AUC CRs between the discovery and validation datasets for all combinations and samples tested (Spearman r = 0.975 and 0.949, respectively; P < 0.0001; Fig. 6B). Given the particularly notable efficacy of combinations involving venetoclax in AML patient specimens from our discovery cohort, a subset of the most effective of these combinations was compared with those of the validation-set AML samples, revealing similar sensitivity distributions in both groups (Fig. 6C). Additionally, expanded synergy analysis of this same subset of combinations was performed for each drug alone over a seven-point dose series and all 49 possible combinations of the two agents on a panel of human leukemia cell lines (MOLM-13, MOLM-14, HL-60, OCI-AML2, OCI-AML3). Overall, cell-line IC50 and AUC CR measures (determined by equimolar series) correlated well with the Bliss synergy score across the full matrix of combinations (Spearman r = 0.649 and 0.787, respectively; P < 0.0001).
Discussion
As the vast molecular heterogeneity of cancer continues to be unraveled, the necessity of defining actionable targeted therapeutic strategies for patients remains paramount to improvement in outcomes. Given difficulties involving the often nondurable responses and rapid development of resistance observed with many single-agent targeted therapies, we sought to identify effective combination strategies for patients with hematologic malignancies. Before this study, we screened more than 1,000 primary patient specimens against a panel of single-agent small-molecule inhibitors. Using these historical drug sensitivity data, we ranked drugs by their IC50 and used these rankings to assemble an initial panel of drug combinations consisting primarily of kinase inhibitors that targeted nonoverlapping pathways. Primary patient samples with various hematologic malignancies were screened, and, based on data from this initial panel, we generated a second iteration including new combinations of kinase inhibitors as well as combinations of inhibitors from select additional drug classes. However, for both panels, broad cytotoxicity was problematic for many combinations, and unsupervised hierarchical clustering of CR values revealed no distinct clusters tracking with diagnosis category. This preliminary work informed the design of the panel of combinations used herein, which added several additional classes of inhibitors as well as an intentional inclusion of FDA-approved drugs or drugs in clinical development wherever possible. To this end, defining drug combinations for ex vivo screening is a highly iterative process of refinement. Although logistical considerations preclude comprehensive evaluation of all possible pairwise combinations of inhibitors, these cumulative data may also prove amenable to applied machine learning-based computational models to predict novel drug target pairings in specific malignancies (28, 29), and may be adaptable as a testing scheme for other tumor types given recent developments to isolate circulating tumor cells (30, 31).
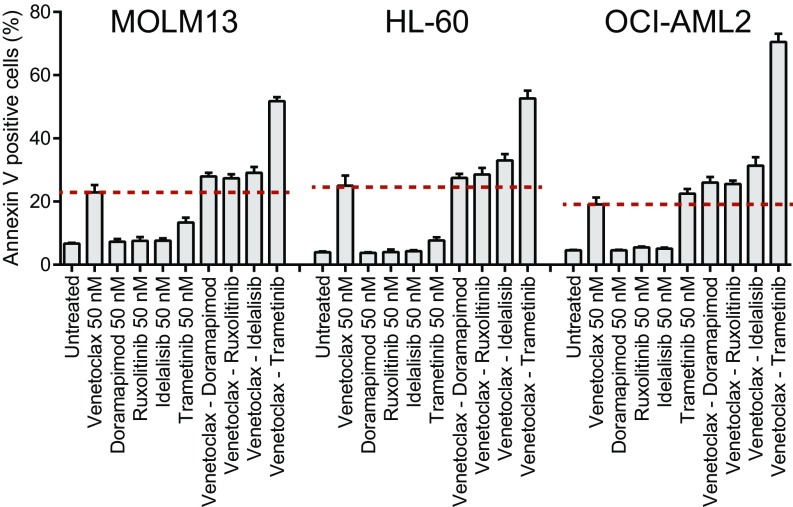
A critical finding in this study is the effectiveness of several combinations of targeted agents that include a kinase inhibitor and venetoclax, a selective inhibitor of BCL2, for myeloid-derived tumors (Fig. 6). The relative sensitivity to these combinations in the ex vivo assay supports the notion that coupling a kinase-derived proliferative signal inhibitor with an antiapoptotic agent improves efficacy. Furthermore, we confirmed the capacity of select venetoclax combinations to augment apoptotic cell death in a panel of human AML cell lines (Fig. S4). Combinations such as dasatinib, doramapimod, sorafenib, or idelalisib with venetoclax are broadly effective on myeloid-derived tumor samples and may be useful for treatment of AML in particular. These combinations appear to be effective across a broad percentage of AML patient samples, irrespective of cohort subtype heterogeneity. Consistent with these observations, recent reports indicate the combined inhibition of BCR-ABL1 and BCL2 is a promising strategy for targeting Philadelphia chromosome-positive ALL as well as the stem cell population in chronic myeloid leukemia (32, 33). We also observed certain combinations with venetoclax to be effective on CLL samples with 13q deletions. Although venetoclax has recently achieved FDA approval for patients with CLL with 17p deletions, our data suggest that venetoclax, even as a single agent, may be more broadly effective in patients with CLL with diverse cytogenetic profiles, and combinations may offer options particularly for disease states in which venetoclax as a single agent is not effective. It is noteworthy that venetoclax is effective for a variety of hematologic malignancy subsets, including CLL, multiple myeloma, and AML (34–37). Additionally, we note that genetic features that are considerably less frequent in AML or CLL will require expanded sample sizes to more robustly interrogate possible associations with inhibitor combination efficacy. Similarly, small sample sizes of other diagnosis groups (ALL, MPN, or MDS/MPN) are a limitation of the present study. We may potentially miss important drug combinations because of small sample sizes and lower power, and, as such, these diagnostic groups will require larger sample sizes to permit the identification of relationships between combination efficacy and disease-specific features.
Fig. S4.
Apoptosis induction for venetoclax combinations in AML cell lines. Three human AML cell lines (MOLM13, HL-60, and OCI-AML2) were cultured in the presence of venetoclax alone or in combination with doramapimod, ruxolitinib, idelalisib, or trametinib. All drug concentrations used were 50 nM. After 48 h, the percentage of cells positive for annexin V staining were measured by Guava Nexin assay (Millipore). Bars indicate the mean of three replicates ± SD.
Developments in functional screening technology have produced several assay platforms for the evaluation of responses of tumor cells to exogenous perturbations. Functional screening efforts in hematologic malignancies have often involved the culture of patient cells in conventional 2D tissue culture platforms, sometimes with conventional culture conditions (38–40) and other times using additives or feeder cell coculture that promote certain phenotypic aspects of the cells, such as preservation of primitive cell differentiation state (41) and cell proliferation (42). A key feature of our ex vivo assay is that it provides drug sensitivity data within 4 d, a time frame that can support and influence clinical decision-making. Furthermore, this approach addresses some of the challenges in deploying effective therapies in which there is a substantial gap between clinical, diagnostic, or genetic markers and available drugs. Ideally, the integration of functional and genomic data types may facilitate more precise insight into the molecular mechanisms contributing to disease. For example, our data suggest that combined targeting of the BTK inhibitor ibrutinib and the multikinase inhibitor quizartinib may represent a promising strategy for patients with CLL. The efficacy of ibrutinib is well established for CLL (reviewed in ref. 43). Although quizartinib is primarily considered to be a FLT3 inhibitor, it is also a potent inhibitor of CSF1R, a target recently implicated for CLL treatment because of its effects on supportive nurse-like monocyte/macrophage cells that express CSF1R (44–46), highlighting the importance of teasing out tumor-intrinsic mechanisms from tumor-extrinsic microenvironment contributions to sensitivity/resistance. Accordingly, simultaneous inhibition of BTK- and CSF1R-mediated signaling pathways may result in further improvement of responses (Fig. 6).
Although we have identified several links between actionable diagnostic and genetic features and effective combinations of targeted agents, we also acknowledge certain limitations to our analyses. For example, the inhibitor screening method requires the use of prospectively collected, freshly isolated patient samples as a consequence of a clinical visit. Variability in the number of mononuclear cells recovered from a specimen limits the scope and number of inhibitors that may be tested. Furthermore, the challenge of overcoming noise in the drug sensitivity data caused by variations in biological response and technique limit the utility of applying conventional methods of determining synergy (25). Additionally, as a result of the prospective nature of sample collection, there are differences in sample size and frequency of genetic parameters surveyed within a given diagnostic group. To address these issues, we used an integrated and robust approach, including the development of a CR to measure relative efficacy of inhibitor combinations, requiring that both effect measures (IC50 and AUC) achieve significance, performing rank-based tests of the median within and between relevant subgroups, and applying multiplicity adjustments to the P values from these tests. We also note that developments in new oncology drugs within various drug classes will present opportunities to increase target/pathway representation in combination screening. Finally, our assay makes use of aggregate readings of whole mononuclear cell population responses to drug combinations and single agents, whereby a readout that offers granularity of responses at the single-cell level would provide additional data for parsing of drug combination efficacy. Recent approaches that make use of single-cell imaging or flow cytometry (47–51), or CRISPR/Cas9 gene silencing (52), will be useful in enhancing our initial view of combination efficacy and in identifying new targets.
In summary, our data identify promising drug combinations that were previously unrecognized but may promote the testing of a select number of these drug combinations in clinical trials. Validation of these combinations will require testing in clinical trials, which may be suitable initially for patients with AML with the relapsed/refractory disease. Accordingly, we formed combinations with FDA-approved drugs to facilitate their translational effect. With respect to the BCL2 inhibitor venetoclax, there is now compelling clinical evidence supporting its evaluation in combination with other agents for AML (53). Independent of the findings reported here, three clinical trials currently in the recruitment phase will provide opportunities to evaluate targeted combinations of the BCL2 inhibitor (venetoclax) with an MEK inhibitor (cobimetinib; ClinicalTrials.gov ID code NCT02670044), an MDM2 inhibitor (idasanutlin; ClinicalTrials.gov ID code NCT02670044), or a BET inhibitor (ABBV-075; ClinicalTrials.gov ID code NCT02391480) on patients with relapsed/refractory AML who are not eligible for induction therapy. Collectively, these findings may yield new therapeutic options for patients while advancing the use of ex vivo functional testing as a valid assay in the clinical decision-making process.
Materials and Methods
Patient Samples.
All patients gave consent to participate in this study, which had the approval and guidance of the institutional review boards at Oregon Health & Science University (OHSU), University of Utah, University of Texas Medical Center (UT Southwestern), Stanford University, University of Miami, and University of Colorado. Mononuclear cells were isolated by Ficoll-gradient centrifugation from freshly obtained bone marrow aspirates or peripheral blood draws and plated into assays within 24 h. All samples were analyzed for clinical characteristics and drug sensitivity. AML and CLL patient samples were additionally analyzed with respect to expanded, disease-specific panels of clinical, prognostic, genetic, cytogenetic, and surface antigen characteristics obtained from patient electronic medical records. Genetic characterization of AML samples included results of a clinical deep-sequencing panel of genes commonly mutated in hematologic malignancies (GeneTrails panel from Knight Diagnostic Laboratories, OHSU; Foundation Medicine reports from UT Southwestern).
Ex Vivo Functional Screen.
Small-molecule inhibitors, purchased from LC Laboratories and Selleck Chemicals, were reconstituted in DMSO and stored at −80 °C. The CSF1R inhibitor ARRY-382 was obtained from Array Biopharma. Inhibitors were distributed into 384-well plates prepared with a single agent per well in a seven-point concentration series ranging from 10 μM to 0.0137 μM for each drug (except dasatinib, which was plated at a concentration range of 1 μM to 0.00137 μM). Similar plates were prepared with the 48 indicated pairwise inhibitor combinations in seven-point fixed molar concentration series identical to those used for single agents. The final concentration of DMSO was ≤0.1% in all wells, and all sets of single-agent and combination destination plates were stored at −20 °C and thawed immediately before use. Primary mononuclear cells were plated across single-agent and combination inhibitor panels within 24 h of collection. Cells were seeded into 384-well assay plates at 10,000 cells per well in RPMI 1640 media supplemented with FBS (10%), l-glutamine, penicillin/streptomycin, and β-mercaptoethanol (10−4 M). After 3 d of culture at 37 °C in 5% CO2, MTS reagent (CellTiter96 AQueous One; Promega) was added, optical density was measured at 490 nm, and raw absorbance values were adjusted to a reference blank value and then used to determine cell viability (normalized to untreated control wells).
Inhibitor Dose–Response Curve Analysis and Effect-Measure Calculations.
Normalized viability values (40) at each dose of a seven-point dilution series for 21 small-molecule inhibitors and 48 pairwise combinations of two of these single agents were analyzed for each of 122 primary leukemia samples. Dose concentrations were log10-transformed, and a probit regression curve was fit to each seven-point drug sensitivity profile by using maximum-likelihood estimation for the intercept and slope. This parametric model was chosen over a polynomial because the probit’s monotonic shape reflects a dose–response curve typically seen in samples incubated with cytotoxic or inhibitory agents (54). Normalized viability values greater than 100%, indicating higher cell viability than the average viability across control wells on a given plate, were truncated to 100% to produce a percentage response variable amenable to probit modeling. From the fitted probit curve for each sample/drug pairing, the IC50 was defined as the lowest concentration to achieve 50% predicted viability and the AUC was computed by integration of the curve height across the tested dose range. If the predicted cell viability (i.e., probit curve height) was ≤50% at the lowest tested dose or >50% across the entire dose range, the IC50 was designated as the lowest dose or highest dose, respectively. For sensitivity profiles with 100% normalized viability at all seven dose points, the IC50 and AUC were designated as the highest tested dose and the maximum possible AUC, respectively. For sensitivity profiles with 0% viability at all seven dose points, the IC50 and AUC were designated as the lowest tested dose and a value (0.5) just below the minimum probit-derived AUC, respectively.
To quantify the efficacy of an equimolar drug combination in comparison with its constituent single agents, a CR effect measure was generated based on the specific IC50 and AUC values for each inhibitor triad (the drug combination and the two single agents). The IC50 CR and AUC CR values were defined as the ratio of the combination’s IC50 or AUC to the minimum IC50 or AUC for the two single agents, respectively. Each sensitivity profile modeled by probit regression was assigned a fit statistic based on the P value for the test of whether the fitted curve’s slope was horizontal. Generally, a smaller fit statistic produced by a decreasing slope indicates a better fit and, by extension, provides a measure of confidence in the curve-derived IC50 and AUC for a particular sample/drug pair. A CR effect measure value less than 1 indicates that a sample is more sensitive to the drug combination than it is to either of the single agents that constitute the combination.
Statistical Analysis.
Unsupervised hierarchical clustering and heat-map displays of inhibitor sensitivity were generated by using GenePattern software (Broad Institute). Inhibitor combination efficacy was compared for all samples (N = 122) across a panel of general clinical variables: diagnostic category (AML, ALL, CLL, MPN or MDS/MPN), age (categorized as <18, 18–39, 40–64, or ≥65 y), sex, and type of specimen (peripheral blood, bone marrow aspirate, leukapheresis). For each CR effect measure, a one-sample Wilcoxon signed-rank test was used to assess whether the median CR value was significantly less than 1 to identify effective drug combinations for each subgroup. Furthermore, differences in median CR across the subgroups of these variables were evaluated with the Kruskal–Wallis test. Additional diagnosis-specific clinical and genetic variables were examined for AML (n = 58) and CLL (n = 42) patient samples. Similar tests were performed on a subgroup’s median CR and on differences across subgroups for each of these clinical or genetic variables. Subgroups for all mutations, cytogenetic abnormalities, and cell surface antigens were defined as positive or negative. Correlations between continuous clinical variables (e.g., WBC count) and CR values were appraised with Spearman correlation coefficients. For all within-subgroup and between-subgroup nonparametric tests, FDR adjustments (55) were applied to P values with an adaptive linear step-up method to account for tests being performed on each of the 48 inhibitor triads.
Supplementary Material
Acknowledgments
We thank the 122 patients who contributed samples to enable this study; David Patterson, David Haussler, and Pierrette Lo for helpful discussions; and Brian Junio for sample coordination. This work was supported by The Leukemia & Lymphoma Society (B.J.D.), National Human Genome Research Institute Grant U54HG007990 (to S.E.K.), the V Foundation for Cancer Research (J.W.T.), the Gabrielle’s Angel Foundation for Cancer Research (J.W.T.), National Cancer Institute Grant 1R01CA183947-01 (to J.W.T.), University of Miami Clinical and Translational Science Institute Award KL2TR000461 (to J.W.), a Howard Hughes Medical Institute Medical Research Fellows Program (to V.K.), and the Biostatistics Shared Resource of the Oregon Health & Science University Knight Cancer Institute via National Cancer Institute Grant 5P30CA69533 (to A.K. and M.M.). B.J.D. is a Howard Hughes Medical Institute investigator.
Footnotes
Conflict of interest statement: D.A.P. serves on the advisory boards for Pharmacyclics and Gilead. J.W.T. receives research support from Agios, Array Biopharma, Aptose, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen R&D, Seattle Genetics, Syros, and Takeda and is a consultant for Leap Oncology. B.J.D. serves on the advisory boards for Gilead and Roche TCRC. B.J.D. is principal investigator or coinvestigator on Novartis and BMS clinical trials. His institution, Oregon Health & Science University, has contracts with these companies to pay for patient costs, nurse and data manager salaries, and institutional overhead. He does not derive salary, nor does his laboratory receive funds from these contracts. M.W.D. serves on the advisory boards and/or as a consultant for Novartis, Incyte, and BMS and receives research funding from BMS and Gilead. The authors certify that all compounds and combinations tested in this study were chosen without input from any of our industry partners.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703094114/-/DCSupplemental.
References
- 1.Dienstmann R, Jang IS, Bot B, Friend S, Guinney J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015;5:118–123. doi: 10.1158/2159-8290.CD-14-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15:747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley TJ, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 5.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Döhner H, et al. European LeukemiaNet Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 9.Oran B, Weisdorf DJ. Survival for older patients with acute myeloid leukemia: A population-based study. Haematologica. 2012;97:1916–1924. doi: 10.3324/haematol.2012.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medeiros BC, et al. Big data analysis of treatment patterns and outcomes among elderly acute myeloid leukemia patients in the United States. Ann Hematol. 2015;94:1127–1138. doi: 10.1007/s00277-015-2351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravandi F, et al. Effective treatment of acute promyelocytic leukemia with all-trans-retinoic acid, arsenic trioxide, and gemtuzumab ozogamicin. J Clin Oncol. 2009;27:504–510. doi: 10.1200/JCO.2008.18.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo-Coco F, Orlando SM, Platzbecker U. Treatment of acute promyelocytic leukemia. N Engl J Med. 2013;369:1472. doi: 10.1056/NEJMc1309895. [DOI] [PubMed] [Google Scholar]
- 13.Nakao M, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 14.Yamamoto Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 15.Man CH, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: Favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood. 2012;119:5133–5143. doi: 10.1182/blood-2011-06-363960. [DOI] [PubMed] [Google Scholar]
- 16.Shah NP, et al. Ponatinib in patients with refractory acute myeloid leukaemia: findings from a phase 1 study. Br J Haematol. 2013;162:548–552. doi: 10.1111/bjh.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker SD, et al. Emergence of polyclonal FLT3 tyrosine kinase domain mutations during sequential therapy with sorafenib and sunitinib in FLT3-ITD-positive acute myeloid leukemia. Clin Cancer Res. 2013;19:5758–5768. doi: 10.1158/1078-0432.CCR-13-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shafer D, Grant S. Update on rational targeted therapy in AML. Blood Rev. 2016;30:275–283. doi: 10.1016/j.blre.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spain L, Julve M, Larkin J. Combination dabrafenib and trametinib in the management of advanced melanoma with BRAFV600 mutations. Expert Opin Pharmacother. 2016;17:1031–1038. doi: 10.1517/14656566.2016.1168805. [DOI] [PubMed] [Google Scholar]
- 20.Chang E, et al. The combination of FLT3 and DNA methyltransferase inhibition is synergistically cytotoxic to FLT3/ITD acute myeloid leukemia cells. Leukemia. 2016;30:1025–1032. doi: 10.1038/leu.2015.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonard JT, et al. Functional and genetic screening of acute myeloid leukemia associated with mediastinal germ cell tumor identifies MEK inhibitor as an active clinical agent. J Hematol Oncol. 2016;9:31. doi: 10.1186/s13045-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxson JE, et al. Identification and characterization of tyrosine kinase nonreceptor 2 mutations in leukemia through integration of kinase inhibitor screening and genomic analysis. Cancer Res. 2016;76:127–138. doi: 10.1158/0008-5472.CAN-15-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxson JE, et al. Therapeutically targetable ALK mutations in leukemia. Cancer Res. 2015;75:2146–2150. doi: 10.1158/0008-5472.CAN-14-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxson JE, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 26.Croce CM, Reed JC. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res. 2016;76:5914–5920. doi: 10.1158/0008-5472.CAN-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J, et al. Target inhibition networks: Predicting selective combinations of druggable targets to block cancer survival pathways. PLOS Comput Biol. 2013;9:e1003226. doi: 10.1371/journal.pcbi.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behinaein B, Rudie K, Sangrar W. Petri net siphon analysis and graph theoretic measures for identifying combination therapies in cancer. IEEE/ACM Trans Comput Biol Bioinformatics. October 3, 2016 doi: 10.1109/TCBB.2016.2614301. [DOI] [PubMed] [Google Scholar]
- 30.Hayes DF, Smerage JB. Circulating tumor cells. Prog Mol Biol Transl Sci. 2010;95:95–112. doi: 10.1016/B978-0-12-385071-3.00005-8. [DOI] [PubMed] [Google Scholar]
- 31.Alix-Panabières C, Pantel K. Circulating tumor cells: Liquid biopsy of cancer. Clin Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 32.Leonard JT, et al. Targeting BCL-2 and ABL/LYN in Philadelphia chromosome-positive acute lymphoblastic leukemia. Sci Transl Med. 2016;8:354ra114. doi: 10.1126/scitranslmed.aaf5309. [DOI] [PubMed] [Google Scholar]
- 33.Carter BZ, et al. Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci Transl Med. 2016;8:355ra117. doi: 10.1126/scitranslmed.aag1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davids MS, Letai A, Brown JR. Overcoming stroma-mediated treatment resistance in chronic lymphocytic leukemia through BCL-2 inhibition. Leuk Lymphoma. 2013;54:1823–1825. doi: 10.3109/10428194.2013.796051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan R, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson MA, et al. The BCL2 selective inhibitor venetoclax induces rapid onset apoptosis of CLL cells in patients via a TP53-independent mechanism. Blood. 2016;127:3215–3224. doi: 10.1182/blood-2016-01-688796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts AW, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374:311–322. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyner JW, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci USA. 2009;106:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pemovska T, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3:1416–1429. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 40.Tyner JW, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabst C, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 42.Klco JM, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2013;121:1633–1643. doi: 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddocks K, Jones JA. Bruton tyrosine kinase inhibition in chronic lymphocytic leukemia. Semin Oncol. 2016;43:251–259. doi: 10.1053/j.seminoncol.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Galletti G, Caligaris-Cappio F, Bertilaccio MT. B cells and macrophages pursue a common path toward the development and progression of chronic lymphocytic leukemia. Leukemia. 2016;30:2293–2301. doi: 10.1038/leu.2016.261. [DOI] [PubMed] [Google Scholar]
- 45.Galletti G, et al. Targeting macrophages sensitizes chronic lymphocytic leukemia to apoptosis and inhibits disease progression. Cell Reports. 2016;14:1748–1760. doi: 10.1016/j.celrep.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 46.Polk A, et al. Colony-stimulating factor-1 receptor is required for nurse-like cell survival in chronic lymphocytic leukemia. Clin Cancer Res. 2016;22:6118–6128. doi: 10.1158/1078-0432.CCR-15-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 48.Kornblau SM, et al. Dynamic single-cell network profiles in acute myelogenous leukemia are associated with patient response to standard induction therapy. Clin Cancer Res. 2010;16:3721–3733. doi: 10.1158/1078-0432.CCR-10-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Gaizo Moore V, Letai A. BH3 profiling–measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett. 2013;332:202–205. doi: 10.1016/j.canlet.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Touzeau C, et al. BH3 profiling identifies heterogeneous dependency on Bcl-2 family members in multiple myeloma and predicts sensitivity to BH3 mimetics. Leukemia. 2016;30:761–764. doi: 10.1038/leu.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman AA, et al. Feasibility of ultra-high-throughput functional screening of melanoma biopsies for discovery of novel cancer drug combinations. Clin Cancer Res. June 27, 2017 doi: 10.1158/1078-0432.CCR-16-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 53.Konopleva M, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov. 2016;6:1106–1117. doi: 10.1158/2159-8290.CD-16-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altshuler B. Modeling of dose-response relationships. Environ Health Perspect. 1981;42:23–27. doi: 10.1289/ehp.814223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25:60–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.