Significance
Epigenetic alterations in nasopharyngeal carcinoma (NPC) are very frequent at the DNA level. Histone modifications are frequently altered in cancers. Because histone modifications are reversible, histone-modifying enzymes or other epigenetic regulators are ideal therapeutic targets, and drugs targeting these enzymes have been proven effective in cancer treatment. Understanding the NPC histone code provides unique insights into NPC pathogenesis and will likely contribute to the identification of unique therapeutics. Using genome-wide analyses of histone modifications, we generated an NPC epigenetic landscape and identified a key oncogene whose expression correlated with patient overall survival, suggesting that epigenetic profiling can effectively identify key oncogenic pathways. These studies provide proof-of-concept strategies for further characterization of the NPC epigenome on a larger scale.
Keywords: nasopharyngeal carcinoma, super-enhancer, H3K27ac, Epstein–Barr virus, ETV6
Abstract
Nasopharyngeal carcinoma (NPC) most frequently occurs in southern China and southeast Asia. Epidemiology studies link NPC to genetic predisposition, Epstein–Barr virus (EBV) infection, and environmental factors. Genetic studies indicate that mutations in chromatin-modifying enzymes are the most frequent genetic alterations in NPC. Here, we used H3K27ac chromatin immune precipitation followed by deep sequencing (ChIP-seq) to define the NPC epigenome in primary NPC biopsies, NPC xenografts, and an NPC cell line, and compared them to immortalized normal nasopharyngeal or oral epithelial cells. We identified NPC-specific enhancers and found these enhancers were enriched with nuclear factor κB (NF-κB), IFN-responsive factor 1 (IRF1) and IRF2, and ETS family members ETS1 motifs. Normal cell-specific enhancers were enriched with basic leucine zipper family members and TP53 motifs. NPC super-enhancers with extraordinarily broad and high H3K27ac signals were also identified, and they were linked to genes important for oncogenesis including ETV6. ETV6 was also highly expressed in NPC biopsies by immunohistochemistry. High ETV6 expression correlated with a poor prognosis. Furthermore, we defined the EBV episome epigenetic landscapes in primary NPC tissue.
Nasopharyngeal carcinoma (NPC) is a leading cancer in southern China. NPC incidence is also high in southeast Asia and northern Africa. In 2008, >84,000 new cases were diagnosed and >51,000 patients died from NPC worldwide (1, 2). While less common in the United States with 0.5–2 cases per 100,000 people annually, NPC is the most frequently occurring malignancy in adults in Guangdong Province, China, where NPC incidence reaches 80 cases per 100,000 people annually and has thus been nicknamed “Guangdong Cancer” (3). Since NPC occurs at a hidden anatomic location and lacks specific symptoms, many patients at initial NPC diagnosis already have metastatic disease. At diagnosis, 60% of patients have lymph node metastases, and 4.2% have distant metastases (4). While NPC is sensitive to radiotherapy and chemotherapy, patients with distant metastatic disease have very poor prognoses, with a median survival of just 13 mo and with 3- and 5-y survival of <15% and <5%, respectively (5). NPC derives from nasopharyngeal epithelial cells and can be classified into keratinizing squamous cell carcinoma, nonkeratinzing carcinoma, and undifferentiated carcinoma (2). In endemic area, >97% of NPCs are undifferentiated carcinoma (2). Epidemiology studies indicate that genetic predisposition, Epstein–Barr virus (EBV) infection, and environmental factors are associated with NPC pathogenesis (6–8).
Genetic and epigenetic alterations play critical roles in NPC pathogenesis. First-degree relatives of NPC patient have greatly elevated risk developing NPC (8). Genome-wide association studies identified NPC susceptibility loci at HLA region, TNFRSF19, MDS1-EVI, and the CDKN2A-CDKN2B gene cluster on 9p21 (9, 10). Chromosome 9q21 and 3p21.3 are also frequently deleted in NPCs (8). Genes encoded in these loci includes p14ARF, p16INK4A, and RASSF1A, and they can also be inactivated by promoter hypermethylation (8). p14ARF and p16INK4A can induce senescence during oncogenic stress. DNA hypermethylation frequently occurs at promoters of tumor suppressors in NPC (11, 12). Genome-wide studies found NPC has more hypermethylation than 11 other cancer types characterized by the Cancer Genome Atlas project (13), similar to EBV-positive gastric cancers (14). EBV-encoded latent membrane protein 1 (LMP1) activates the expression of DNA methyl transferase (DNMT) 1, 3A, and 3B to down-regulate CDH1 (E-Cadherin) expression (15). In addition to DNA methylation, histone-modifying enzymes including epigenetic writers, EZH2, EP300, and cofactors BMI1 have been implicated in NPC pathogenesis and prognosis. EZH2 is a member of the polycomb repressive complex 2 (PRC2) that methylates H3K27 and causes transcription repression. EZH2 is frequently expressed at a high level in NPC, and its expression correlates with poor prognosis (16). EP300, on the other hand, acetylates H3K27 and is correlated with active enhancers or promoters. High EP300 expression correlates with poor overall survival (16). BMI1 is a member of the polycomb repressive complex 1 (PRC1) and is a cofactor for H2AK119 monoubiquitination upon PRC1 binding to H3K27me3. BMI1 can repress PTEN tumor suppressor in nasopharyngeal epithelial cells (17). NPC whole-exon sequencing found that chromatin modification pathway has the highest mutation frequency (18). The genes mutated in this pathway include ARID1A, BAP1, MLL2, MLL3, TET1, TET2, TET3, and TSHZ3. ARID1A is a member of the SWI/SNF family chromatin remodeling protein with ATPase and helicase activities. MLL2 and MLL3 methylate H3K4 and TETs are important for 5 mC modifications (18). These mutations are associated with poor overall survival and higher EBV burden. NPCs have mutations in chromatin modification pathway also have higher mutation rates in other genes (18). Histone modifications are frequently altered in different cancers (19). H3K4 methylations mark active promoters or enhancers, while H3K27 methylation marks repressed genes. H3K27 acetylation marks both active enhancers and promoters (19). No genome-wide NPC histone modification profiles have been reported.
Transcription profiling using microarray and RNA-seq identified many cell genes up-regulated in NPC (20–22). Some of these genes can be used as biomarker to predict patient’s response to treatments or prognosis. These genes include CD147, Caveolin-1, and matrix metalloproteinase 11. Little is known about how these genes are regulated in NPC.
EBV is the first human DNA tumor virus, discovered more than 50 y ago (23). EBV infects both B lymphocytes and oral epithelial cells and establishes lifelong persistence (2). EBV is associated with posttransplant lymphoproliferative disease (PTLD), AIDS lymphomas, ∼50% of Hodgkin’s lymphoma, 10% of gastric carcinoma, and almost all undifferentiated NPC (2). Three different types of EBV latency are associated with different cancers. In Burkitt’s lymphoma, EBV type I latency program is predominant. These cancer cells express EBV nuclear antigen (EBNA) 1, noncoding RNA EBER, and miRNAs. EBV type II latency is seen in NPC and Hodgkin’s lymphoma (24, 25). These cancer cells express LMP1 and 2, EBNA1, EBERs, and miRNAs. EBV type III latency is often seen in PTLD where all EBV latency products, EBNAs (1, LP, 2, 3A, 3B, and 3C), LMPs, EBERs and viral miRNAs, are expressed.
Active EBV replication precedes NPC development (6). Detection of elevated IgA antibody titers against EBV early antigen and viral capsid antigens has been used to screen for early NPC patients (26). Peripheral-blood EBV DNA levels correlate with NPC occurrence, relapse, metastasis, and prognosis (27). Noncoding RNAs are the most consistent and abundant viral products in NPC cells. The two main categories of these noncoding RNAs are the EBERs, single-strand RNAs of about 170 pb with a complex secondary structure, and the viral miRNAs of the BART family, which often account for as much as one-third of all miRNAs contained in malignant NPC cells (2). A few EBV-encoded proteins are consistently expressed in NPCs. However, their expression is highly heterogeneous from one tumor to another and inside a given tumor. EBNA1, which is required for the persistence of the EBV episome in proliferating malignant cells, is the most consistently expressed gene. The latent membrane proteins, LMP1, LMP2A, and LMP2B are less consistently expressed and often at a low abundance. LMP1 cytoplasmic tail binds to tumor necrosis factor receptor-associated factors to constitutively activate NF-κB (28). NF-κB activates the expression of many cell genes to promote cell growth, survival, and metastasis (29–31). LMP1 expression can be detectable in some premalignant precursor lesions of NPC, for example in monoclonal carcinoma in situ (32). Elevated LMP1 expression levels correlate with poorer prognosis (21). LMP2A is more ubiquitously expressed in NPCs and can transform epithelial cell lines and activate phosphatidylinositol 3-kinase (PI3-K)/Akt survival pathway (2, 33). LMP2A can affect NF-κB activation with LMP1 in B cell but negatively affect NF-κB in NPC cells (34, 35).
EBV infection of primary resting B cells causes dramatic epigenetic reprogramming and the establishment of immortalized lymphoblastoid cell lines (LCLs) (36, 37). Despite EBV’s well-defined roles in B-cell lymphomas and the likelihood that it is similarly important in NPC oncogenesis (38), there is considerably less knowledge of EBV’s role in NPC histone modifications. Epigenetic changes are reversible. Epigenetic modifiers are proven “druggable” targets in cancer treatment (39). Understanding of the NPC epigenetic landscape will likely allow us to identify unique therapeutics.
Results
NPC Epigenetic Landscapes.
To identify active NPC promoters and enhancers, H3K27ac chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) was used. Two untreated primary NPC biopsies from north Africa, EBV-positive NPC cell line C666-1 from untreated patient (40), and NPC xenografts C15, C17, and C18 were included in the study. C15 was from an untreated primary NPC, and C17 and C18 were from NPC metastases that had been treated with radiotherapy or chemotherapy (41). SV40 T antigen immortalized normal nasopharyngeal epithelial cell line NP69 and hTERT immortalized normal oral epithelial cell line NOK were used as normal controls (42, 43).
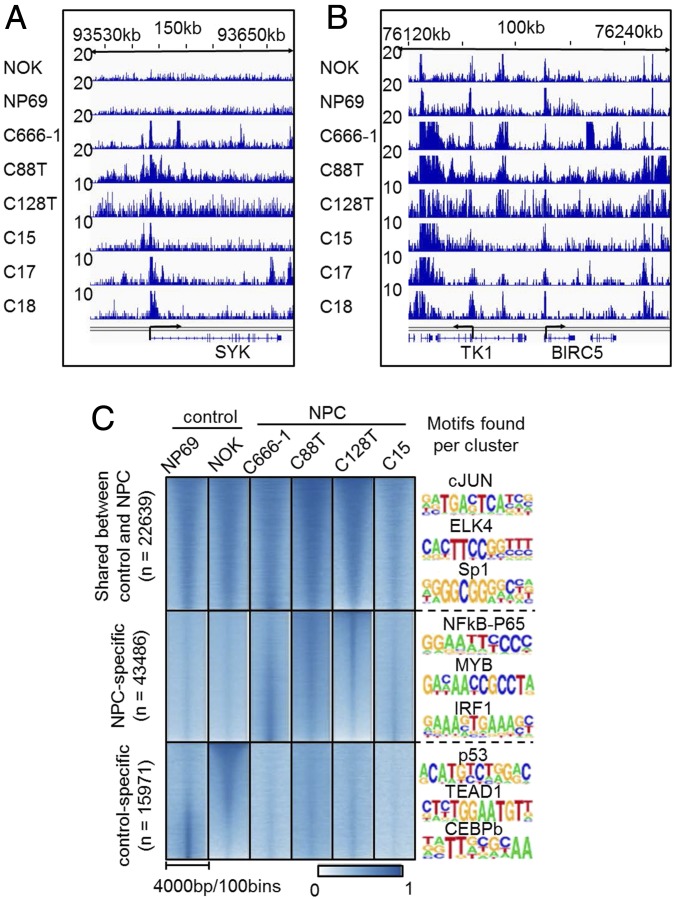
We initially surveyed the H3K27ac signals at genes known to be important for NPC. Significant H3K27ac signals were found at the genes previously shown to be important for NPC prognosis. For example, significant H3K27ac signals were found near the transcription start site (TSS) of SYK in NPC biopsies, NPC cell line, and NPC xenografts. In contrast, only background level signals were present at the control cell lines (Fig. 1A). SYK, a member of the nonreceptor tyrosine kinase family, is induced by LMP2A in NPC and epithelial cells (33). SYK signaling promotes epithelial cell proliferation. Abundant H3K27ac signals were also found at the BIRC5 (Survivin) promoter, even though the signals were also evident in the control cell lines (Fig. 1B). The H3K27ac signals at the gene body and neighboring region were also abundant. BIRC5 is an LMP1-induced prosurvival gene (44).
Fig. 1.
NPC epigenetic landscape. H3K27ac ChIP-seqs were done in control cells, NOK and NP69; NPC cell line, C666-1; primary NPC biopsies, C88T and C128T; and NPC xenografts, C15, C17, and C18. ChIP-seq reads were mapped to the human genome and significant H3K27ac peaks were identified using MACS. (A and B) H3K27ac ChIP-seq signals at SYK and BIRC5 (Survivin) loci. SYK and BIRC5 TSSs are indicated. The height of ChIP-seq signals are indicated on the Left of each track. (C) Heatmap of H3K27ac signals at sites shared by NPC samples and controls, sites unique for NPC samples, and sites unique for controls. Homer was used to identify TF motifs enriched for each cluster. Enriched motifs for each cluster are listed on the Right of the heatmaps.
Model-based analysis for ChIP-seq (MACS) was used to systemically identify significant peak from the NPC samples and the control samples. We compared the primary NPC tumors C128T, C88T, C666-1, and C15 with the controls NOK and NP69. In total, 22,639 peaks were shared with NPC and controls; 43,486 peaks were unique to NPC samples; and 15,971 peaks were unique to controls (Fig. 1C). HOMER was then used to identify the motifs enriched in these sites with high H3K27ac signals. De novo motifs enriched in shared peaks included AP1, ELK4, Sp1, NRF1, EGR1, YY1, and ZNF143 (Fig. 1C and Table S1, motifs shared by NPC and controls). De novo motifs enriched in NPC unique peaks include NF-κB subunit RELA, p50, p52; ETS family members ETS1; IFN-responsive factors 1 and 2; MYB; and insulator CTCT (Fig. 1C and Table S1, motifs enriched in NPC samples). De novo motifs enriched in the control unique peaks include AP-1 family basic leucine zipper transcription factors (TFs), and more importantly, TP53 tumor suppressor (Fig. 1C and Table S1, motifs enriched in control samples). The enrichment of NF-κB motifs in NPC unique peaks reflected the high NF-κB state in NPCs, likely activated by LMP1 or mutations in its activation pathways (45).
Table S1.
Motifs shared by NPC and controls, motifs enriched in NPC samples, and motifs enriched in control samples
 |
NPC Super-enhancers.
Super-enhancers (SEs) are enhancer clusters critically important in development, differentiation, and oncogenesis (46). SEs are cooccupied by many TFs, cofactors such as mediator MED1, histone modification-related proteins such as epigenetic reader bromodomain- containing protein 4 (BRD4), and basal TFs, accompanied by extraordinarily broad and high ChIP-seq signals for active enhancer marks including H3K27ac (46, 47). SE formation is driven by master TF’s binding to their recognition sites and the recruitment of their associated cofactors. We recently reported that EBV-encoded oncogenic TFs and virus-activated NF-κB can form SEs in lymphoblastoid cells (38). Fusion TFs resulting from chromosome translocation can also form SEs (48). Genomic amplifications, mutations in enhancer region, changes in 3D genome organization, or tumor virus integration all can lead to cancer cells acquiring new SEs (49–51). SEs are more sensitive to perturbations than typical enhancers (TEs) (47, 52). Small-molecule inhibitors including BRD4 inhibitor JQ1, TFIIH subunit CDK7 inhibitor THZ1, and mediator-associated CDK8 inhibitor cortistatin A all can preferentially inactivate SEs and stop cell growth (38, 47, 53, 54). SEs control the expression of key oncogenes including MYC (38, 47).
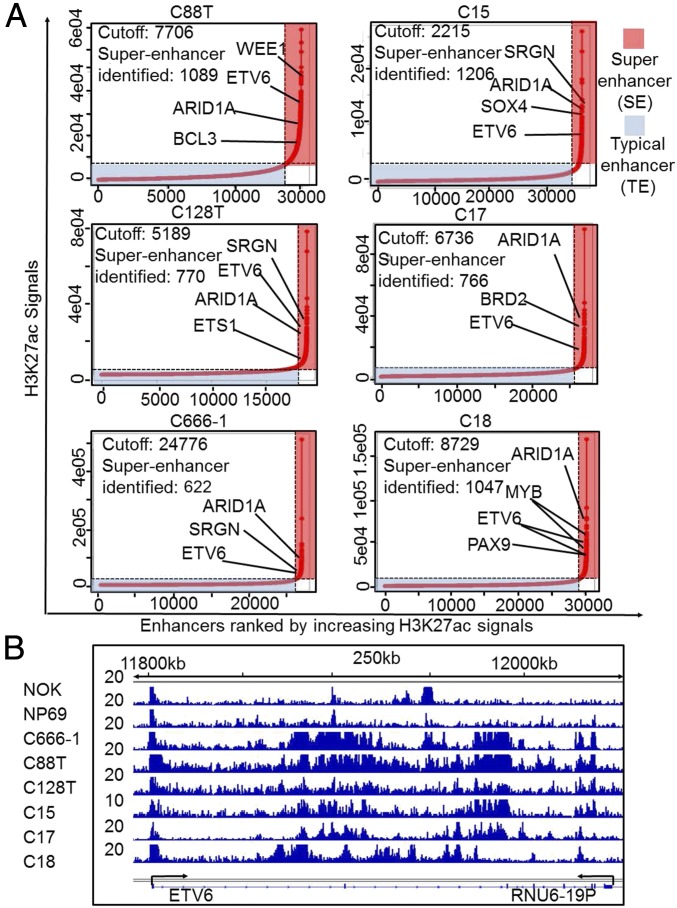
To identify NPC-specific SEs, H3K27ac signals from sliding windows of 12.5-kb genomic regions containing enhancers for NPC biopsies, NPC cell line, NPC xenografts, and control cell lines were ranked (38, 46). Enhancers with greater than fourfold H3K27ac ChIP-seq signals than the rest of the enhancers were assigned as SEs. In total, 1,089 and 770 SEs were identified for two NPC biopsies; 622 SEs were identified for C666-1; 1,206, 766, and 1,047 SEs were identified for C15, C17, and C18 NPC xenografts; and 767 and 622 SEs were identified for NOK and NP69 cells (Fig. 2A). These SEs were then assigned to their nearest genes. We identified 54 common SEs in four untreated NPC samples. These 54 NPC SE-associated genes were compared with the SEs identified from NP69 and NOK cells. Twenty NPC SEs were absent in normal immortalized epithelial cells. These genes include ETV6 (Fig. 2B), a member of the ETS transcription factor family which functions as a transcription repressor. ETV6 is frequently involved in chromosomal translocations in various hematopoietic malignancies (55).
Fig. 2.
NPC SEs. (A) H3K27ac signals within 12.5-kb window at significant peaks were ranked for each of the NPC samples. Peaks with four times higher H3K27ac signals than the rest of the peaks were assigned as SEs (marked in red box). The rest of the enhancers were marked in blue box. Each enhancer was assigned to its nearest gene. Genes important for growth and survival are indicated. (B) NPC ETV6 SE. H3K27ac signals at ETV6 are shown.
NPC SEs Were Sensitive to BRD4 Inhibition.
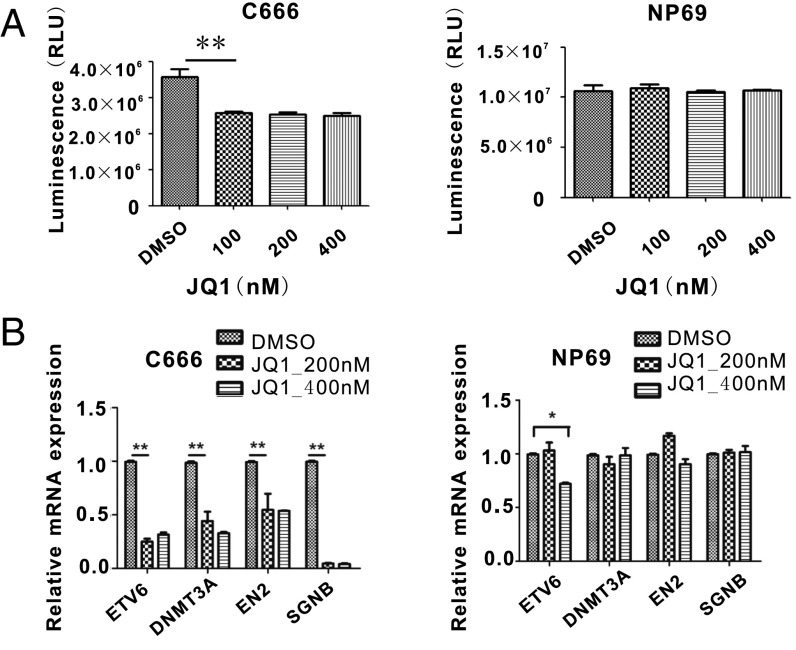
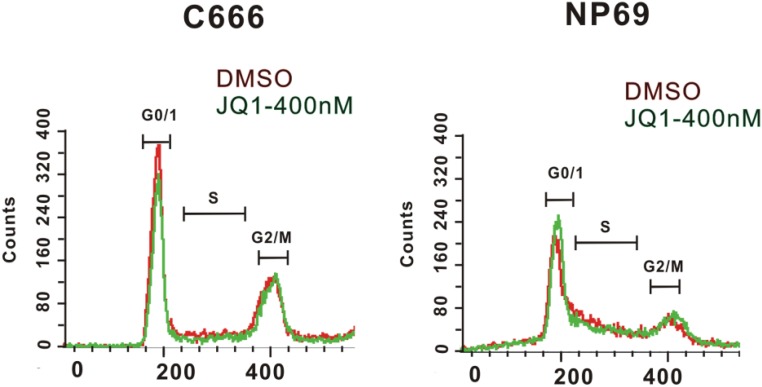
Chromatin reader BRD4 binds to H3K27ac and recruits CCNT1/CDK9 to phosphorylate RNA Pol II C-terminal serine 5 residue, thus releasing the paused Pol II to enable transcription elongation. BRD4 is an important component of SEs in many cancers (47). JQ1, a small-molecule inhibitor, blocks BRD4 binding to H3K27ac. SEs are more sensitive to JQ1 perturbation than TEs (47). JQ1 treatment greatly reduces LCL growth and MYC expression (38). We tested whether JQ1 treatment has similar effects on C666-1. JQ1 treatment of C666-1 cell reduced cell growth while it had no effect on NP69 cell growth (Fig. 3A). JQ1 treatment reduced the C666-1 cells in G0/G1 and S phase (Fig. S1). JQ1 treatment significantly down-regulated the expression of NPC SE-associated genes ETV6, DNMT3A, EN2, and SGNB in C666-1 cells (Fig. 3B). JQ1 treatment had no effect on DNMT3A, EN6, and SGNB expression in control NP69 cells. Higher dose of JQ1 treatment also reduced the ETV6 gene expression in the control NP69 cells. However, the JQ1 effect on C666-1 for ETV6 was stronger. At lower dosage, JQ1 treatment had no effect on ETV6 in NP69 cells but had a significant effect in C666-1 cells.
Fig. 3.
NPC cells are susceptible to BRD4 inhibition. (A) C666-1 and NP69 cells were treated with indicated JQ1 with vehicle as control for 3 d. CellTiter-Glo luminescent was used to determine the cell viability. C666-1 cells treated with JQ1 grow significant slower than DMSO-treated cell (P < 0.01). JQ1 does not slow NP69 cell growth in the same time course. (B) Total RNAs from C666-1 and NP69 cells treated with JQ1 or vehicle were analyzed by qRT-PCR for ETV6, DNMT3A, EN2, and SGNB expression, normalized against GAPDH. DMSO-treated cells were set to 1. *P < 0.05, **P < 0.01.
Fig. S1.
JQ1 treatment decreased C666-1 cells in G0/G1 and S phase. C666-1 and NP69 cells were treated with 400 nM JQ1 for 3 d. Cells were stained with propidium iodide and analyzed by FACS.
NPC SE-Driven ETV6 Correlated with Poor Prognosis.
SE was found in ETV6 introns and coding region in all NPC samples. In contrast, no SE was found in control cell lines. Since ETV6 is known to be critical in oncogenesis, its expression level was further evaluated in NPC patient samples.
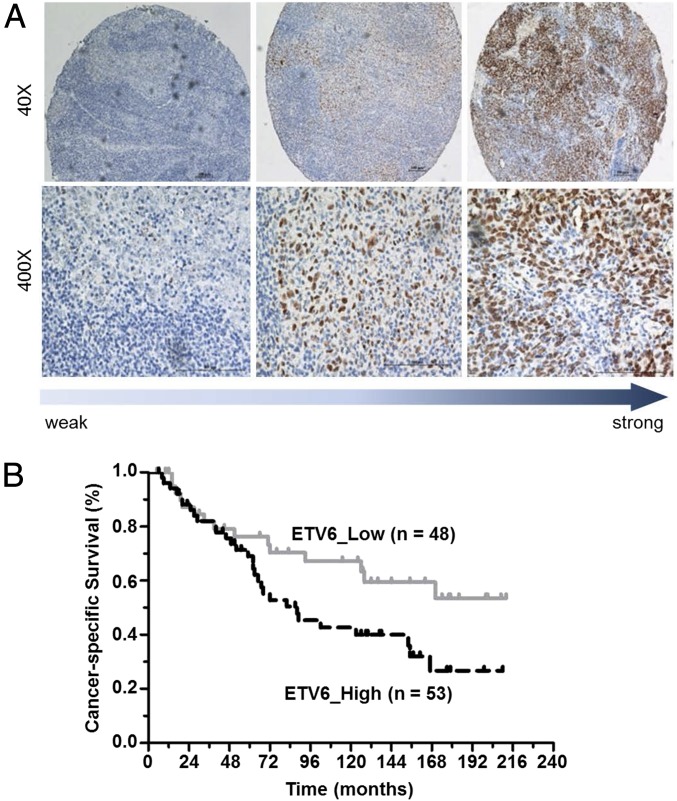
ETV6 immunohistochemistry (IHC) staining was used to determine ETV6 expression levels in 101 primary NPC biopsies. ETV6 protein was detected in 85 of 101 (85%) primary NPC tissues. Most of the ETV6 staining was found in the nuclei (Fig. 4A). The intensity of ETV6 staining was determined based on the medium value of IHC score. High ETV6 expression was found in 53 tumor tissues (52%). To correlate ETV6 expression with prognosis, patients were classified into ETV6 high and low groups accordingly. The clinical and demographic data between the two groups were not significantly different (Table S2). However, the level of ETV6 was negatively correlated with cancer-specific survival rate of NPC patients (P = 0.041). Moreover, multivariate analysis showed that age, T stage, N stage, TNM stage, and ETV6 expression were independent prognostic factors of overall survival for patients with NPC (Table S3).
Fig. 4.
ETV6 expression correlates with poor survival in NPC patients. ETV6 immunohistochemistry staining was used to determine the ETV6 protein expression levels in primary NPC samples. (A) Expression levels of ETV6 in low-, medium-, and high-expression group. (B) Kaplan–Meier cancer-specific survival curve for NPC patients with high or low ETV6 expression. High expression, n = 53; low expression, n = 48.
Table S2.
Distribution of patient demographic and clinical characteristics before treatment
| Characteristics | ETV6_Low (n = 48) | ETV6_High (n = 53) | P* | ||
| No. | % | No. | % | ||
| Age, y | 0.153 | ||||
| Median | 49.1 | 45.6 | |||
| Range | 24–70 | 18–71 | |||
| Sex | 0.143 | ||||
| Male | 41 | 85.4 | 39 | 73.6 | |
| Female | 7 | 14.6 | 14 | 26.4 | |
| Histology, WHO type† | 1.000 | ||||
| II | 3 | 6.3 | 3 | 5.7 | |
| III | 45 | 93.7 | 50 | 94.3 | |
| T stage‡ | 0.543 | ||||
| 1 | 5 | 10.4 | 8 | 15.1 | |
| 2 | 24 | 50.0 | 19 | 35.8 | |
| 3 | 12 | 25.0 | 17 | 32.1 | |
| 4 | 7 | 14.6 | 9 | 17.0 | |
| N stage‡ | 0.383 | ||||
| 0 | 12 | 25.0 | 12 | 22.6 | |
| 1 | 22 | 45.8 | 26 | 49.1 | |
| 2 | 13 | 27.1 | 10 | 18.9 | |
| 3 | 1 | 2.1 | 5 | 9.4 | |
| Clinical stage‡ | 0.879 | ||||
| I | 1 | 2.1 | 1 | 1.9 | |
| II | 20 | 41.7 | 19 | 35.8 | |
| III | 18 | 37.5 | 20 | 37.7 | |
| IVa-b | 9 | 18.8 | 13 | 24.5 | |
| Dose of RT, Gy, mean ± SD | |||||
| NPDT | 69.94 ± 3.8 | 69.87 ± 4.2 | 0.264 | ||
| LNDT | 59.68 ± 5.5 | 61.43 ± 7.2 | 0.842 | ||
| NACT | 0.482 | ||||
| 0 | 39 | 81.2 | 40 | 75.5 | |
| 1 | 9 | 18.8 | 13 | 24.5 | |
| CCRT | 0.057 | ||||
| 0 | 41 | 85.4 | 51 | 96.2 | |
| 1 | 7 | 14.6 | 2 | 3.8 | |
Abbreviations: CCRT, concurrent chemoradiotherapy; NACT, new adjuvant chemotherapy; RT, radiotherapy.
P values were calculated using χ2 test or independent Student’s t test based on types of variables.
II, poorly differentiated nonkeratinizing carcinoma; III, undifferentiated nonkeratinizing carcinoma.
According to the seventh edition of the AJCC staging system; I, T1N0M0; II, T2N0M0, T1N1M0; II, T3N0-2M0, T1-2N2M0; IVa-b, T4N0-2M0, T1-4N3M0.
Table S3.
Multivariate analysis of prognostic factors on cancer-specific survival of patients with nasopharyngeal carcinoma
| Variable | HR | 95% CI for HR | P* |
| Age | 1.031 | 1.003–1.060 | 0.032 |
| Gender | 1.214 | 0.539–2.734 | 0.640 |
| NPDT | 1.023 | 0.967–1.083 | 0.422 |
| LNDT | 0.965 | 0.910–1.023 | 0.226 |
| NACT | 1.114 | 0.487–2.550 | 0.798 |
| CCRT | 0.578 | 0.152–2.194 | 0.420 |
| Type of histopathology | 0.443 | 0.102–1.917 | 0.276 |
| T stage† | 2.305 | 1.121–4.741 | 0.023 |
| N stage† | 2.039 | 1.085–3.830 | 0.027 |
| TNM stage† | 0.381 | 0.163–0.893 | 0.026 |
| ETV6 | 2.310 | 1.204–4.433 | 0.012 |
P values less than 0.05 are indicated in bold. Abbreviations: 95% CI, 95% confidence interval; HR, hazard ratio; LNDT, radiotherapy dose to lymph nodes; NPDT, radiotherapy dose to nasopharyngeal.
P values were calculated using adjusted Cox proportional hazards model. The following parameters were included in the Cox proportion hazard model by enter method: age (continuous variable), gender (male vs. female), NPDT (continuous variable), LNDT (continuous variable), use of induction chemotherapy (with vs. without), use of chemoradiotherapy (with vs. without), type of histology (WHO III vs. II), T stage (T1–4), N stage (N0–3), TNM stage (I–IVb), particularly, ETV6 expression (high vs. low) was included in the Cox proportion hazard model.
According to the seventh edition of the Union for International Cancer Control/American Joint Committee on Cancer staging system.
Epigenetic Profiles of EBV Genome in NPC.
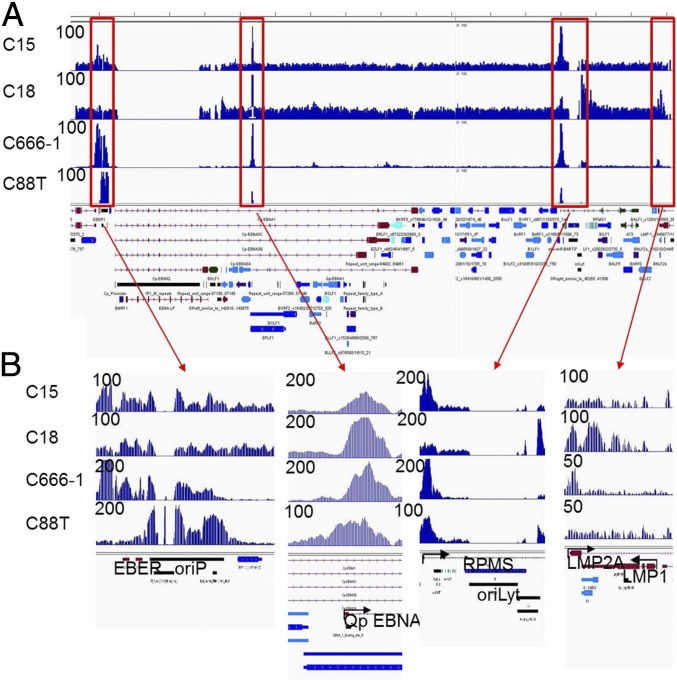
Histone modifications of EBV genome are evident in LCLs. Very high H3K4me1, H3K4me3, and H3K27ac signals are present at the LMP1, LMP2, Cp, Ori-P, and the BART miRNA promoters (36). Enrichment of H3ac, H4ac, and H3K4me2 was found at the LMP2A promoter in C666-1 cells (56). However, no genome-wide histone modification profiles of EBV genome in primary human tumors were reported. It has also been shown that the EBV gene expression profiles vary greatly in NPC (20). The most consistently expressed genes include EBNA1, LMP2A, EBERs, and the BART miRNAs (20). To determine the NPC EBV H3K27ac profiles, ChIP-seq reads from NPC samples were mapped to the EBV genome. In total, 0.12% (27,529), 0.19% (44,728), 0.08% (16,042), and 0.05% (18,508) reads from C15, C18, C666-1, and C88T were mapped to the EBV genome. Significantly fewer reads were mapped to C128T. Among the four NPCs with abundant EBV reads, the EBNA1 Q-promoter was the most consistently activated in all four cancers (Fig. 5). Abundant H3K27ac signals were also found near the EBV BART miRNAs. In C15, H3K27ac signals were also high near ori-P and EBER. Only a small peak was found near the LMP1 promoter. C18, in contrast, had high H3K17ac signals at lytic origin of replication, ori-Lyt. This is very interesting because among the NPC xenografts, C18 is the one where EBV latency is the less stringent with easy induction of BZLF1 and BRLF1 expression (57). C666-1 had very high H3K27ac signals at ori-P and EBERs and lower signals at LMP2A and ori-Lyt, in agreement with LMP2A message level and gp350 protein expression by immune chemistry (40). C88T had abundant H3K27ac signals at ori-P and very little signals at LMP1 and LMP2A. These data indicated that, even though it is believed that NPCs are very homogeneous population-wise, the EBV epigenetic landscapes varied significantly from patient to patient.
Fig. 5.
Epigenetic landscape of EBV genome in NPC. H3K27ac ChIP-seq reads were mapped to the genome of Akata EBV stain. (A) Overall view of the EBV genome. Sample names are indicated on the Left of the tracks. Peak heights are listed next to each track on the Left. EBV genome annotation is under the tracks. (B) Zoom-in view of major peaks.
Discussion
NPC patients respond well to radiation therapy and chemotherapy, especially patients with early-stage cancer. However, early diagnosis is difficult to achieve. Patients with late-stage cancer or metastases have much poorer prognosis. Even among patients who are cured, late secondary effects remain a major concern, especially xerostomia. New therapies are therefore needed to improve NPC patient overall survival. Epigenetic alterations in cancer cause oncogene overexpression and tumor suppressor silencing, without affecting the DNA sequences. These epigenetic alterations are usually reversible, possibly by small-molecule inhibitors targeting epigenetic enzymes. These epigenetic enzyme inhibitors may inactivate oncogenes or restore the expression of tumor suppressor to halt cancer cell growth. The cancer cell’s addiction to epigenetic alterations can also be targeted to selectively kill these cancerous cells. Currently, histone deacetylase inhibitors are used in clinical treatment of various cancers and many other unique inhibitors are in clinical trials.
Expression profilings using microarrays or RNA-seqs identified global NPC-specific expression changes that were critical for NPC pathogenesis. Here, we used a unique approach to assess the NPC global histone epigenetic landscapes. Our findings not only further confirmed many known NPC biomarkers but also identified unique NPC markers. A large number of NPC unique enhancers were identified by this approach. Many of them are located in the intergenic regions. Their directly associated genes will be identified by assays interrogating the 3D genome organization in future analyses.
It is believed that master pioneering TFs first bind to DNA and subsequently recruit histone-modifying enzymes to alter the epigenetic landscapes. These TFs are likely to be critically important for the formation of active enhancer sites. The TF motifs enriched at the enhancers shared by NPC and normal controls cell lines included TFs involved in diverse biologic processes. These TFs are likely to be important for nasopharyngeal epithelial cells. Interestingly, both YY1 and ZNF143 motifs were enriched in the group. These proteins are important for the 3D genome organization, suggesting that long-range enhancer promoter interactions are also likely important for these cells. Strikingly, TP53 tumor suppressor motif was highly enriched in the control cells. TP53 mutations are detected in a subset of NPCs, especially at late stages, but their frequency is low by comparison with most other head and neck carcinomas (about 10% in contrast with more than 60%) (45, 58). However, in malignant NPC cells, the TP53 protein is likely to be functionally inactivated by mechanisms which are not yet elucidated. The motifs enriched at the NPC unique cluster included NF-κB, IRF1, and MYB. Activation of NF-κB pathways is now recognized as the most consistent alteration in the signaling landscape of NPC cells. Interestingly, it is achieved by various mechanisms which are either dependent on viral factors (mainly LMP1 expression) or dependent on cellular gene alterations (45). Mutations in NF-κB negative regulators including CYLD, TRAF3, and NFKBIA were found in 41% of NPC samples, and these mutations and LMP1 expression were mutually exclusive (45). EBV LMP2 is more consistently expressed in NPC and may be involved in NF-κB activations. IRF1 is involved in the oncogenesis of various cancers (59). MYB is involved in the pathogenesis of several different cancers including some leukemia, colon, and breast cancers (60). MYB can be activated by SEs formed by translocations in adenoid cystic carcinoma (61).
SEs have great impact on cancer cell growth and survival (47). SEs are much larger complexes than TEs and are more susceptible to perturbations. Several small-molecular inhibitors of SE constituents are effective in killing different cancer cells (38, 47, 53). Here, we found NPC SEs were susceptible to perturbation and targeted an important oncogene, ETV6. ETV6 is an ETS family TF. ETV6 is frequently involved in different chromosome translocations that result in the formation of different fusion genes in different cancers. For example, ETV6-NTRK3 is found in salivary gland tumor and secretory breast carcinoma (62, 63). ETV6-RUNX1 fusion can promote the survival of acute lymphoblastic leukemia cells (64). Overexpression of ETV6 also correlates with poor prognoses for non–small-cell lung cancer (65).
EBV genomes persist as episomes in cancer cells. In LCLs, EBV genomes are chromatinized, and histone modifications related to active transcription, including K3K4me1, H3K4me3, and H3K27ac, are evident at EBV episomes in regions where active transcription occur (36). Similarly, Karposi sarcoma-associated herpes virus persists in Karposi sarcoma as chromatinized episome (66). In NPC, the most consistent H3K27ac signals were EBNA1 Qp, EBERs, ori-P, and miRNA, consistent with their near ubiquitous expression or activity among NPC samples. Lack of H3K27ac signals at the LMP1 promoter or gene body in C666-1 is consistent with previous findings that LMP1 is not detectable at protein level in these cells (67).
Our study suggested that epigenetic profiling can be used to effectively identify NPC biomarkers. We also found that NPC cell line was susceptible to histone modification reader, BRD4 inhibition. Identification of altered histone modification enzymes (writers, readers, and erasers) in NPC will likely enable the development of unique therapeutics.
Materials and Methods
H3K27ac ChIP-seq (38) was used to identify NPC SEs from NPC cell line C666-1, normal oral keratinocytes (NOKs) and nasopharynx keratinocytes (NP69), NPC xenografts, and primary NPC tissues. For IHC, the primary NPC tissues was stained (68) using the anti-ETV6 antibody. The intensity and percentage of IHC staining was scored by two pathologists independently. The primary NPC tissues for tissue microarray were obtained from the Department of Pathology of Sun Yat-sen University Cancer Center (SYSUCC), with the approval of SYSUCC IRB and prior patient consents. More details are provided in SI Materials and Methods.
SI Materials and Methods
Cell Lines, NPC Xenografts, and Primary NPC Tissues.
C666-1 cells were grown in RPMI medium 1640 supplemented with 10% (vol/vol) FBS (Gibco) and 2 mM l-glutamine. Human telomerase (hTERT)-immortalized normal oral keratinocytes (NOKs) and SV40 T antigen immortalized nasopharynx keratinocytes (NP69) cells were cultured in keratinocyte serum-free medium supplemented with human epidermal growth factor and bovine pituitary extract (Life Technologies). NPC xenografts were grafted in athymic mice. Primary NPC tissues from Tunisian patients were collected under the supervision of the scientific board of the “South Tunisian Association for Investigations of Nasopharyngeal Carcinomas” and were histopathologically confirmed by a certified pathologist.
H3K27ac ChIP-Seq.
C666-1, NOK, and NP69 cells were cross-linked with 1% formaldehyde and lysed. Xenografts and primary NPC tissues were pulverized in liquid nitrogen with mortar. Tissue powder was cross-linked and lysed. Genomic DNA was then sonicated into 500-bp fragments using ultrasonicator in lysis buffer. After clearing the debris, the supernatants were diluted and the lysates were incubated the anti-H3K27ac antibody (ab4729; Abcam) overnight at 4 °C. Protein A agarose beads were used to capture protein–DNA complexes. After extensive wash, protein–DNA complexes were eluted and reverse cross-linked. DNA was purified with Qiagen PCR purification kit. Ten nanograms of DNA were used for library-prep using NEBNext ChIP-Seq library preparation kit for Illumina (NEB). The sequencing libraries were analyzed by bioanalyzer (Agilent Technologies) and quantified by qPCR for quality control. The libraries were sequenced using an Illumina HiSeq 2000. Inputs from each samples were sequenced together using different bar codes. Sequencing data are available upon request.
ChIP-Seq Analyses and Identification of SEs.
ChIP-seq reads were mapped using Botie (38). MACS was used to identify the H3K27ac peaks (38). SEs were identified as previously described (38).
JQ1 Treatment of C666-1 Cells.
C666-1 cells were seeded on day 0 and treated with JQ1 or DMSO for indicated concentration and time on day 1. CellTiter-Glo Luminescent Cell Viability Assay was used to detect the cell viability.
qRT-PCR.
RNAs of C666-1 cells treated with JQ1 or DMSO for 24 h were isolated using Purelink RNA Mini Kit (Life Technologies), followed by qRT-PCR using RNA-TO-CT 1-STEP SYBR kit (Applied Biosystems). The expression of DMSO-treated cells was set to 1.
IHC Staining.
The tissue microarray was constructed and stained as previously described (68). Briefly, antigen retrieval of tissue microarray was achieved in pH 8.0 EDTA solution after deparaffinating and hydrating; the anti-ETV6 antibody (H00002120-M01; Abnova) in a concentration of 1:50 was used to detect the expression of ETV6 in primary NPC tissues. The intensity and percentage of IHC staining was scored by two pathologists independently using semiquantitative immunoreactive score as previously described (68). The intensity of the color reaction was scored as follows: no staining, 0; weak staining, 1; moderate staining, 2; strong staining, 3. The percentage of stained cells was scored as follows: 0%, 0; 1–10%, 1; 11–50%, 2; 51–80%, 3; 81–100%, 4. The average value from the two referees was used as the final score. The IHC score was then generated by the following: intensity score × percentage score.
Statistics Analysis.
Comparisons between groups were performed using χ2 analysis or independent Student’s t test as indicated. Kaplan–Meier survival analysis was performed and log-rank test was used to calculate accurate survival rate using Statistical Product and Service Solutions (SPSS 23.0). Multivariate analyses with an adjusted Cox proportional hazards model were used to identify significant independent variables.
Acknowledgments
We thank Dr. Zhen Lin (Tulane University) for the Akata EBV annotation gtf file. B.Z. is supported by National Institute of Allergy and Infectious Diseases Grant R01AI123420; E.K. is supported by National Cancer Institute Grants R01CA047006, R01CA170023, and R01CA085180; X.G. is supported by International Cooperation Project of Science and Technology Plan of Guangdong Province 2014A050503033 and 2016A050502011; P.B. is supported by the Bristol-Myers Squibb Foundation for Immunology Research (2017); and M.Z. is supported by National Natural Science Foundation of China Grant 81520108022.
Footnotes
The authors declare no conflict of interest.
Data deposition: The samples were collected before 2015; therefore, we do not have patient consent to deposit the data into a database. Data are available upon request.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705236114/-/DCSupplemental.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Raab-Traub N. Nasopharyngeal carcinoma: An evolving role for the Epstein-Barr virus. Curr Top Microbiol Immunol. 2015;390:339–363. doi: 10.1007/978-3-319-22822-8_14. [DOI] [PubMed] [Google Scholar]
- 3.Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:421–429. doi: 10.1016/s1044579x02000858. [DOI] [PubMed] [Google Scholar]
- 4.Spano JP, et al. Nasopharyngeal carcinomas: An update. Eur J Cancer. 2003;39:2121–2135. doi: 10.1016/s0959-8049(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 5.Loong HH, Ma BB, Chan AT. Update on the management and therapeutic monitoring of advanced nasopharyngeal cancer. Hematol Oncol Clin North Am. 2008;22:1267–1278. doi: 10.1016/j.hoc.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 7.Raab-Traub N. Novel mechanisms of EBV-induced oncogenesis. Curr Opin Virol. 2012;2:453–458. doi: 10.1016/j.coviro.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 9.Bei JX, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42:599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 10.Lu SJ, et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346:470–471. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 11.Niller HH, Wolf H, Minarovits J. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin Cancer Biol. 2009;19:158–164. doi: 10.1016/j.semcancer.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Lo KW, et al. Hypermethylation of the p16 gene in nasopharyngeal carcinoma. Cancer Res. 1996;56:2721–2725. [PubMed] [Google Scholar]
- 13.Dai W, et al. Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma. Cancer Med. 2015;4:1079–1090. doi: 10.1002/cam4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai CN, Tsai CL, Tse KP, Chang HY, Chang YS. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc Natl Acad Sci USA. 2002;99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong ZT, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 17.Song LB, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin DC, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 19.Suvà ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta S, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 21.Wang HY, et al. Eight-signature classifier for prediction of nasopharyngeal [corrected] carcinoma survival. J Clin Oncol. 2011;29:4516–4525. doi: 10.1200/JCO.2010.33.7741. [DOI] [PubMed] [Google Scholar]
- 22.Hu L, et al. Comprehensive profiling of EBV gene expression in nasopharyngeal carcinoma through paired-end transcriptome sequencing. Front Med. 2016;10:61–75. doi: 10.1007/s11684-016-0436-0. [DOI] [PubMed] [Google Scholar]
- 23.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 24.Frappier L. Role of EBNA1 in NPC tumourigenesis. Semin Cancer Biol. 2012;22:154–161. doi: 10.1016/j.semcancer.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Marquitz AR, Raab-Traub N. The role of miRNAs and EBV BARTs in NPC. Semin Cancer Biol. 2012;22:166–172. doi: 10.1016/j.semcancer.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Y, et al. Serological mass survey for early detection of nasopharyngeal carcinoma in Wuzhou City, China. Int J Cancer. 1982;29:139–141. doi: 10.1002/ijc.2910290204. [DOI] [PubMed] [Google Scholar]
- 27.Tang LQ, et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2015;108:djv291. doi: 10.1093/jnci/djv291. [DOI] [PubMed] [Google Scholar]
- 28.Mosialos G, et al. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 29.Cahir-McFarland ED, et al. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol. 2004;78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meckes DG, Jr, Menaker NF, Raab-Traub N. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol. 2013;87:1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tworkoski K, Raab-Traub N. LMP1 promotes expression of insulin-like growth factor 1 (IGF1) to selectively activate IGF1 receptor and drive cell proliferation. J Virol. 2015;89:2590–2602. doi: 10.1128/JVI.02921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–698. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- 33.Fotheringham JA, Coalson NE, Raab-Traub N. Epstein-Barr virus latent membrane protein-2A induces ITAM/Syk- and Akt-dependent epithelial migration through αv-integrin membrane translocation. J Virol. 2012;86:10308–10320. doi: 10.1128/JVI.00853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guasparri I, Bubman D, Cesarman E. EBV LMP2A affects LMP1-mediated NF-kappaB signaling and survival of lymphoma cells by regulating TRAF2 expression. Blood. 2008;111:3813–3820. doi: 10.1182/blood-2007-03-080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart S, et al. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc Natl Acad Sci USA. 2004;101:15730–15735. doi: 10.1073/pnas.0402135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arvey A, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA. 2011;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, et al. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe. 2015;17:205–216. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: The challenge of hitting the mark. J Clin Invest. 2014;124:64–69. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheung ST, et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer. 1999;83:121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 41.Busson P, et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer. 1988;42:599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 42.Tsao SW, et al. Establishment of two immortalized nasopharyngeal epithelial cell lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochim Biophys Acta. 2002;1590:150–158. doi: 10.1016/s0167-4889(02)00208-2. [DOI] [PubMed] [Google Scholar]
- 43.Piboonniyom SO, et al. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 2003;63:476–483. [PubMed] [Google Scholar]
- 44.Ai MD, et al. Regulation of survivin and CDK4 by Epstein-Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cell lines. Cell Res. 2005;15:777–784. doi: 10.1038/sj.cr.7290347. [DOI] [PubMed] [Google Scholar]
- 45.Li YY, et al. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat Commun. 2017;8:14121. doi: 10.1038/ncomms14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomazou EM, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015;10:1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dooley KE, Warburton A, McBride AA. Tandemly integrated HPV16 can form a Brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. MBio. 2016;7:e01446-16. doi: 10.1128/mBio.01446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelish HE, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Braekeleer E, et al. ETV6 fusion genes in hematological malignancies: A review. Leuk Res. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Gerle B, et al. Acetylated histone H3 and H4 mark the upregulated LMP2A promoter of Epstein-Barr virus in lymphoid cells. J Virol. 2007;81:13242–13247. doi: 10.1128/JVI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng WH, Israel B, Raab-Traub N, Busson P, Kenney SC. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 2002;62:1920–1926. [PubMed] [Google Scholar]
- 58.Effert P, et al. Alterations of the p53 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3768–3775. doi: 10.1128/jvi.66.6.3768-3775.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, et al. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 2007;67:2535–2543. doi: 10.1158/0008-5472.CAN-06-3530. [DOI] [PubMed] [Google Scholar]
- 60.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 61.Drier Y, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48:265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunt JL. An update on molecular diagnostics of squamous and salivary gland tumors of the head and neck. Arch Pathol Lab Med. 2011;135:602–609. doi: 10.5858/2010-0655-RAIR.1. [DOI] [PubMed] [Google Scholar]
- 63.Tognon C, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 64.Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood. 2011;118:4910–4918. doi: 10.1182/blood-2011-05-354266. [DOI] [PubMed] [Google Scholar]
- 65.Liang JZ, Li YH, Zhang Y, Wu QN, Wu QL. Expression of ETV6/TEL is associated with prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:2937–2945. [PMC free article] [PubMed] [Google Scholar]
- 66.Sun R, et al. Epigenetic landscape of Kaposi’s sarcoma-associated herpesvirus genome in classic Kaposi’s sarcoma tissues. PLoS Pathog. 2017;13:e1006167. doi: 10.1371/journal.ppat.1006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shair KH, Schnegg CI, Raab-Traub N. Epstein-Barr virus latent membrane protein-1 effects on junctional plakoglobin and induction of a cadherin switch. Cancer Res. 2009;69:5734–5742. doi: 10.1158/0008-5472.CAN-09-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li XJ, et al. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 2011;71:3162–3172. doi: 10.1158/0008-5472.CAN-10-3557. [DOI] [PubMed] [Google Scholar]