Significance
Increases in human life expectancy over the next century will be accompanied by increased frailty and massive and unsustainable health care costs. Developing means to extend the time that individuals remain healthy and free of age-related infirmities, called healthspan, has therefore become a critical goal of aging research. We show that small molecules produced by the microbiota and related to indole extend healthspan in geriatric worms, flies, and mice, without attendant effects on lifespan. Indoles act via the aryl hydrocarbon receptor and cause animals to retain a youthful gene expression profile. Indoles may represent a new class of therapeutics that improve the way we age as opposed to simply extending how long we live.
Keywords: C. elegans, aging, frailty, aryl hydrocarbon receptor, microbiota
Abstract
Multiple studies have identified conserved genetic pathways and small molecules associated with extension of lifespan in diverse organisms. However, extending lifespan does not result in concomitant extension in healthspan, defined as the proportion of time that an animal remains healthy and free of age-related infirmities. Rather, mutations that extend lifespan often reduce healthspan and increase frailty. The question arises as to whether factors or mechanisms exist that uncouple these processes and extend healthspan and reduce frailty independent of lifespan. We show that indoles from commensal microbiota extend healthspan of diverse organisms, including Caenorhabditis elegans, Drosophila melanogaster, and mice, but have a negligible effect on maximal lifespan. Effects of indoles on healthspan in worms and flies depend upon the aryl hydrocarbon receptor (AHR), a conserved detector of xenobiotic small molecules. In C. elegans, indole induces a gene expression profile in aged animals reminiscent of that seen in the young, but which is distinct from that associated with normal aging. Moreover, in older animals, indole induces genes associated with oogenesis and, accordingly, extends fecundity and reproductive span. Together, these data suggest that small molecules related to indole and derived from commensal microbiota act in diverse phyla via conserved molecular pathways to promote healthy aging. These data raise the possibility of developing therapeutics based on microbiota-derived indole or its derivatives to extend healthspan and reduce frailty in humans.
Recent advances in health care have contributed to a significant increase in life expectancy of individuals, especially in developed countries, which predict an expansion of geriatric populations by as much as 350-fold over the next 40 y (1). However, extension of lifespan is often accompanied by increased frailty, and attendant increases in global healthcare expenditures are expected to be both massive and unsustainable (2). Such data highlight the need to develop means to extend healthspan, which is broadly defined as the length of time that an individual remains healthy and free of age-related infirmities (3, 4).
Healthspan has often been convolved with lifespan, and extended healthspan has been associated with slowed onset of normal age-related changes (e.g., sarcopenia). Thus, mutations that extend lifespan might be expected to likewise extend healthspan. Recent studies in Caenorhabditis elegans indicate that, relative to wild-type animals, mutations that extend lifespan do indeed extend the period of youthfulness, in which animals are motile and resistant to bacterial infection (healthspan), but also extend the period of decrepitude or frailty, where animals are relatively immobile (5, 6) Other studies in C. elegans that take into account multiple measures of health, each normalized to maximal lifespan, indicate that mutations or conditions that extend lifespan minimally impact or even reduce healthspan, depending on the particular measure; however, all long-lived mutants increased the proportion of time spent in a frail state (1). Together, these data suggest that a comprehensive assessment of healthspan requires evaluation of survival curves together with additional criteria such as motility, tolerance to stressors, stem cell resilience, and fecundity, among others (3). Moreover, such criteria must be evaluated longitudinally since changes occur in each of these processes at different times and with different rates during normal aging.
Whereas significant information is available about the factors and mechanisms controlling lifespan, much less information is available regarding regulation of healthspan. The observation that extension of lifespan results in reduced periods of health (1) suggests that these processes are coordinately regulated. The question arises as to whether mechanisms exist that uncouple healthspan from lifespan and extend the former without affecting the latter.
Commensal microbiota facilitate nutrient metabolism, augment integrity of the intestinal epithelial barrier, enhance host immunity, and limit pathogen colonization (7–9). Shifts in commensal microbiota occur with aging and may contribute to infirmity (10). However, little is known about the mechanisms by which the microbiota regulates healthspan and frailty. Using C. elegans as a biosensor, we identified indole and several metabolites [e.g., indole-3-carboxaldehyde (ICA), indole acetic acid (IAA)] as molecules secreted by Escherichia coli that induce hormetic protection against stressors in worms, an effect mediated by factors controlling innate immunity and lifespan (11–14). In mammals, indoles derived from plant-based dietary sources, or produced by intestinal microbiota via tryptophanase (TnaA)-mediated catalysis of dietary tryptophan, attain millimolar concentrations in the intestinal tract, and derivatives can be detected throughout the body (15). Indoles regulate virulence in pathogenic bacteria, protect hosts from infection, and limit colitis induced by pathogens or chemical stressors (12, 16, 17).
Here, we show that indoles from commensal bacteria extend healthspan in diverse species and alter patterns of gene expression in aged animals to more closely resemble those seen in the young, effects mediated in part by the aryl hydrocarbon receptor (AHR).
Results
Indoles Derived from Commensal E. coli Extend Healthspan of C. elegans.
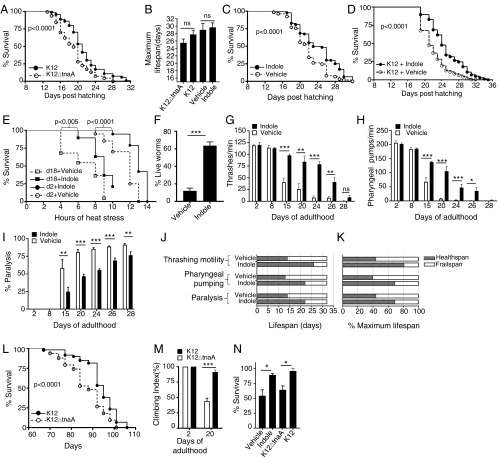
To investigate effects of indoles secreted by commensal bacteria on survival, wild-type C. elegans (N2) were grown on plates seeded with either an E. coli K12 variant (called K12) that produces and secretes indole and indole derivatives, or an isogenic mutant E. coli strain that contains a deletion in the tryptophanase gene (tnaA), which is required to convert tryptophan into indole (called K12ΔtnaA). K12 was selected for its capacity to colonize the intestinal tract and grow as a commensal in mice (18). Growing C. elegans on K12ΔtnaA, compared with K12, shifted the Kaplan–Meier survival curve to the left (Fig. 1A) but was without significant effect on maximal lifespan (Fig. 1B). E. coli OP50, the normal food source for C. elegans, which also produces indoles, induced a survival curve similar to that seen with K12 (Fig. S1A). Growth of animals on K12ΔtnaA supplemented with 250 μM indole, compared with K12ΔtnaA supplemented with vehicle (methanol), shifted the Kaplan–Meier survival curve to the right (Fig. 1C) but was without significant effect on maximal lifespan (Fig. 1B). The shift of the population survival curves with K12 or indole, even without an attendant increase in maximal lifespan, raised the possibility that indole produced by K12 augments healthspan. Notably, growth of animals on K12 supplemented with 100 μM indole further shifted the survival curve to the right compared with animals grown on K12 alone, and marginally extended maximal lifespan, suggesting that exogenous indole may provide a therapeutic effect even beyond that of commensal bacteria (Fig. 1D).
Fig. 1.
Commensal E. coli-derived indole extends lifespan and healthspan in C. elegans and Drosophila melanogaster. (A) Kaplan–Meier lifespan curves of wild-type C. elegans N2 on E. coli K12, which produces indole, or E. coli K12ΔtnaA, which does not. Graph represents seven independent experiments, n > 500 worms per condition). (B) Maximal lifespan obtained from at least three independent experiments comparing K12 vs. K12ΔtnaA or K12ΔtnaA plus indole (Indole) vs. K12ΔtnaA plus vehicle (Vehicle). (C) Lifespan curves of N2 on K12ΔtnaA supplemented with either 250 μM indole or vehicle (Methanol). Graph combines three independent experiments with n > 490 worms per condition. (D) Kaplan–Meier lifespan curves of N2 on K12 supplemented with either 100 μM indole or vehicle (n > 150 animals per condition). (E) Survival curves of N2 at 35 °C (day 2 or day 18) from K12ΔtnaA plus vehicle (Vehicle) or K12ΔtnaA plus indole (Indole). n = 25 animals per condition. (F) Survival of day 4 adult N2 animals at 35 °C grown in vehicle or indole without bacteria. (G) Thrashing motility in liquid in N2 adults grown on either K12ΔtnaA plus vehicle (Vehicle) or K12ΔtnaA plus indole (Indole), and monitored throughout life (n = 8–12 worms); (H) pharyngeal pumping rate per minute in N2 animals grown as in G (n = 8–12 worms); (I) paralysis events in N2 adults grown as in G and H (n = 200 worms per condition). (J) Healthspan and frail span measurements of N2 adults grown either on K12ΔtnaA-vehicle (Vehicle) or K12ΔtnaA-indole (Indole) conditions. Values for motility in liquid, pharyngeal pumping rate, and paralysis events were obtained from G, H, and I, respectively. (K) Healthspan values normalized with maximum lifespan set at 100%. (L) Lifespan of germ-free wild-type Drosophila melanogaster w1118 monoassociated with either K12 or K12ΔtnaA (n > 60 animals per condition). (M) Climbing assay of young (2 d), and older (20 d) adult germ-free w1118 flies monoassociated with either K12 or K12ΔtnaA (n > 30 animals per condition). (N) Heat stress resistance assay with conventionally raised streptomycin-treated w1118 flies, fed either K12, or K12ΔtnaA, or vehicle (Methanol) or 250 μM indole (n > 30 animals per condition). G–I and L–N are representative of at least two independent experiments. P values of percent survival curves were calculated with log-rank test. Values of B, F–I, and M and N represent mean values ± SEM, and t tests were performed to calculate P values. *P < 0.05; **P < 0.01; ***P < 0.001. Summary of the lifespan experiments are presented in Table S3.
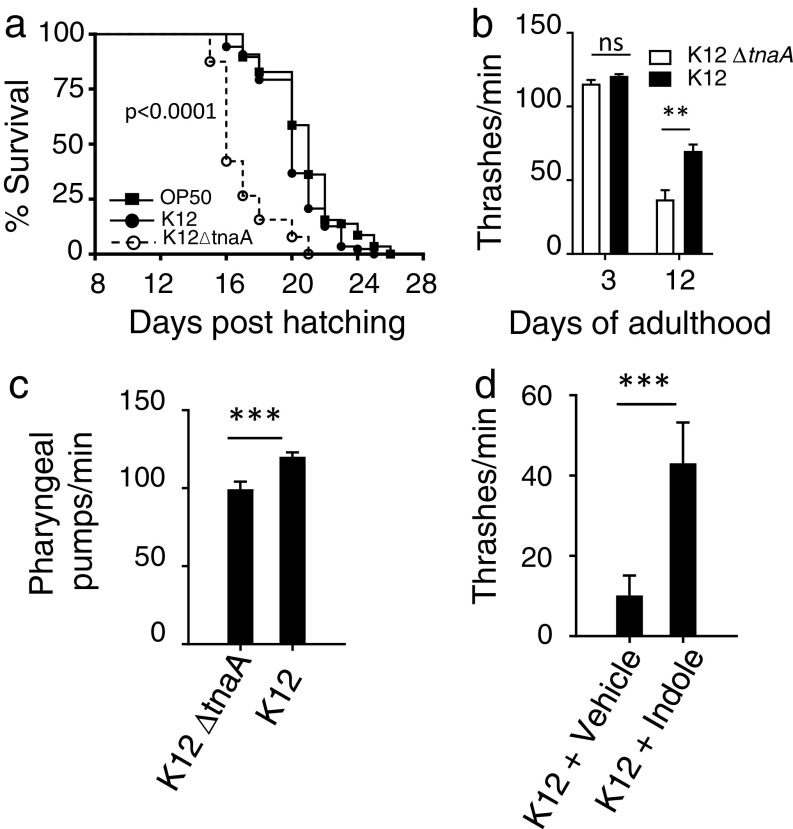
Fig. S1.
Effects of indoles on healthspan. (A) Lifespan curve of N2 with K12, K12ΔtnaA, or OP50 (n > 60 worms per condition; P < 0.0001 for K12ΔtnaA vs. other strains; differences between K12 and OP50 were not significant). (B) Thrashing motility in liquid (n = 8–12 worms per condition); (C) pharyngeal pumping rate per minute (day 12 adults, n = 8–12 worms per condition) of N2 adults grown on K12 or K12ΔtnaA. (D) Thrashing motility of N2 animals grown on K12 animals supplemented with 100 µM indole or vehicle (day 20 adults, n = 12 worms).
Table S3.
Summary of the lifespan experiments
| Figures | Conditions | Maximum lifespan + SEM, d | Median lifespan + SEM, d | N | Censored | P value |
| Fig. 1A | N2 + K12ΔtnaA | 25.5 ± 1.0 | 17.2 ± 0.8 | 534 | 15 | <0.0001 |
| N2 + K12 | 27.7 ± 1.1 | 20.2 ± 0.7 | 677 | 15 | ||
| Fig. 1C | N2 + K12ΔtnaA + vehicle | 29.0 ± 1.7 | 18.2 ± 0.5 | 542 | 3 | <0.0001 |
| N2 + K12ΔtnaA + indole | 29.6 ± 1.2 | 22.4 ± 1.2 | 492 | 5 | ||
| Fig. 1D | N2 + K12 + vehicle | 32.5 ± 0.5 | 22 ± 1 | 226 | 0 | <0.0001 |
| N2 + K12 + indole | 36.5 ± 1.5 | 25.5 ± 1.5 | 243 | 0 | ||
| Fig. 1L | W1118 + K12ΔtnaA | 103.0 ± 2.0 | 90.5 ± 2.5 | 116 | 0 | <0.0001 |
| W1118 + K12 | 110.0 ± 4.0 | 100.0 ± 5.0 | 60 | 0 | ||
| Fig. 2A | ahr-1(ju145) + K12ΔtnaA | 31.3 ± 0.6 | 22.8 ± 1.5 | 117 | 12 | 0.15 |
| ahr-1(ju145) + K12 | 32.0 ± 0.6 | 22.3 ± 1.2 | 112 | 5 | ||
| Fig. 2B | ahr-1(ia3) + K12ΔtnaA | 33.0 ± 0.7 | 24.8 ± 0.8 | 95 | 0 | 0.065 |
| ahr-1(ia3) + K12 | 31.5 ± 0.5 | 23.0 ± 0.9 | 104 | 0 | ||
| Fig. 2D | ahr-1(ju145) + K12ΔtnaA + vehicle | 28.7 ± 2.0 | 24.0 ± 2.0 | 98 | 1 | 0.051 |
| ahr-1(ju145) + K12ΔtnaA + indole | 29.0 ± 1.7 | 22.7 ± 2.3 | 111 | 1 | ||
| Fig. 2E | ahr-1(ia3) + K12ΔtnaA + vehicle | 30.5 ± 1.5 | 21.5 ± 1.5 | 368 | 3 | 0.36 |
| ahr-1(ia3) + K12ΔtnaA + indole | 30.5 ± 1.5 | 20 ± 2 | 258 | 2 | ||
| Fig. S2B | sir-2.1(ok434) + K12ΔtnaA | 22.0 ± 0.0 | 18.0 ± 0.0 | 79 | 0 | <0.0001 |
| sir-2.1(ok434) + K12 | 24.0 ± 0.0 | 21.0 ± 1.0 | 93 | 0 | ||
| Fig. S2C | skn-1(ju169) + K12ΔtnaA | 19.0 ± 0.0 | 12.5 ± 0.5 | 80 | 0 | <0.0001 |
| skn-1(ju169) + K12 | 19.0 ± 0.0 | 15.0 ± 1.0 | 79 | 0 | ||
| Fig. S2D | daf-16(m26) + K12ΔtnaA | 23.0 ± 0.0 | 18.0 ± 0.0 | 88 | 7 | <0.0001 |
| daf-16(m26) + K12 | 23.0 ± 0.0 | 20.3 ± 0.3 | 59 | 5 | ||
| Fig. S2E | cat-2(e1112) + K12ΔtnaA | 21.3 ± 0.7 | 17.6 ± 1.55 | 71 | 0 | <0.0001 |
| cat-2(e1112) + K12 | 24.0 ± 1.0 | 20.0 ± 0.0 | 56 | 2 | ||
| Fig. S2F | dop-3(vs.106) + K12ΔtnaA | 30.0 ± 0.0 | 22.0 ± 1.0 | 91 | 0 | <0.0001 |
| dop-3(vs.106) + K12 | 30.0 ± 0.0 | 25.0 ± 0.0 | 90 | 0 |
P values were obtained from log-rank (Mantel–Cox) test.
Aging in C. elegans is accompanied by a decline in locomotion and pharyngeal pumping (19), and decreased resistance to thermal stress (20), which are direct measures of healthspan. Motility, pharyngeal pumping, and resistance to thermal stress were thus assessed over the lifespan of animals grown on K12ΔtnaA supplemented with either indole or vehicle. Exposure to indole increased survival of both young (2 d) and old (18 d) adult worms following thermal stress (Fig. 1E). Moreover, brief preexposure (72 h) of young (1 d) adults to indole in the absence of K12ΔtnaA still rendered animals more resistant to heat stress compared with vehicle-treated controls (Fig. 1F), suggesting that indole acts directly on the host rather than via an intermediate produced by bacteria. Worms grown until day 8 on K12ΔtnaA with or without indole exhibited similar rates of thrashing motility and pharyngeal pumping, and exhibited no paralysis. However, differences were evident after day 15; animals grown with indole exhibited more thrashing motility and pharyngeal pumping, and exhibited delayed onset of paralysis, compared with animals grown without indole (Fig. 1 G–I). Similar effects were evident when animals were grown on K12 compared with K12ΔtnaA (Fig. S1 B and C), or on K12 supplemented with indole (Fig. S1D). As a means of quantifying overall healthspan, the percentage of animals displaying at least 50% of the maximal response for each parameter at each time point were counted as healthy, whereas those falling below this mark were considered frail (1) (Fig. 1J). When normalized to maximum lifespan, these data indicate that indole increased healthspan and concomitantly decreased frail span (Fig. 1K). Taken together, these data suggest that indole provided either exogenously, or via K12, extends healthspan of C. elegans.
Effects of Indole on Healthspan Are Conserved in Drosophila melanogaster.
To test whether indole effects on healthspan were conserved across phyla, germ-free D. melanogaster w1118 were gnotobiotically monoassociated with either K12 or K12ΔtnaA. Flies grown on K12 showed a significant extension of the Kaplan–Meier curve compared with animals grown on K12ΔtnaA (P < 0.0001; Fig. 1L). Whereas no difference in climbing motility was evident in very young flies (day 2) grown on K12 or K12ΔtnaA, day 20 flies grown on K12 were approximately twofold more motile than those grown on K12ΔtnaA (Fig. 1M). To determine whether indole regulated responses to thermal stress, conventionally raised flies were treated with streptomycin to reduce commensal flora, and transferred to media containing either K12 or K12ΔtnaA, or, alternatively, media supplemented with 250 μM indole or vehicle for 5–7 d. Flies exposed to K12 or indole displayed increased resistance to heat stress compared with those exposed to K12∆tnaA or vehicle (Fig. 1N). These data indicate that exposure to indole or indole-producing bacteria augments healthspan in Drosophila. Taken together, survival, motility, and stressor assays in flies and worms, indicate that K12 or indoles augment healthspan across a broad range of evolutionarily diverse species from different phyla.
Indoles Extend Healthspan in C. elegans and Drosophila via AHR.
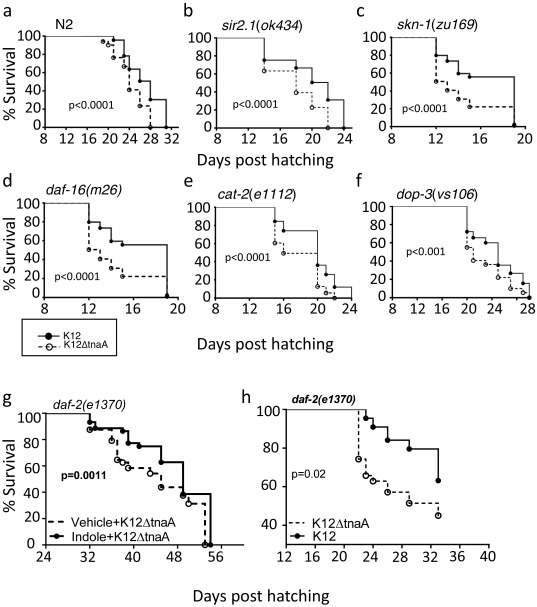
To identify genes mediating the effects of indoles on healthspan, we assessed the effects of indoles on loss-of-function alleles in genes known to mediate conditioning, regulate lifespan, or modulate stress responses in C. elegans (11, 13, 21). These included the FOXO homolog DAF-16, the insulin receptor homolog DAF-2, the sirtuin homolog SIR2.1, the Nrf-2 homolog SKN-1, the dopamine receptor DOP-3, the tyrosine hydroxylase CAT-2, and AHR-1 (22), which encodes an ortholog of the mammalian AHR, a xenobiotic receptor that can bind to a variety of small molecules including indole (23). Similar to N2, shift in the survival curves was still evident with K12 compared with K12ΔtnaA in daf-16(m26), daf-2(e1370), sir2.1(ok434), skn-1(zu169), dop-3(vs.106), and cat-2(e1112) animals (Fig. S2), ruling out these genes as mediators of indole effects. However, although ahr-1 mutants ahr-1(ia3) and ahr-1(ju145) both displayed longer maximal lifespan compared with wild-type (N2) animals, irrespective of presence of indole or K12 (compare Figs. 1B and 2C), neither K12 nor indole significantly shifted the Kaplan–Meier curves compared with K12ΔtnaA (Fig. 2 A–E). Furthermore, ahr-1(ia3) mutants exposed to indole showed no improvement in thrashing motility, pharyngeal pumping, or resistance to thermal stress compared with those treated with vehicle (Fig. 2 F–H). Quantification of overall healthspan scores indicated that indoles did not extend healthspan; thus, ahr-1(ia3) animals grown on K12 or indole had an identical healthspan/frail span ratio to that seen with ahr-1(ia3) mutants or N2 worms grown on K12ΔtnaA plus vehicle (Fig. 2 I and J; compare with Fig. 1 J and K).
Fig. S2.
Genes associated with lifespan and stress response regulation in C. elegans do not mediate healthspan effect of indole. (A–F) Kaplan–Meier lifespan curves of wild-type C. elegans, N2 (A), sir2.1(ok434) (B), skn-1(zu169) (C), daf-16(m26) (D), cat-2(e1112) (E), dop-3(vs.106) (F), daf-2(e1370) (G) on E. coli K12 or K12ΔtnaA mutant; daf-2(e1370) on K12ΔtnaA plus vehicle or K12ΔtnaA plus indole (H) (n > 50 worms/condition). Graphs are representatives of at least two independent experiments. Details of the lifespan experiments are listed in Table S3.
Fig. 2.
The AHR mediates effects of indoles on healthspan. (A and B) Kaplan–Meier lifespan curves of ahr-1(ju145) and ahr-1(ia3) on K12 and K12ΔtnaA, respectively. (C) Maximum lifespan obtained from the lifespan experiments comparing K12 vs. K12ΔtnaA with at least six biological replicates. (D and E) ahr-1(ju145) and ahr-1 (ia3) lifespan on K12ΔtnaA supplemented with 250 μM indole or vehicle (Methanol). Lifespan curves are representative of at least three independent experiments (n > 60 worms per condition in each experiment). (F) Heat stress assays of day 2 ahr-1(ia3) at 35 °C grown on K12ΔtnaA plus vehicle (Vehicle) or K12ΔtnaA plus indole (Indole) (n = 25 animals/condition). (G) Thrashing motility in liquid (n = 8–12 worms/condition); (H) pharyngeal pumping rate per minute (n = 8–12 worms per condition) of ahr-1(ia3) adults grown on K12ΔtnaA plus vehicle (Vehicle) or K12ΔtnaA plus indole (Indole), and monitored throughout their lifespan. (I and J) Healthspan and frail span measurements of ahr-1(ia3) adults grown on K12ΔtnaA plus vehicle (Vehicle) or K12ΔtnaA plus indole (Indole). Values for motility in liquid and pharyngeal pumping rate were obtained from G and H, respectively, to calculate healthspan, and then normalized to maximal lifespan in J. (K and L) Climbing assays (K) and heat stress assays (L) of day 20 germ-free w1118 and ss1 Drosophila fed K12 or K12ΔtnaA (n > 30 animals per condition). P values of percent survival curves were calculated by a log-rank test. Values in C, G, H, K, and L represent mean ± SEM. Summary of the lifespan experiments are presented in Table S3.
To determine whether AHR mediates effects of indoles in Drosophila, climbing assays were carried out with w1118 animals or with the isogenic ss1 mutant, which harbors a loss-of-function allele for the Drosophila ahr-1 homolog (24). Whereas K12 increased climbing motility and resistance to heat stress in wild-type animals compared with K12ΔtnaA, ss1 mutants fed either K12 or K12ΔtnaA displayed similar levels, which resembled that seen with w1118 flies grown with K12∆tnaA (Fig. 2 K and L). Thus, the effects of indoles on healthspan in C. elegans and Drosophila depend upon functional AHR.
Indole Limits Age-Dependent Changes in Gene Expression and Regulates a Distinct Set of Healthspan Genes.
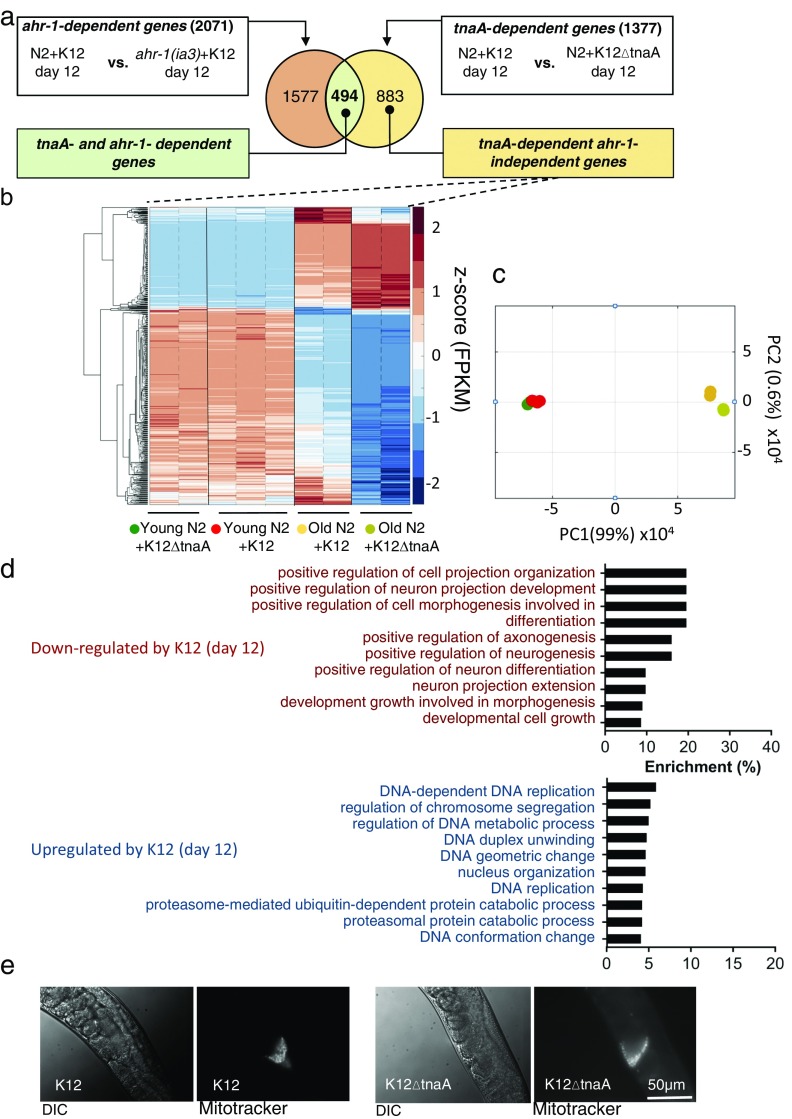
We next identified genes regulated by indole and AHR-1 that are associated with healthspan, and determined how indole impacted the expression of these genes during aging. To do this, transcript levels were assessed by RNA sequencing (RNA-Seq) in RNA samples derived from young (day 2) or older (day 12) adult N2 or ahr-1(ia3) animals grown in either K12 or K12ΔtnaA. Analysis of expression levels by RT-PCR in several growth conditions of two genes, ahr-1 and fmo-2, validated expression levels obtained from RNA-Seq (Fig. S3 and Table S1). Comparison of RNA-Seq data from N2 animals from day 12 grown in either K12 or K12ΔtnaA identified TnaA-dependent genes (Fig. 3A), and comparison of N2 and ahr-1(ia3) animals from day 12 grown on K12 yielded Ahr-dependent genes (Fig. 3A). Comparison of these two groups yielded 494 TnaA- and Ahr-dependent genes (blue box, Fig. 3A), and 883 TnaA-dependent and Ahr-independent genes (beige box, Fig. 3A). Following z-score normalization, expression levels of TnaA- and Ahr-dependent genes were compared across four conditions: young N2 animals with K12 or K12ΔtnaA, and older N2 animals with K12 or K12ΔtnaA (Fig. 3B). Growth in K12 versus K12ΔtnaA had little effect on expression levels in young animals (compare Fig. 3B, columns 1–2 vs. 3–5). Comparison of levels in young animals grown in K12ΔtnaA or K12 with old animals grown in K12ΔtnaA (Fig. 3B, compare columns 1–5 with 8–9) showed that genes expressed at low levels in young animals increased with age (blue → red), whereas those expressed at high levels decreased with age (red → blue). In contrast to young animals, indole treatment in aged animals limited the effects of age, such that genes expressed at low levels in young animals showed less increase with age, and genes expressed at high levels showed less decrease (Fig. 3B; compare columns 6–7 with 8–9). Principal-component analysis (PCA) confirmed that the variance in expression from young animals in K12 or K12ΔtnaA was similar, and that the variance of expression in young animals more closely resembled old animals grown on K12 than old animals grown in K12ΔtnaA (Fig. 3C). Although the effects were not as robust, similar results were evident with TnaA-dependent and Ahr-independent genes (Fig. S4 A–C). Thus, indole delays or limits changes in expression of Ahr-dependent and Ahr-independent genes associated with healthspan.
Fig. S3.
Expression of ahr-1 and fmo-2 increases with age but not with indole. (A and B) Real-time PCR analysis of expression levels of ahr-1 (A) and fmo-2 (B) in young and old N2 animals grown in K12 or K12ΔtnaA. (C) fmo-2 expression level in young and old ahr-1(ia3) animals grown in K12 or K12ΔtnaA. act-1 expression levels were used to normalize the ahr-1 and fmo-2 mRNA levels.
Table S1.
FPKM values of ahr-1 and fmo-2 in young and old N2 animals grown in K12 or K12ΔtnaA and FPKM values of fmo-2 in young and old ahr-1(ia3) animals grown in K12 or K12ΔtnaA
| Gene | Conditions | FPKM values |
| ahr-1 | N2-young + K12ΔtnaA | 7.1, 3.2 |
| ahr-1 | N2-young + K12 | 4.7, 14.3 |
| ahr-1 | N2-old + K12ΔtnaA | 28.9, 33.9, 23.1 |
| ahr-1 | N2-old + K12 | 47, 31.3 |
| fmo-2 | N2-young + K12ΔtnaA | 32.5, 25.8 |
| fmo-2 | N2-young + K12 | 55.65, 48.5 |
| fmo-2 | N2-old + K12ΔtnaA | 505.8, 375.4, 518.5 |
| fmo-2 | N2-old + K12 | 413.8, 397.2 |
| fmo-2 | ahr-1(ia3)-young + K12ΔtnaA | 83.5, 63.9, 74.5 |
| fmo-2 | ahr-1(ia3)-young + K12 | 54.9, 62.8, 83.1 |
| fmo-2 | ahr-1(ia3) + K12ΔtnaA | 1,251.3, 681.5, 756.2 |
| fmo-2 | ahr-1(ia3)-old + K12 | 439.5, 951.5, 357.2 |
Fig. 3.
Indole limits age-associated changes in gene expression in C. elegans. (A) Venn diagram indicating the schema for identification of TnaA- and Ahr-dependent genes and TnaA-dependent and Ahr-independent genes. (B) Hierarchical clustering of 494 z-score–normalized TnaA- and Ahr-dependent genes in young and old animals grown in K12 or K12ΔtnaA. Replicates in each condition are demarcated by dotted black line, while each of the conditions is demarcated by solid black line. (C) PCA using FPKM values of 494 TnaA- and Ahr-dependent genes in young and old animals grown in K12 or K12ΔtnaA. (D) Hierarchical clustering of 1,254 aging genes (aging and lifespan-associated genes from ref. 25 in young and old animals grown in K12 or K12ΔtnaA). (E) PCA using FPKM values of 1,254 aging genes in young and old animals grown in K12 or K12ΔtnaA.
Fig. S4.
Characterization of transcriptional responses from C. elegans. (A) Schematic for the identification TnaA-dependent and Ahr-independent genes. (B) Expression profile of TnaA-dependent and Ahr-independent genes in young and old animals grown in K12 or K12ΔtnaA through hierarchical clustering of genes based on their corresponding z-score values. The replicates in each condition are separated by dotted black line; conditions are demarcated by solid black line. (C) PCA with FPKM values of TnaA-dependent and Ahr-independent genes in young and old animals grown in K12 or K12ΔtnaA. (D) GO output with TnaA-dependent and Ahr-independent genes performed using GOAmigo software. (E) Images of spermatheca of day 12 adult N2 hermaphrodites grown in K12 or K12ΔtnaA, after mating with males whose sperm had been labeled with MitoTracker. (Scale bar, 50 µm.)
We next investigated the effects of indole on genes specifically associated with aging and longevity. Previous studies with C. elegans comparing young and old N2 worms with long- or short-lived mutants identified 1,254 genes associated with aging and longevity (25). These genes are regulated by the GATA transcription factors ELT-5 and ELT-6, whose expression increases with age, and by ELT-3, whose expression declines with age (25). TnaA- and Ahr-dependent genes showed only ∼7% overlap with these genes. Moreover, indole was without effect on expression of elt-3, elt-5, or elt-6, and had little impact on the expression profile of this gene set overall (Fig. 3D). PCA confirmed that with respect to this gene set, variance in young animals grown with K12 or K12ΔtnaA were clustered, and distinct from that of old animals grown with K12 or K12ΔtnaA, which were likewise clustered (Fig. 3E). These data suggest that indoles regulate a set of genes associated with healthspan but distinct from GATA-regulated genes associated with aging and longevity.
Indole Extends Reproductive Span of C. elegans.
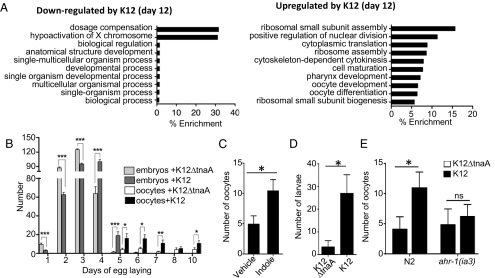
Gene ontology (GO) analysis of TnaA- and Ahr-dependent genes indicated that, by day 12, indole caused significant down-regulation of genes associated with sex determination (dosage compensation), and up-regulation of genes associated with meiosis, cell cycle, cytokinesis, and egg shell formation (Fig. 4A). Analysis of TnaA-dependent and Ahr-independent genes likewise showed up-regulation by day 12 of genes associated with meiosis, mitosis, cell division, sex organ formation, and larval development (Fig. S4D). These processes contribute to maturation of oocytes and reproduction, which was highly unusual because C. elegans normally cease reproduction by day 4 and oocyte generation by day 8 (26, 27). Prompted by the expression profile data, we confirmed that indole had a positive effect on the production of eggs and viable offspring. Although N2 animals grown on K12 or K12ΔtnaA produced similar numbers of fertilized eggs through day 5 (average of 273 and 287, respectively; n = 12), animals exposed to K12 exhibited a slightly delayed onset in egg production, and produced offspring for a day longer (Fig. 4B). Notably, animals grown in K12 produced significantly more unfertilized oocytes after day 4 compared with animals grown on K12ΔtnaA (Fig. 4B). Similar results were obtained with animals grown to day 10 on K12ΔtnaA plus indole compared with K12ΔtnaA plus vehicle (Fig. 4C). To determine whether oocytes produced at day 10 could still be fertilized, day 9–11 hermaphrodites were mated with young males. Using this scheme, more viable progeny were produced by hermaphrodites grown on K12 compared with those grown on K12ΔtnaA (Fig. 4D). Labeling of males with MitoTracker indicated that sperm was, in fact, deposited, and that K12∆tnaA did not cause mating anomalies (Fig. S4E). Therefore, indoles induced an extension of reproductive span, possibly owing to an increase in oocyte quality or viability (28). As with other healthspan measures, no increase in oocyte production was evident in day 10 ahr-1(1a3) animals grown on K12 compared with K12ΔtnaA (Fig. 4E). Although ahr-1 appeared necessary for increased fecundity, GO analysis suggested an additional role for ahr-1–independent genes (Fig. S4D). These observations suggest that indoles increase reproductive span, which is consistent with the induction of a more youthful state.
Fig. 4.
Indole extends reproductive span of C. elegans. (A) GO analysis of 494 TnaA- and Ahr-dependent genes using GOAmigo software. (B) Average number of embryos and oocytes (unfertilized) produced per day by N2 adults grown on K12 or K12ΔtnaA over time (n = 12 adult worms per condition). (C) Average number of oocytes produced by day 10 N2 adults in K12ΔtnaA plus indole or K12ΔtnaA plus vehicle (n = 12 worms per condition). (D) Average number of larvae obtained from mating day 10 N2 hermaphrodites grown in K12 and K12ΔtnaA with young males. (E) Average number of oocytes produced by day 10 N2 and ahr-1(ia3) adults in K12 and K12ΔtnaA conditions (n = 12 worms per condition). B–E are representative of at least two independent experiments.
Effects of Indole on Healthspan Are Conserved in Mammals.
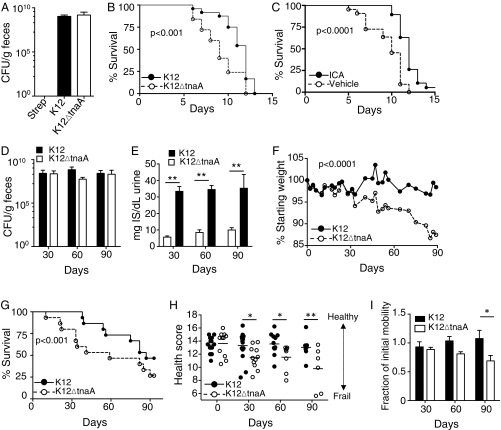
To assess effects of indoles on healthspan in mammals, we assessed sensitivity to stressors in young mice, and multiple health metrics in geriatric animals. Twelve- to 16-wk-old C57BL/6 mice were treated with streptomycin and recolonized with either K12 or K12ΔtnaA (Fig. 5A). After 1 wk, the animals were subjected to lethal total body irradiation (TBI) (12 Gy), and their survival was monitored. Colonization with K12 enhanced survival following TBI, compared with animals colonized with K12ΔtnaA (Fig. 5B). Alternatively, animals were administered ICA [150 mg⋅kg−1⋅d−1 per os (p.o.); ref. 12] or vehicle beginning 1 d before TBI. ICA was chosen rather than indole because of the difficulties in providing the latter compound to mice other than by slow dissolving capsule or enema (17). Like K12, ICA proved protective against TBI (Fig. 5C). Together, these data indicate that indoles produced by commensal bacteria increase the tolerance of young mice to TBI, and exogenous ICA provided as a supplement to indoles produced by commensal bacteria enhance protection. These data are in accordance with previous reports from our laboratory and others showing protective effects of indoles against stressors in mice (12, 16).
Fig. 5.
Indole from commensal E. coli augments lifespan and extends healthspan of mice. (A) Colony-forming units per gram of bacteria in feces of C57BL/6 mice colonized with streptomycin- and nalidixic acid-resistant K12 or K12ΔtnaA. Mice were colonized 1 wk before TBI (12 Gy), and bacterial counts were assessed 1 d before TBI on plates containing streptomycin and nalidixic acid. (B) Survival of C57BL/6 mice (n = 15) following colonization with either K12 or K12ΔtnaA, and exposure to TBI. (C) Survival of C57BL/6 mice (n = 20) following TBI. Mice were treated with either ICA (150 mg⋅kg−1⋅d−1) or vehicle, daily, beginning 1 d before irradiation. (D) Colony-forming units per gram of streptomycin/nalidixic acid-resistant K12 or K12ΔtnaA in feces of geriatric BALB/c mice (28 mo, n = 15). Bacterial counts were assessed at 30, 60, and 90 d postcolonization. (E) Urinary 3-indoxyl sulfate levels from geriatric mice colonized with either K12 or K12ΔtnaA. (F) Time course of weight changes in geriatric mice colonized with K12 or K12ΔtnaA. (G) Survival of geriatric mice colonized with K12 or K12ΔtnaA. (H) Composite health scores in geriatric mice colonized with K12 or K12ΔtnaA for 30, 60, or 90 d. (I) Average fraction of initial motility in geriatric BALB/c animals inoculated with K12 or K12ΔtnaA for 30, 60, or 90 d. Student’s t test only showed significant differences between groups at the 90-d time point.
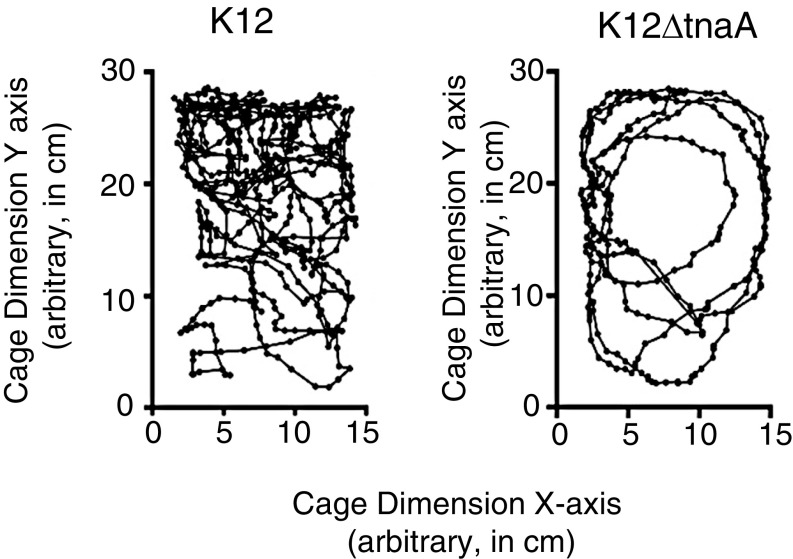
We next assessed whether indoles extended healthspan in mammals. Aged BALB/c mice (28 mo old) were treated with streptomycin and then recolonized with either K12 or K12∆tnaA, and health score parameters were assessed over a 3-mo period. Stable and equivalent colonization was achieved with these strains over this time period (Fig. 5D). The indole metabolite 3-indoxyl sulfate, an indirect measure of indole production by intestinal microbes (29, 30), was readily detected in urine from animals colonized with K12, but appeared at significantly lower levels in animals colonized with K12∆tnaA (Fig. 5E). Over the 3-mo period, animals colonized with K12 were better able to maintain their weight (Fig. 5F) and showed improved survival compared with those colonized with K12ΔtnaA (Fig. 5G). Based on assessment of hunching, skin and coat condition, activity level, facial grimacing, curiosity, and grip strength, animals colonized with K12 had significantly greater composite health scores (Fig. 5H). Measurement of total displacement in the cage (Fig. S5) confirmed that older animals colonized with K12 exhibited greater mobility than those colonized with K12ΔtnaA, although a statistically significant difference was only apparent after 90 d (Fig. 5I). No change in dwell time or total number of rearing events was evident over this time period (data not shown). Thus, indoles provided by commensal bacteria increase healthspan in both young and geriatric mice, and indicate that its effects on healthspan are broadly conserved across evolutionarily diverse species from different phyla.
Fig. S5.
Mouse motility in geriatric BALB/c animals colonized with K12 or K12ΔtnaA. Motility tracking of geriatric animals for 2 min. The y axis represents the long side of a cage in centimeters, and the x axis represents the short side of the cage in centimeters. The images show representative tracks from two geriatric animals one colonized with K12 (Left), and the other with K12ΔtnaA (Right) for 90 d. From these data, the total distance traveled was calculated.
Discussion
Youth is characterized by the capacity to move without impairment, to reproduce, to tolerate diverse environmental stressors with minimal attendant damage (damage tolerance), and to regenerate following damage, conditions that define healthspan (31–33). As animals age, health decreases and frailty increases. Dietary factors, as well as dysregulation of the microbiota (dysbiosis), have been implicated in various disease states, including inflammatory bowel disease, obesity, and diabetes (34–37). Such maladies are often associated with dysregulated inflammatory responses, or with susceptibility to damage caused by such responses. As such, it has been postulated that altering the diet or the microbiota may augment damage tolerance and limit pathology (37).
We and others have implicated indoles and other products of bacterial TnaA in protective responses to damage by stressors and pathogens in both C. elegans and mammals (11–13, 16) (Fig. 5 A–C). Data presented here show that indoles also augment other healthspan readouts associated with youth and with healthy aging, such as motility (Figs. 1 and 5). Indole-regulated genes required to augment healthspan in C. elegans are generally distinct from those associated with longevity per se (Fig. 3 D and E), and by shifting gene expression in the aged toward “healthy” patterns seen in the young (Fig. 3C and Fig. S4C), indoles in effect uncouple effects of aging on health. While readouts such as stressor tolerance and motility define healthspan, the transcriptomic data suggest that this description is nevertheless incomplete. Thus, indoles up-regulate genes associated with reproduction in aged animals, and accordingly extend reproductive span, a phenotype normally only expressed in young adults (Fig. 4).
Previous reports suggested that AHR-1 limits lifespan in C. elegans grown on OP50, which, like K12, produces indoles (38). We likewise found that both ahr-1(ia3) and ahr-1(ju145) exhibited longer lifespan compared with N2 (Figs. 1B and 2C); however, characterization of nematode locomotion, pharyngeal pumping, and heat stress in ahr-1 mutant animals, and normalization of these measures to lifespan, indicated that AHR-1 mediates effects of indoles on these aspects of healthspan, as well as on reproductive span (Figs. 2 I and J and 4E), an effect corroborated in Drosophila (Fig. 2 K and L).
While AHR-1 appears necessary for the healthspan measures assessed herein, Ahr-dependent genes represent only 36% of indole-regulated genes while Ahr-independent genes comprise 64% (Fig. 3A). Notably, genes from both the Ahr-dependent and Ahr-independent groups map to pathways controlling meiosis and oogenesis, suggesting both may contribute to reproductive span (Fig. 4A and Fig. S4D). These data also raise the additional possibility that receptors besides AHR-1 mediate effects of indole on reproduction. Moreover, although Ahr-independent genes do not contribute to differences in survival curves with indole (Fig. 2 A–E), the possibility exists that they may do so when animals are grown under conditions that pose different exigencies. Regulation of AHR-1 expression may control healthspan responses during normal aging in C. elegans. Expression of ahr-1 increases with age (Fig. S3A), and although no changes in expression are evident with indole, age-dependent increases in AHR-1 expression may amplify responses to indole in older animals, and so favor gene expression patterns associated with youthfulness.
In mammals, indoles and AHR have been implicated in repair and/or protection of intestinal epithelia via IL-22 produced by type 3 innate lymphoid cells (ILC3) (16, 39). We were unable to evaluate a direct role for AHR in mammalian healthspan because Ahr−/− mice lack ILC3s (40), which themselves may mediate indole effects in aged mice. Notably, Ahr−/− mice also exhibit diminished reproductive capacity, which is at least in part due to deficiencies in sperm production, motility, and sensitivity to oxidative stress (41). Although, indole and ICA bind AHR and activate AHR-dependent transcriptional responses (23), AHR-independent mechanisms may also mediate effects of indole or its derivatives. In the context of aging, epithelial barrier integrity is important in limiting dissemination of bacterial products (42, 43), and the subsequent hyperinflammatory responses that can cause widespread tissue damage, a process termed “inflammaging” (44, 45). Barrier integrity limits enteritis and extends survival following TBI (46, 47). Whether augmenting intestinal barrier integrity or systemic antiinflammatory effects contribute to indole effects on healthspan and motility in geriatric animals is currently under investigation.
Whereas data presented here show protective effects of E. coli K12 versus K12ΔtnaA, recent data suggest that other intestinal eubacteria, including many Bacteroides and Lactobacillus species, likewise use TnaA to produce protective indole derivatives, including ICA and indole-3-carbinol (I3C)) (16, 48–50). Responsiveness to particular indoles may differ and be tuned to the precise complement of indoles produced by the microbiota. Alternatively, different indole species may offer different levels of protection depending on the stressor, or affect different aspects of healthspan. Testing of combinations of indoles is thus warranted and may offer the most comprehensive means to extend healthspan.
Production of indoles may be a general property of eubacteria, and coevolution of indole-producing bacteria with animals over the last ∼500 My may explain how indoles coordinate responses to a myriad of stressors in such diverse organisms. Plants, particularly cruciferous vegetables such as kale and broccoli, also produce indoles including ICA and IAA, which is also known as auxin (51). Certain indoles [e.g., I3C or 3,3′-diindolylmethane (DIM)] are available as dietary supplements or plant extracts (52), although it remains to be determined whether indoles provided in this fashion have sufficient bioavailability to provide protection against diverse stressors. Fecal transplants, or oral probiotic treatments with known indole-producing bacteria, offer potential advantages in limiting infirmity associated with dysbiosis (53, 54). However, durable repopulation with the transplanted species requires an open niche within the intestinal tract, which may necessitate coordinate administration of antibiotics, and the procedure is less well tolerated in cases of severe colitis (55). High levels of tryptophan in the diet can likewise augment production of indoles by intestinal bacteria, and increasing consumption of tryptophan may represent a viable therapeutic strategy in the context of a microbiota with adequate TnaA activity (56). Notably, in worms and mice, exogenous administration of indole or ICA alone can supplement that provided by the microbiota, and further augment healthspan (12, 16) (Fig. 1D). These data raise the possibility that exogenous administration of bioactive indoles might both compensate for a suboptimal microbiota, and bypass limitations associated with fecal transplants, probiotic treatments, or dietary interventions.
Materials and Methods
Bacterial Strains.
Bacterial strains used include a nalidixic acid and streptomycin-resistant E. coli variant selected for its capacity to grow in mice (MG1655* referred to as K12) obtained from Dr. P. Cohen, University of Rhode Island, Kingston, RI, that is an efficient colonizer of the murine intestinal tract (18), and a MG1655*ΔtnaA constructed by us from MG1655* (called here K12ΔtnaA) by deletion of TnaA gene (tnaA) using lambda red-recombinase system (57), and E. coli OP50 (58). All MG1655* strains were cultured in Luria Broth (LB) (Difco) containing streptomycin (100 μg/mL), whereas OP50 was cultured in LB.
C. elegans Strains.
C. elegans strains obtained from the Caenorhabditis Genetics Center include wild-type Bristol strain N2, ahr-1(ju145), ahr-1(ia3), daf-16(m26), daf-16(mu86), skn-1(zu169), sir2.1(ok434), cat-2(e1112), and dop3(vs.106). All strains were maintained on nematode growth medium (NGM) at 16 °C under standard culturing conditions with E. coli OP50 as food source (26).
C. elegans Lifespan Assays.
Lifespan assays were performed at 16 °C on NGM plates. Bacterial strains grown overnight in LB broth at 37 °C to an OD600 of 0.8–1.0 were used to seed the plates. Gravid C. elegans adult worms were transferred to assay plates with bacteria and allowed to lay eggs for 4–5 h at room temperature. Plates with synchronized eggs were then transferred and maintained in 16 °C (to ensure slow bacterial growth). Lifespan assays were performed without 5-fluorodeoxyuridine, which has been used previously to limit appearance of the progeny, because the compound itself affected lifespan. Hence, adult worms developing from the synchronized eggs were transferred daily for the first 8–10 d and every other day thereafter so as to avoid mixing with next generation. For some assay plates, indole (Sigma-Aldrich) dissolved in methanol (Sigma-Aldrich) was added to a final concentration of 250 µM. Methanol was added to control plates, which were then seeded with K12ΔtnaA bacteria. The concentration of indole was reduced to ∼100 µM in the assay plates used to transfer older adult (post-day 5) worms.
C. elegans Motility and Stress Assays.
Thrashing motility assay.
Individual worms were picked and transferred to a drop of M9 buffer [3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, 1 mL of 1 M MgSO4 in 1 L of water (26)] on fresh NGM plates followed by a 30-s recovery time. Their C-shape body-bending movement was then counted for 1 min under microscope.
Pharyngeal pumping assay.
A worm’s pharyngeal area was videorecorded, and the video was analyzed to obtain the number of pharyngeal muscle pumps in a 1-min period. Only motile worms were considered for this assay.
C. elegans paralysis assay.
Worms were counted as paralyzed when they failed to move upon repeated prodding with a platinum wire.
C. elegans heat stress assay.
Worms from each condition were transferred to NGM plates without bacteria (∼20 worms per plate) and subsequently transferred to 36 °C incubator. Plates were monitored every hour. Worms were considered dead if they failed to respond to gentle prodding with a platinum wire and showed no indication of pharyngeal pumping.
C. elegans Reproductive-Span Assays.
Oocyte and embryo assays.
Worms were grown in desired condition from the embryonic stage to the L4 stage. Each L4 animal was then transferred to a separate NGM plate seeded with either K12 or K12ΔtnaA, and each worm was transferred to a fresh plate daily. Plates were seeded with bacteria a few hours before the experiment to ensure a thin bacterial spot, which facilitated visualization of eggs or oocytes. Worms were incubated at 16 °C, and oocyte or embryo numbers were counted each day. Fertilized oocytes that resulted in larvae were scored as embryos, whereas those that remained were scored as oocytes.
C. elegans mating and sperm staining.
One day-10 hermaphrodite grown previously on either K12 or K12ΔtnaA was incubated with eight males and then transferred to NGM plates seeded with either 5 µL of K12 or K12ΔtnaA. After 2 d, the number of larvae was counted. To confirm mating, MitoTracker Deep Red FM (Invitrogen) was used to label male sperm using a modification of a method described previously (59). Sperm were labeled by soaking the males in 50 µM MitoTracker Deep Red FM in M9 buffer for 2 h at 16 °C. Following mating of labeled males with hermaphrodites, labeled sperm in the spermatheca of the hermaphrodites were imaged with a fluorescence microscope (Zeiss), indicating successful mating.
D. melanogaster Assays.
Lifespan assays.
For gnotobiotic experiments with Drosophila, germ-free flies were prepared as previously described (60). Briefly, Drosophila embryos were collected and transferred to a cell strainer. Under a sterile hood, embryos were washed three times with sterile PBS, soaked in 50% bleach (1:1 dilution of Clorox) for 5 min, before washing again with sterile PBS. The mesh of the cell strainer was cut with a sterile blade and transferred into sterile vials containing sterilized food (dextrose, 50 g/L; sucrose, 25 g/L; yeast extract, 15 g/L; cornmeal, 60 g/L; agar, 6.5 g/L; tryptone, 30 g/ L; molasses, 65 g/L) seeded with either K12 or K12ΔtnaA bacteria (50 µL of saturated culture/5 mL of food) and incubated at 25 °C. Flies were transferred every 2–3 d to avoid overgrowth of bacteria.
Climbing assays.
Adult germ-free flies were transferred to a glass cylinder with diameter of 2.5 cm and height of 24 cm and were allowed to acclimatize for 2 min. The cylinder was then tapped to bring flies to the bottom. Their climbing was monitored for 30 s. Flies that climb up one-half of the cylinder (12 cm) height within 30 s were scored as successful climbers. A climbing index was calculated as (successful climbers/total number of flies used for assay) × 100.
Heat stress assays.
Conventionally raised adult flies (not germ-free) were transferred to food containing streptomycin (100 μg/mL) and incubated at room temperature for a further 4–5 d. Animals were subsequently transferred to food with different additives including K12, K12ΔtnaA, methanol, or indole and incubated for an additional 4–5 d. Flies from different conditions were transferred to empty transparent fly vials with wet plugs. The vials were then shifted to 38 °C. Flies were monitored every hour. When at least one of the conditions showed more than ∼80% paralyzed flies, all flies were removed from 38 °C and transferred to fresh vials with food and subsequently kept at room temperature overnight for recovery. Live flies were counted next day. Drosophila lines used include w1118, and Dmel\ss1 stock #2973 obtained from the Bloomington Drosophila Stock Centre.
Murine Lifespan, Health Score, Motility, and Radiation Assays.
Animals.
C57BL/6 mice were purchased from The Jackson Laboratory, and aged BALB/c mice were obtained from the Aged Rodent Colonies maintained by the National Institute on Aging, a division of the National Institutes of Health. Mice were allowed to acclimate for at least 1 wk following shipment and before experiments. Animal-handling and experimental procedures were in accordance with the Guide for the Care and Use of Laboratory Animals (61) and approved by the Emory University Institutional Animal Care and Use Committee.
ICA administration.
ICA (Sigma) was delivered daily by oral gavage at a dose of 150 mg⋅kg−1⋅d−1 in a vehicle of DMSO/PEG400/5% citric acid (1:4.5:4.5). ICA treatment was started 24 h before total body irradiation (TBI).
Colonization with K12 or K12ΔtnaA.
Mice were given streptomycin in their drinking water (5 g/L) starting 24 h before colonization to clear commensal flora. E. coli K12 and K12ΔtnaA strains were grown to saturation at 37 °C in LB containing streptomycin (100 µg/mL) and were introduced to mice by a single oral gavage (450 µL of culture per mouse, pelleted and resuspended in 200 µL of sterile PBS) 24 h after initiation of streptomycin treatment. Mice remained on streptomycin for the duration of the experiment. Colonization levels in the gut were indirectly assessed by serial dilution plating of fecal samples on McConkey Agar containing streptomycin (100 µg/mL) and nalidixic acid (50 µg/mL). The resulting colonies were checked for indole production using Kovacs reagent (Sigma) following overnight growth in LB at 37 °C.
Indoxyl sulfate analysis.
Urine samples from mice were diluted 1:10 in water and analyzed for indoxyl sulfate (Indican) by spectrophotometric assay using an Indican Assay Kit (Sigma).
Lethal TBI.
Mice were exposed to 12-Gy lethal TBI using a 137Cs source at a rate of ∼1.7 Gy/min. Mice were weighed daily following irradiation and tracked for survival.
Mouse health scoring.
Health scores were determined by assessing six parameters known to be indicative of health status in mice: hunching, skin and coat condition, activity level, facial grimace, curiosity, and grip strength. Mice were assessed in a blinded fashion by personnel familiar with healthy mouse appearance and behavior, and in comparison with control healthy mice of a similar age and strain. Each parameter was assigned a score on a scale of 1–3, with 3 being normal or healthy, and 1 being extremely impaired or unhealthy, for a total possible score of 18 for each group of six mice. This total score was multiplied by the percentage weight loss (percent of starting weight) to calculate the final score. For motility scoring, elderly animals (2 y old), inoculated with either K12 or K12ΔtnaA, were moved to clean cages and monitored by video for 2 min. Videos were analyzed using NIH ImageJ, 32-bit, version 1.47, and the tracking plug-in MTRACKJ (62). Mice were tracked individually over a 3-mo period, and total distance traveled in 2 min, the amount of time the animals were at rest (dwell time), and the number of rearing events were recorded.
RNA-Seq and Transcriptomic Data Analysis.
RNA sample preparation and sequencing.
Worms were grown in respective conditions since embryo stage as described in the lifespan assays. RNAs either from young (approximately day 2) or old adults (approximately day 12) grown in different conditions were isolated in TRIzol and purified using RNeasy Plus Mini kit (Qiagen). At least two biological replicates for each condition were considered for the RNA-Seq with each replicate containing purified RNA from ∼150 adult worms. Sequencing was performed at Yerkes Nonhuman Primates Genomics Core, Emory University, using Illumina Trueseq.
RNA-Seq data analysis.
The quality of the raw sequence data were inspected with FASTQC. Trimming of the adapters were performed using cutadapt. The reads with low QC values (Phred score < 30, i.e., 99.9% accurate base call) and smaller than 30 bases of mRNA were discarded after trimming process (63, 64). Using the software package STAR aligner (version 2.5.0), sequencing reads were subsequently mapped to the C. elegans reference genome (ce10) (University of California, Santa Cruz, 2016) with default parameters allowing only one alignment per read (65). Overall, 95–98% of total sequenced fragments was mapped contiguously or to exon–exon junctions of the genome. A summary of the results produced by this approach is shown in Table S2. Mapped reads were further sorted using SAMtools software run with generic parameters (66). The total number of mapped reads for each library ranged from 11 to 21 million. Mapped reads that overlap with coding gene features were counted using htseq-count (mode = union) (67). Reads were not counted if they map to more than one genomic position or to a position with overlapping exons from more than one gene. The per-gene counts were used as an input for the DESeq2 R package for differential gene expression analysis (68). Genes with >1 reads mapped to them in all replicates were called “expressed,” and these transcript counts were used to test for differential expression. Mapped sequencing fragment counts per gene were size-factor normalized and variance estimated, which has been shown to reduce bias when comparing gene expression between samples. DESeq2 was also used as quality control, and the variance stabilization-transformed data were used to perform PCA and clustering analysis. Differentially expressed genes were called at a false-discovery rate (FDR) of 0.05% by Benjamini–Hochberg multiple testing adjustment using the DESeq2 in R, which models count data by a negative binomial distribution (68). GO analysis for overexpressed and underexpressed genes was carried out using GoAmigo (amigo.geneontology.org/amigo) (69). A category containing at least three genes and a corrected P value of <0.05 (Benjamini–Hochberg method) was considered significant.
Table S2.
Alignment summary of total RNA-Seq reads to C. elegans reference genome (ce10)
| Sample ID | Read length | No. of reads | % Total aligned | % Unaligned | % Stranded |
| 14_N2tnaA_LATE2 | 1 × 151 | 17,077,925 | 98.53 | 1.47 | 99.57 |
| 15_N2tnaA_LATE3 | 1 × 151 | 11,568,564 | 91.67 | 8.33 | 99.57 |
| 16_N2K12_LATE1 | 1 × 151 | 17,635,082 | 98.41 | 1.59 | 99.67 |
| 18_N2K12_LATE3 | 1 × 151 | 16,765,410 | 97.66 | 2.34 | 99.66 |
| 2_N2tnaA_EARLY2 | 1 × 151 | 17,265,225 | 98.98 | 1.02 | 99.82 |
| 37_AHRK12_LATE1 | 1 × 151 | 13,349,441 | 96.24 | 3.76 | 99.58 |
| 39_AHRK12_LATE3 | 1 × 151 | 14,609,013 | 96.54 | 3.46 | 99.65 |
| 3_N2tnaA_EARLY3 | 1 × 151 | 16,956,473 | 98.84 | 1.16 | 99.81 |
| 4_N2K12_EARLY1 | 1 × 151 | 16,414,814 | 98.70 | 1.30 | 99.81 |
| 5_N2K12_EARLY2 | 1 × 151 | 16,813,260 | 98.88 | 1.12 | 99.76 |
| 6_N2K12_EARLY3 | 1 × 151 | 16,406,106 | 98.79 | 1.21 | 99.80 |
Heatmap and PCA analysis.
Clustergram module of Matlab R2016a was used to hierarchical cluster the genes based on z-scores calculated for each gene across all samples, where z-scoregeneA,condition = ((FPKMgeneA,conditionX) − (Mean_FPKMgeneA All conditions))/(standard-deviation_FPKMgeneAAll conditions). A Euclidian distance metric was used to assess clustering. PCA was carried out on the fragments per kilobase of transcript per million mapped reads (FPKM) values of the samples using Matlab R2016a.
Statistical Analysis.
All Kaplan–Meier lifespan curves were analyzed by log-rank Mantel–Cox test. For most comparisons, experiments were repeated at least three times, and comparisons were made using a t test or ANOVA. P values less than 0.05 were considered significant.
Acknowledgments
We thank T. Cleverley and W. Kelly for helpful discussions, and C. Moreno, P. O’Lague, B. Shur, and B. Weiss for helpful discussions and for reviewing the manuscript. This work was supported by grants from Bio-Merieux Foundation and NIH (2R01DK074731-04A1) (to D.K.). The Bloomington Drosophila Stock Centre, which provided Drosophila lines, is supported by NIH Grant P40OD018537. The authors wish to acknowledge the memory of Vladimir Brezina (1958–2016), an old and dear friend.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw RNA-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE101910).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706464114/-/DCSupplemental.
References
- 1.Bansal A, Zhu LJ, Yen K, Tissenbaum HA. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc Natl Acad Sci USA. 2015;112:E277–E286. doi: 10.1073/pnas.1412192112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meara E, White C, Cutler DM. Trends in medical spending by age, 1963–2000. Health Aff (Millwood) 2004;23:176–183. doi: 10.1377/hlthaff.23.4.176. [DOI] [PubMed] [Google Scholar]
- 3.Tissenbaum HA. Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2012;67:503–510. doi: 10.1093/gerona/gls088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatar M. Can we develop genetically tractable models to assess healthspan (rather than life span) in animal models? J Gerontol A Biol Sci Med Sci. 2009;64:161–163. doi: 10.1093/gerona/gln067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podshivalova K, Kerr RA, Kenyon C. How a mutation that slows aging can also disproportionately extend end-of-life decrepitude. Cell Rep. 2017;19:441–450. doi: 10.1016/j.celrep.2017.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang WB, et al. Extended twilight among isogenic C. elegans causes a disproportionate scaling between lifespan and health. Cell Syst. 2016;3:333–345. doi: 10.1016/j.cels.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Langille MG, et al. Microbial shifts in the aging mouse gut. Microbiome. 2014;2:50. doi: 10.1186/s40168-014-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anyanful A, Easley KA, Benian GM, Kalman D. Conditioning protects C. elegans from lethal effects of enteropathogenic E. coli by activating genes that regulate lifespan and innate immunity. Cell Host Microbe. 2009;5:450–462. doi: 10.1016/j.chom.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bommarius B, et al. A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PLoS One. 2013;8:e54456. doi: 10.1371/journal.pone.0054456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anyanful A, et al. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones RM, et al. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 2015;12:1217–1225. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Wood TK, Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Shimada Y, et al. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leatham MP, et al. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun. 2005;73:8039–8049. doi: 10.1128/IAI.73.12.8039-8049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 20.Keith SA, Amrit FRG, Ratnappan R, Ghazi A. The C. elegans healthspan and stress-resistance assay toolkit. Methods. 2014;68:476–486. doi: 10.1016/j.ymeth.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 22.Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zoolog A Comp Exp Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbard TD, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melnick MB, Noll E, Perrimon N. The Drosophila stubarista phenotype is associated with a dosage effect of the putative ribosome-associated protein D-p40 on spineless. Genetics. 1993;135:553–564. doi: 10.1093/genetics/135.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budovskaya YV, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Vol 17 Cold Spring Harbor Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 27.McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- 28.Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenq RR. How’s your microbiota? Let’s check your urine. Blood. 2015;126:1641–1642. doi: 10.1182/blood-2015-08-661504. [DOI] [PubMed] [Google Scholar]
- 30.Weber D, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood. 2015;126:1723–1728. doi: 10.1182/blood-2015-04-638858. [DOI] [PubMed] [Google Scholar]
- 31.Richardson A, et al. Measures of healthspan as indices of aging in mice—a recommendation. J Gerontol A Biol Sci Med Sci. 2016;71:427–430. doi: 10.1093/gerona/glv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lithgow GJ. Stress response and aging in Caenorhabditis elegans. Results Probl Cell Differ. 2000;29:131–148. doi: 10.1007/978-3-540-48003-7_7. [DOI] [PubMed] [Google Scholar]
- 33.Huffman DM, et al. Evaluating health span in preclinical models of aging and disease: Guidelines, challenges, and opportunities for geroscience. J Gerontol A Biol Sci Med Sci. 2016;71:1395–1406. doi: 10.1093/gerona/glw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou JK, Lee D, Lewis J. Diet and inflammatory bowel disease: Review of patient-targeted recommendations. Clin Gastroenterol Hepatol. 2014;12:1592–1600. doi: 10.1016/j.cgh.2013.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50:495–507. doi: 10.1007/s00535-015-1064-1. [DOI] [PubMed] [Google Scholar]
- 36.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34:39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Richards JL, Yap YA, McLeod KH, Mackay CR, Mariño E. Dietary metabolites and the gut microbiota: An alternative approach to control inflammatory and autoimmune diseases. Clin Transl Immunology. 2016;5:e82. doi: 10.1038/cti.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckers A, et al. The aryl hydrocarbon receptor promotes aging phenotypes across species. Sci Rep. 2016;6:19618. doi: 10.1038/srep19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen DA, Esakky P, Drury A, Lamb L, Moley KH. The aryl hydrocarbon receptor is important for proper seminiferous tubule architecture and sperm development in mice. Biol Reprod. 2014;90:8. doi: 10.1095/biolreprod.113.108845. [DOI] [PubMed] [Google Scholar]
- 42.Saltzman JR, Kowdley KV, Perrone G, Russell RM. Changes in small-intestine permeability with aging. J Am Geriatr Soc. 1995;43:160–164. doi: 10.1111/j.1532-5415.1995.tb06382.x. [DOI] [PubMed] [Google Scholar]
- 43.Saltzman JR, Russell RM. The aging gut. Nutritional issues. Gastroenterol Clin North Am. 1998;27:309–324. doi: 10.1016/s0889-8553(05)70005-4. [DOI] [PubMed] [Google Scholar]
- 44.Franceschi C, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 45.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69:S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 46.Booth C, Tudor G, Tudor J, Katz BP, MacVittie TJ. Acute gastrointestinal syndrome in high-dose irradiated mice. Health Phys. 2012;103:383–399. doi: 10.1097/hp.0b013e318266ee13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciorba MA, Stenson WF. Probiotic therapy in radiation-induced intestinal injury and repair. Ann N Y Acad Sci. 2009;1165:190–194. doi: 10.1111/j.1749-6632.2009.04029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouse M, Singh NP, Nagarkatti PS, Nagarkatti M. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br J Pharmacol. 2013;169:1305–1321. doi: 10.1111/bph.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho HJ, et al. 3,3′-Diindolylmethane suppresses the inflammatory response to lipopolysaccharide in murine macrophages. J Nutr. 2008;138:17–23. doi: 10.1093/jn/138.1.17. [DOI] [PubMed] [Google Scholar]
- 50.Devlin AS, et al. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe. 2016;20:709–715. doi: 10.1016/j.chom.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weijers D, Wagner D. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol. 2016;67:539–574. doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- 52.Huang H, Arocho GMR, Davis C, Yu L, Wang TTY. Consumption of selected cruciferous vegetables and soy phytochemical dietary supplements can alter gut microbiome composition. FASEB J. 2016;30(Suppl 1):1176.26. [Google Scholar]
- 53.Wischmeyer PE, McDonald D, Knight R. Role of the microbiome, probiotics, and “dysbiosis therapy” in critical illness. Curr Opin Crit Care. 2016;22:347–353. doi: 10.1097/MCC.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y, et al. Fecal microbiota transplantation for ulcerative colitis: A systematic review and meta-analysis. PLoS One. 2016;11:e0157259. doi: 10.1371/journal.pone.0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunde S, et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr. 2013;56:597–601. doi: 10.1097/MPG.0b013e318292fa0d. [DOI] [PubMed] [Google Scholar]
- 56.Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology. 2013;159:402–410. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 57.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hill KL, L’Hernault SW. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev Biol. 2001;232:105–114. doi: 10.1006/dbio.2000.0136. [DOI] [PubMed] [Google Scholar]
- 60.Luo L, Reedy AR, Jones RM. Detecting reactive oxygen species generation and stem cell proliferation in the Drosophila intestine. Methods Mol Biol. 2016;1422:103–113. doi: 10.1007/978-1-4939-3603-8_10. [DOI] [PubMed] [Google Scholar]
- 61.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 62.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 63.Andrews S. 2010 FastQC: A quality control tool for high throughput sequence data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed June 20, 2016.
- 64.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 65.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carbon S, et al. AmiGO Hub; Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]