Significance
Necroptosis is a programmed form of necrotic cell death which is implicated in a wide range of human pathological conditions. It is controlled by receptor-interacting protein kinase 3 (RIPK3) and its substrate mixed-lineage kinase domain-like protein (MLKL). Phosphorylated MLKL forms tetramers and translocates to membrane fractions to induce cell death. Here we report that MLKL tetramers further polymerize to form disulfide bond-dependent amyloid-like fibers, which are required for necroptosis. Furthermore, induced polymerization of the MLKL N-terminal domain is sufficient to activate necroptosis without RIPK3. This work reveals a mechanism for MLKL activation and generates exciting directions for necroptosis regulation.
Keywords: necroptosis, MLKL, disulfide bond, amyloid-like, polymer
Abstract
Mixed-lineage kinase domain-like protein (MLKL) is essential for TNF-α–induced necroptosis. How MLKL promotes cell death is still under debate. Here we report that MLKL forms SDS-resistant, disulfide bond-dependent polymers during necroptosis in both human and mouse cells. MLKL polymers are independent of receptor-interacting protein kinase 1 and 3 (RIPK1/RIPK3) fibers. Large MLKL polymers are more than 2 million Da and are resistant to proteinase K digestion. MLKL polymers are fibers 5 nm in diameter under electron microscopy. Furthermore, the recombinant N-terminal domain of MLKL forms amyloid-like fibers and binds Congo red dye. MLKL mutants that cannot form polymers also fail to induce necroptosis efficiently. Finally, the compound necrosulfonamide conjugates cysteine 86 of human MLKL and blocks MLKL polymer formation and subsequent cell death. These results demonstrate that disulfide bond-dependent, amyloid-like MLKL polymers are necessary and sufficient to induce necroptosis.
Necroptosis is a form of programmed necrotic cell death, morphologically characterized by organelle swelling and plasma membrane rupture (1, 2). Recent studies have implicated necroptosis in a broad range of pathological conditions including infection, inflammation, sepsis, and ischemia injury as well as neurodegeneration (3–8). Understanding the regulation of necroptosis will have profound impact on human health.
The molecular mechanism of necroptosis has been under intensive investigation in recent years. The best-studied pathway is TNF-induced necroptosis. Upon TNF treatment and apoptosis inhibition, a series of events leads to the formation of the necrosome, which contains RIPK1, RIPK3, and MLKL (9–16). RIPK3 phosphorylates at least two important sites in the activation loop region of the kinase-like domain of MLKL (14, 17, 18). Phosphorylated MLKL homo-oligomerizes and translocates to cellular membranes to trigger cell death (17, 19, 20). However, how MLKL oligomers kill cells is still under debate. Recombinant MLKL was shown to bind phospholipids and cause liposome leakage in vitro, leading to the model that oligomerized MLKL could form pores to permeabilize cell membrane (17, 21–23). Nonetheless, membrane translocation alone is not sufficient for cell death induction, because some MLKL mutants can form oligomers and translocate to the cell membrane but cannot kill cells (24). Others reported that MLKL binds to a calcium transporter or sodium transporters on the cell membrane to induce ion influx (19, 20). Recently it was reported that recombinant MLKL can form cation channels (25, 26). How to reconcile these results and clarify the execution step of necroptosis remains a big challenge.
One exciting development in the field is that RIPK1 and RIPK3 form amyloid fibrils to activate necroptosis (27). Traditionally, amyloids are viewed as loss-of-function filamentous protein aggregates associated with a multitude of neurodegenerative diseases (28, 29). Recently, they have received a lot attention due to their ability to serve as polymeric platforms capable of recruiting and nucleating signaling protein complexes (30). For example, the death effector domain-containing proteins FADD and caspase 8 were reported to form filamentous structures to activate apoptosis (31). More recently, prion-like polymerization has been shown to be responsible for mitochondrial antiviral signaling protein (MAVS)-dependent antiviral innate immune response (32), and apoptosis-associated spec-like protein (ASC)-dependent inflammasome assembly (33–35).
In this study, we report that upon necroptosis induction MLKL forms disulfide bond-dependent amyloid-like polymers that are distinct from RIPK1 and RIPK3 polymers. Recombinant MLKL N-terminal domain (NTD) forms fibers in vitro. NTD with all cysteines mutated fails to form polymers and is unable to induce necroptosis efficiently. Furthermore, compound necrosulfonamide (NSA) conjugates cysteine 86 of human MLKL to block MLKL polymer formation and subsequent cell death.
Results
MLKL Forms Polymers upon Necroptosis Induction in both Human and Mouse Cells.
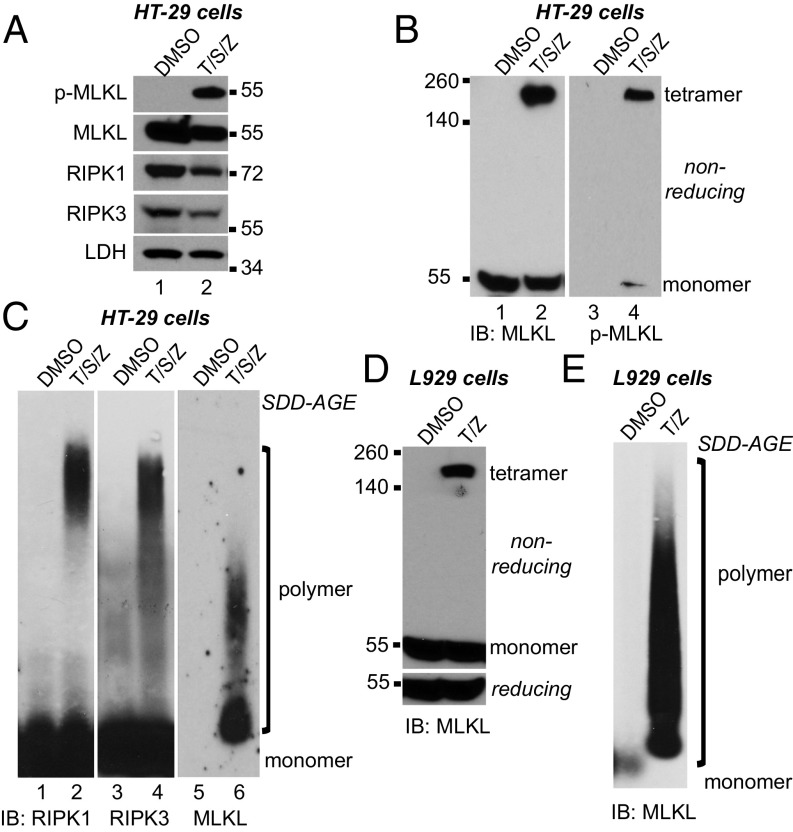
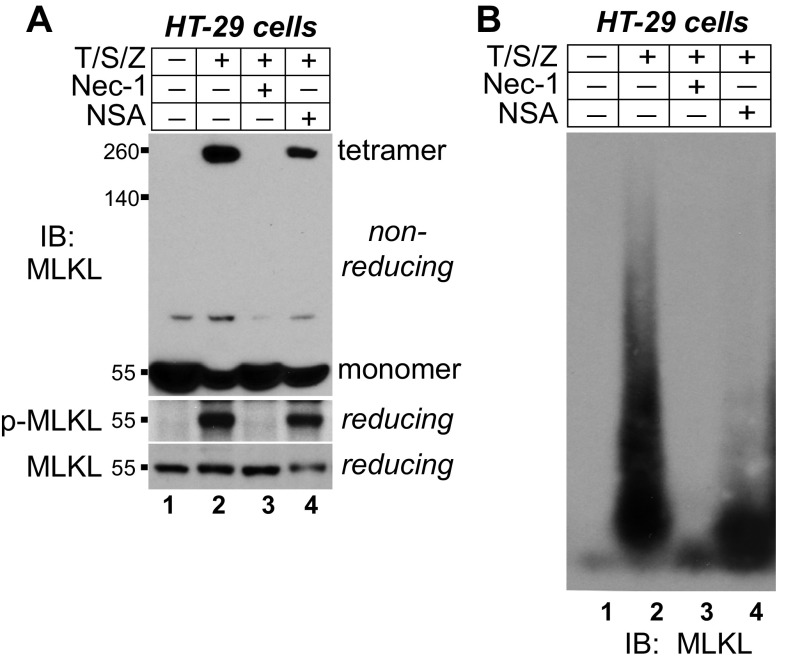
RIPK1 and RIPK3 form amyloid complexes during necroptosis (27). Because MLKL interacts with RIPK3 in the necrosome, we asked if MLKL was also in amyloid-like complexes. HT-29 human colon cancer cells underwent necroptosis upon TNF (T), Smac-mimetic (S), and Z-VAD-FMK (Z) treatment. MLKL was phosphorylated (Fig. 1A) and formed tetramers on a nonreducing gel (Fig. 1B, lane 2). As expected, the majority of phosphorylated MLKL was in the tetramer (Fig. 1B, lane 4).
Fig. 1.
MLKL forms large polymers during necroptosis in human and mouse cells. (A) HT-29 cells were treated with DMSO or 20 ng/mL TNF (T), 100 nM Smac-mimetic (S), and 20 μM Z-VAD-FMK (Z). Cell lysates were subjected to Western blotting with the indicated antibodies. P-MLKL antibody detects phospho-S358 of human MLKL. (B) HT-29 cell lysates were separated by nonreducing SDS/PAGE and detected with antibodies against MLKL or p-MLKL. (C) HT-29 cell lysates were separated by SDD-AGE and detected with RIPK1, RIPK3, or MLKL antibodies. (D) L929 cells were treated with DMSO or 2 ng/mL TNF (T) and 20 μM Z-VAD-FMK (Z), and cell lysates were subjected to nonreducing or reducing SDS/PAGE and detected with MLKL antibody. (E) L929 cell lysates were separated by SDD-AGE and detected with MLKL antibody. IB, immunoblot.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) detects large protein polymers resistant to 2% SDS treatment (36). After necroptosis induction, RIPK1 and RIPK3 formed clear polymers, which clustered at the top (Fig. 1C, lanes 2 and 4). The migration patterns of RIPK1 and RIPK3 are almost identical, confirming the earlier report that RIPK1 and RIPK3 are presented in the amyloid complexes in a 1:1 ratio (27). Interestingly, MLKL also formed polymers with a unique migration pattern (Fig. 1C, lane 6). MLKL polymers were more heterogeneous and spread out on the gel, and the majority of MLKL polymers migrated faster than RIPK1/3 polymers, suggesting that the size of MLKL polymers is smaller. Under this condition, the monomer form of MLKL was barely detected (Fig. 1C, lane 5), possibly because the antigen (amino acids 1–226) was not fully exposed. MLKL tetramers and polymers were also detected in mouse L929 cells (Fig. 1 D and E), suggesting that polymer formation of MLKL is conserved across species.
MLKL Polymers Are Protease K-Resistant, High Molecular Weight Complexes.
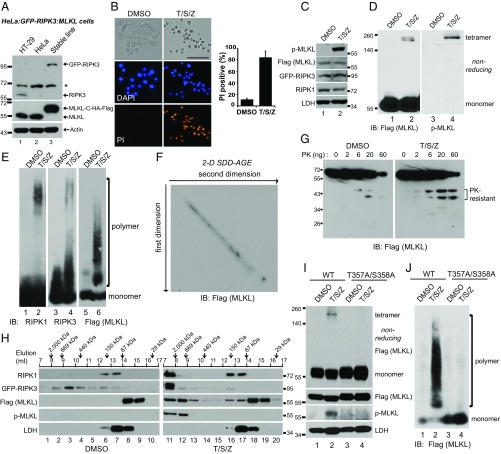
To facilitate purification of MLKL polymers, we engineered a HeLa:GFP-RIPK3:MLKL cell line in which endogenous MLKL was knocked out using CRISPR-Cas9, and C-terminal HA and Flag-tagged MLKL and GFP-RIPK3 were stably expressed (Fig. 2A). With T/S/Z treatment these cells underwent necroptosis and positively stained by propidium iodide (PI), which is a cell-impermeable DNA dye (Fig. 2B). Moreover, MLKL was phosphorylated and formed tetramers (Fig. 2 C and D). As in HT-29 cells, phosphorylated MLKL was mostly in tetramers. On nonreducing gels, MLKL was sometimes detected as doublets, possibly because of intramolecular disulfide bonds (Fig. 2D). Importantly, RIPK1, RIPK3, and MLKL all formed SDS-resistant polymers (Fig. 2E). To address the possibility that the size heterogeneity of the MLKL polymers came from the degradation of large polymers during the running of the SDD-AGE, we ran a 2D SDD-AGE. MLKL showed a perfect diagonal pattern, indicating that there is no degradation during SDD-AGE running (Fig. 2F). Some amyloid polymers are more resistant to protease digestion (29). As shown in Fig. 2G, after necroptosis induction, two proteinase K-resistant MLKL fragments were more prominent, suggesting that there is a conformational shift when MLKL forms polymers.
Fig. 2.
MLKL polymers are proteinase K-resistant, high molecular weight complexes. (A) Establishment of the HeLa:GFP-RIPK3:MLKL cell line. Endogenous MLKL was knocked out with CRISPR-Cas9 (lane 3), and C-terminal HA and Flag-tagged MLKL, as well as GFP-RIPK3, were stably expressed. HeLa cells do not express endogenous RIPK3. The asterisk denotes a nonspecific band. (B) HeLa:GFP-RIPK3:MLKL cells were stained with DAPI and PI. (Scale bar, 100 μm.) Quantification is shown on the right. Data are presented as mean ± SD. (C) Cell lysates were separated on SDS/PAGE and detected with the indicated antibodies. (D) Cell lysates were separated by nonreducing SDS/PAGE and detected with antibodies against Flag or p-MLKL. (E) Cell lysates were separated by SDD-AGE and detected with antibodies against RIPK1, RIPK3, or Flag. (F) Lysates from T/S/Z-treated cells were separated by 2D SDD-AGE and detected with Flag antibody. (G) DMSO- or T/S/Z-treated cell lysates (20 μg) were incubated with different amounts of proteinase K at 30 °C for 10 min and subjected to Western blotting with Flag antibody. (H) DMSO- or T/S/Z-treated cell lysates were subjected to Superdex 200 gel filtration fractionation. Fractions were analyzed by Western blotting with the indicated antibodies. The elution positions of the molecular mass markers are identified at the top. LDH was used as a control. (I and J) C-terminal HA- and Flag-tagged wild-type or T357A/S358A mutant MLKL was expressed at low level in MLK-knockout HeLa cells that were then treated with DMSO or T/S/Z. Cell lysates were subjected to nonreducing or reducing SDS/PAGE (I) or SDD-AGE (J) and detected with the indicated antibodies. IB, immunoblot.
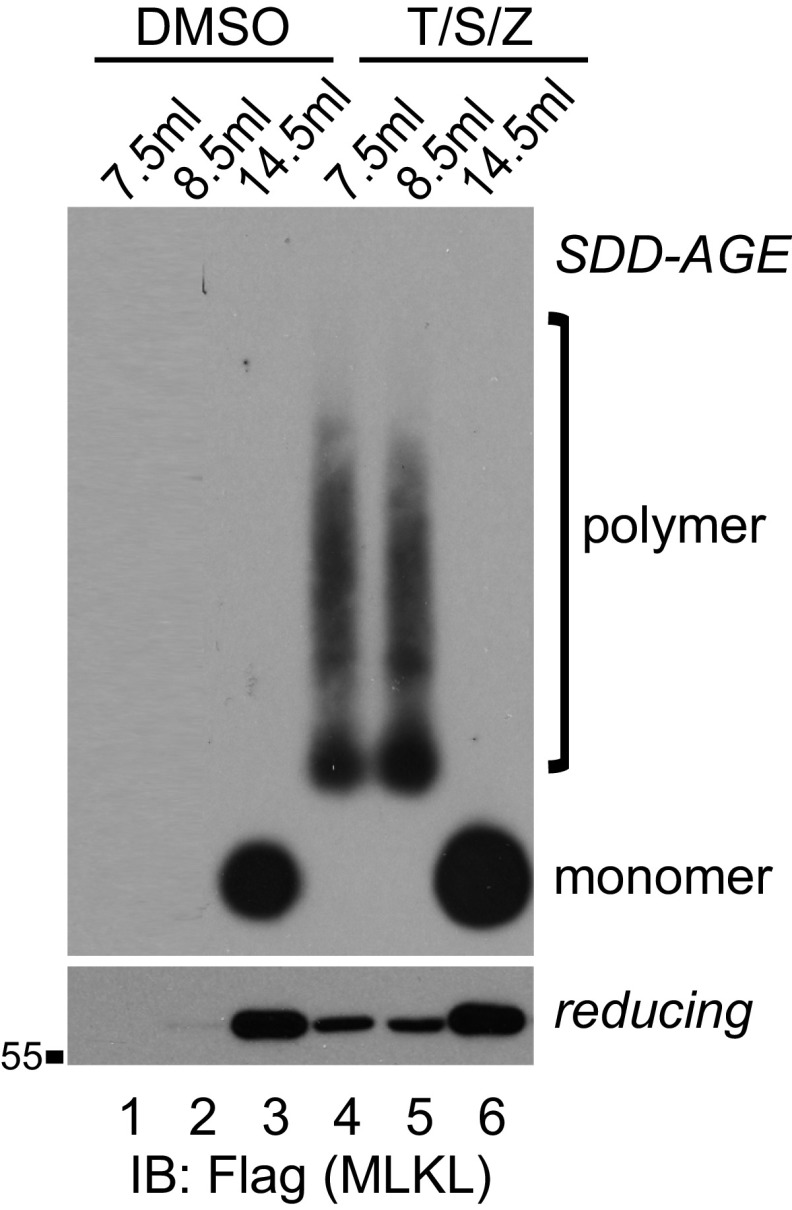
We then examined the size of RIPK1/3 and MLKL complexes through gel filtration. In the DMSO-treated sample, RIPK1 eluted at around 150 kDa, possibly in a mixture of monomer and dimer forms. Interestingly, RIPK3 spread out on the column, while MLKL mainly eluted as monomers at below 67 kDa (Fig. 2H, Left). In the T/S/Z-treated sample, RIPK1 and RIPK3 migrated to the void volume in complexes larger than 2 million Da, while lactate dehydrogenase (LDH) did not change (Fig. 2H, Right). MLKL formed complexes of more than 669 kDa, much larger than a tetramer (∼240 KDa) or the recently reported octamer (∼480 kDa) (26). Phosphorylated MLKL eluted solely in the large complexes (lanes 11 and 12), which contained MLKL polymers (Fig. S1, lanes 4 and 5).
Fig. S1.
MLKL polymers are high molecular weight complexes. DMSO- or T/S/Z-treated cell lysates were subjected to Superdex 200 gel filtration fractionation. Fractions eluted at 7.5 mL, 8.5 mL, and 14.5 mL were analyzed by SDD-AGE and SDS/PAGE and detected with Flag antibody. IB, immunoblot.
Phosphorylation of MLKL at T357 and S358 by RIPK3 has been shown to be required for MLKL activation (14, 17). We examined if phosphorylation of those sites is required for MLKL polymer formation. Consistent with a previous report (17), the T357A/S358A mutant of MLKL could not form tetramers (Fig. 2I, lane 4). It also failed to form polymers (Fig. 2J, lane 4), suggesting that phosphorylation of MLKL by RIPK3 at T357 and S358 is required for MLKL polymer formation.
MLKL Polymers Are Disulfide Bond-Dependent, Amyloid-Like Fibers.
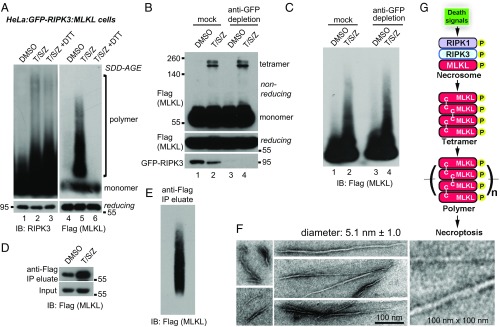
Multiple groups have reported that MLKL forms disulfide bond-linked oligomers (17, 19, 20, 27). Therefore, we tested if MLKL polymers were also linked by disulfide bonds. As shown in Fig. 3A, after incubation with 10 mM DTT for 30 min, RIPK3 polymers remained intact (lane 3). However, MLKL polymers completely dissociated and migrated to the monomer position (lane 6), indicating that disulfide bonds are required to maintain MLKL polymers. This result also suggests that RIPK3 polymers and MLKL polymers are different entities. Indeed, after GFP-RIPK3 was depleted from the whole-cell lysates with an anti-GFP antibody, MLKL tetramers were preserved (Fig. 3B, lane 4), and MLKL polymers were intact (Fig. 3C, lane 4).
Fig. 3.
MLKL polymers are disulfide bond-dependent, amyloid-like fibers that are distinct from RIPK3 polymers. (A) DMSO- or T/S/Z-treated lysates or T/S/Z-treated lysates incubated with 10 mM DTT for 30 min were subjected to SDD-AGE and detected with RIPK3 or Flag antibody. (B and C) DMSO- or T/S/Z-treated cell lysates were depleted with a control antibody or an anti-GFP antibody and subjected to nonreducing or reducing SDS/PAGE (B) or SDD-AGE (C) and were detected with Flag or RIPK3 antibody. (D) Cell lysates were subjected to immunoprecipitation (IP) with anti-Flag M2 beads, eluted with pH 3 buffer, and analyzed by Western blotting. (E) The eluate from immunoprecipitation of the T/S/Z-treated lysates was analyzed by SDD-AGE. (F) The eluate from immunoprecipitation of the T/S/Z-treated lysates was subjected to negative staining and imaged by EM. At least 10 fibers were measured to calculate the average diameter. (G) Working model. MLKL is recruited to the necrosome to be phosphorylated and then forms disulfide bond-dependent tetramers. MLKL tetramers further polymerize to form amyloid-like polymers separated from the RIPK1/RIPK3 fibers to promote necroptosis. C–C, disulfide bond. IB, immunoblot.
MLKL polymers were isolated from cells by anti-Flag immunoprecipitation and pH 3 buffer elution (Fig. 3D). The polymers were preserved after elution, as demonstrated by SDD-AGE (Fig. 3E). The polymers were then subjected to negative-staining EM. Fibers with a consistent diameter (5.1 nm) but of various lengths were observed (Fig. 3F). The variation in the length might reflect the heterogeneity seen by SDD-AGE. The diameter of MLKL fibers is different from that reported for RIPK1/3 fibers, which have a diameter of 11–12 nm (27). We propose a model in which phosphorylated MLKL forms tetramers with stabilizing disulfide bonds and tetramers serve as building blocks that polymerize to form amyloid-like fibers to induce necroptosis (Fig. 3G).
Recombinant MLKL NTD Forms Disulfide Bond-Dependent Amyloid-Like Fibers in Vitro.
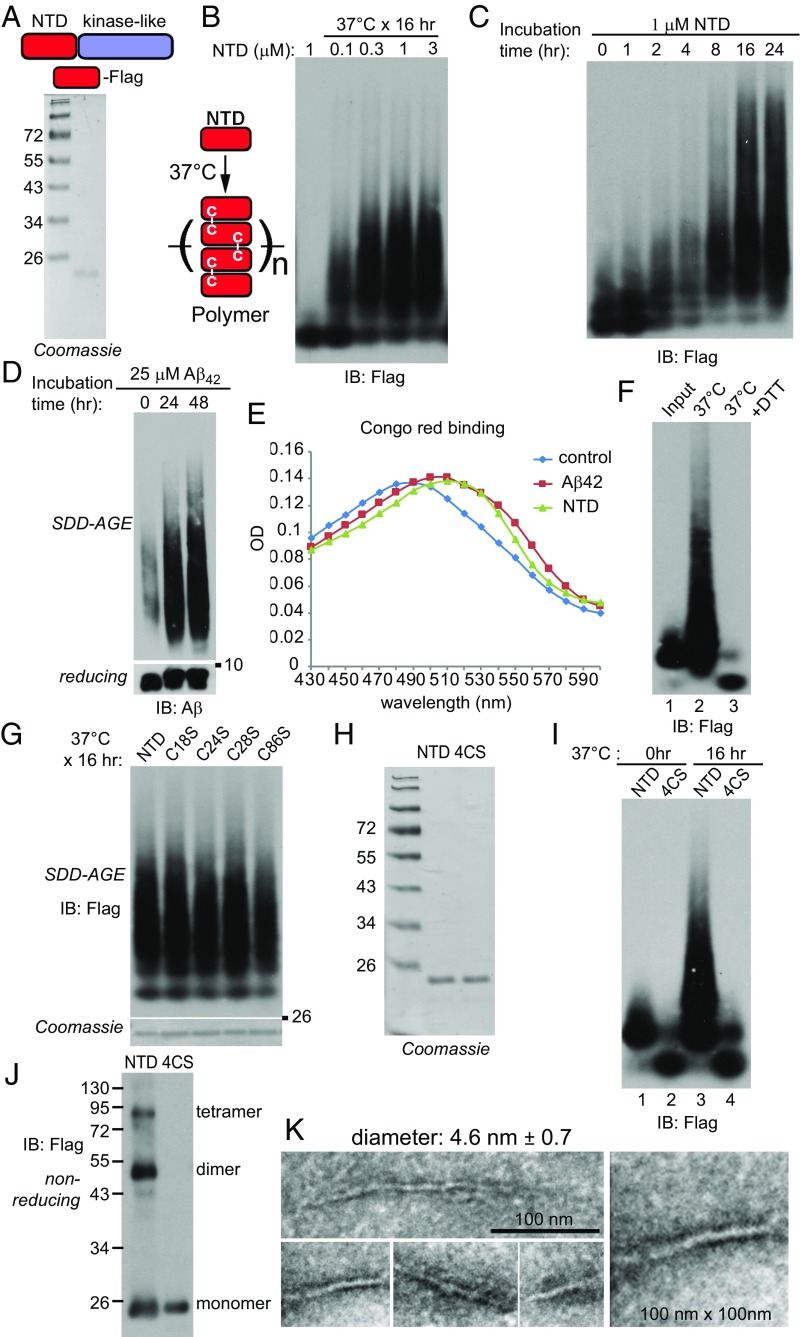
To exclude the possibility that the polymers purified from the cells might still contain other proteins, we turned to a recombinant system. The NTD of MLKL is sufficient to induce necroptosis when overexpressed (20, 23, 24). Moreover, recombinant NTD could permeabilize liposomes (17, 21, 22). We purified recombinant human MLKL-NTD with a Flag tag at the C terminus (Fig. 4A). Incubation of different concentrations of MLKL-NTD at 37 °C resulted in polymer formation (Fig. 4B) in a time-dependent manner (Fig. 4C). As a positive control, β-amyloid peptide (amino acids 1–42, Aβ42) also formed polymers (Fig. 4D). Many amyloid-like polymers bind dyes such as Congo red or thioflavin T. Both MLKL polymers and Aβ42 polymers showed Congo red binding and right shifted the maximal absorbance wavelength (Fig. 4E). DTT incubation totally broke down the NTD polymers on SDD-AGE (Fig. 4F), suggesting that disulfide bonds are also important for holding recombinant polymers together.
Fig. 4.
The recombinant MLKL NTD forms disulfide bond-dependent amyloid-like polymers in vitro. (A) Coomassie staining of recombinant C-terminal Flag-tagged MLKL NTD. (B) Different concentrations of the MLKL NTD were incubated at 37 °C for 16 h. For each condition, 50 ng of protein was loaded for SDD-AGE. (C) Recombinant NTD (1 μM) was incubated at 37 °C for different durations. At each time point, 50 ng of protein was loaded for SDD-AGE. (D) Aβ42 peptide (25 μM) was incubated at 37 °C for 24 or 48 h. At each time point, 100 ng protein was loaded for SDD-AGE and SDS/PAGE. (E) Aβ42 polymers (10 μM) or NTD polymers (2.5 μM) were incubated with 50 μM Congo red for 10 min at room temperature, and absorptions were measured at the indicated wavelengths. (F) NTD (1 μM) was incubated at 37 °C for 16 h (lane 2), and half of the sample was further incubated with 10 mM DTT for 30 min (lane 3). For each lane, 50 ng of protein was loaded for SDD-AGE. (G) Recombinant C18S, C24S, C28S, and C86S proteins were purified (Coomassie staining, Lower), and 1 μM of each was incubated at 37 °C for 16 h. Then 50 ng of each was loaded for SDD-AGE (Upper). (H) Coomassie staining of recombinant NTD and NTD with all four cysteines mutated to serines (4CS). (I) Recombinant NTD or 4CS (1 μM) was incubated at 37 °C for 0 h or 16 h. Then 50 ng of each sample was loaded for SDD-AGE. (J) Purified NTD or 4CS (50 ng) was analyzed by nonreducing SDS/PAGE. (K) Recombinant NTD polymers were subjected to negative staining and imaged by EM. At least 10 fibers were measured to calculate the average diameter. IB, immunoblot.
There are four cysteines in the NTD: C18, C24, C28, and C86. We mutated each individual cysteine to a serine, and the recombinant proteins still formed polymers (Fig. 4G). Disulfide bonds are known to rearrange and shuffle (37), which might compensate for the loss of a single cysteine. We then mutated all four cysteines. The mutant 4CS was no longer able to form polymers (Fig. 4 H and I). Even before incubation, the 4CS mutant migrated faster than the wild type (compare lane 2 and lane 1), suggesting that the wild-type NTD might have formed disulfide bond-linked oligomers without incubation. Indeed, dimers and tetramers of NTD were detected on a nonreducing gel, while 4CS only had monomers (Fig. 4J). The NTD polymers were then subjected to negative staining and EM analysis. The diameter of the fibers was slightly smaller than the fibers isolated from cells, at about 4.6 nm (Fig. 4K). The difference in the diameter might be because the recombinant protein lacked the C-terminal kinase-like domain.
Disulfide Bonds Are Important for NTD-DmrB Polymer Formation and Necroptosis Induction in Vivo.
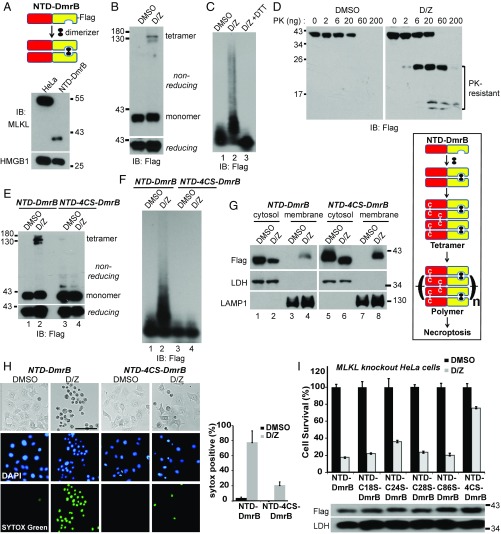
To examine the functional importance of the disulfide bond in the NTD, we used the dimerization system from Clontech, in which the DmrB domain forms a dimer with dimerizer (D) addition. The NTD of human MLKL was fused to the DmrB domain with a C-terminal Flag tag and was stably expressed in MLKL-knockout HeLa cells (Fig. 5A). With D and Z treatment, the fusion protein formed tetramers (Fig. 5B) and polymers (Fig. 5C). Importantly, DTT completely dissociated the polymers (Fig. 5C, lane 3). Again, the D/Z-treated sample had prominent PK-resistant fragments (Fig. 5D).
Fig. 5.
Disulfide bonds are necessary for NTD-DmrB polymer formation and necroptosis induction in vivo. (A) Establishment of the NTD-DmrB cell line. C-terminal Flag-tagged NTD-DmrB fusion protein was stably expressed in MLKL-knockout HeLa cells. Cell lysates were subjected to Western blotting and detected with MLKL or HMGB1 antibody. (B) NTD-DmrB cells were treated with DMSO or 20 nM dimerizer (D) and 20 μM Z-VAD-FMK (Z). Cell lysates were separated by nonreducing or reducing SDS/PAGE. (C) DMSO- or D/Z-treated lysates or D/Z-treated lysates incubated with 10 mM DTT were subjected to SDD-AGE. (D) Cell lysates (20 μg) were incubated with different amounts of proteinase K at 30 °C for 10 min and then subjected to Western blotting. (E and F) NTD-4CS-DmrB was stably expressed in MLKL-knockout HeLa cells. DMSO- or D/Z-treated lysates were subjected to nonreducing or reducing SDS/PAGE (E) or SDD-AGE (F). (G) DMSO- or D/Z-treated cells were fractionated into cytosol and crude membrane fractions. (Left) The samples were then subjected to Western blotting with the indicated antibodies. LDH is a cytosol marker, and LAMP1 is a membrane marker. (Right) Working model. Dimerizer induces the formation of disulfide bond-dependent NTD-DmrB tetramers that further polymerize to induce necroptosis. (H) NTD-DmrB or NTD-4CS-DmrB cells were treated with DMSO or D/Z and were stained with DAPI and SYTOX Green. (Scale bar, 100 μm.) Quantification is shown on the right. Data are presented as mean ± SD. (I, Upper) NTD-DmrB cysteine mutants were expressed in MLKL-knockout HeLa cells, and cell survival was determined by CellTiter-Glo assay. Data are presented as mean ± SD. (Lower) Western blotting with Flag or LDH antibodies was performed to assess protein expression. IB, immunoblot.
The four-cysteine mutant NTD-4CS-DmrB did not form tetramers on a nonreducing gel (Fig. 5E, lane 4) and failed to form polymers on SDD-AGE (Fig. 5F, lane 4). MLKL translocates to membrane fractions during necroptosis (17, 19, 20). As shown in Fig. 5G, a portion of the NTD-4CS-DmrB mutant translocated to the crude membrane fractions (lane 8), similar to the NTD-DmrB (lane 4). Importantly, the NTD-4CS-DmrB mutant was severely compromised in its ability to induce necroptosis (Fig. 5 H and I), while the single-cysteine mutants retained most of that ability. These results highlight the importance of disulfide bond-dependent MLKL polymer formation for necroptosis induction.
NSA Blocks NTD Polymer Formation in a Cysteine 86-Dependent Manner to Block Necroptosis.
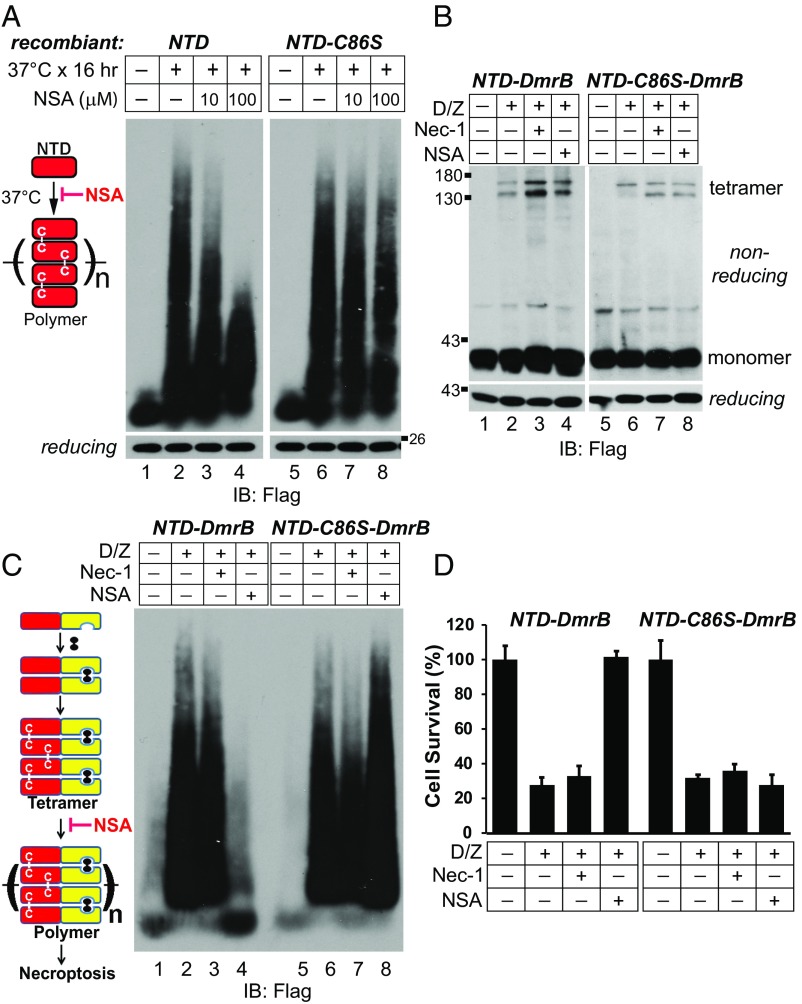
Compound NSA conjugates cysteine 86 of human MLKL to block necroptosis (14). We examined the effect of NSA on MLKL polymer formation. As shown in Fig. 6A, NSA inhibited recombinant NTD polymerization in vitro in a dose-dependent manner (lanes 3 and 4). However, it had no effect on NTD-C86S (lanes 7 and 8), indicating that NSA needs to conjugate cysteine 86 of MLKL to block polymer formation.
Fig. 6.
NSA blocks NTD polymer formation in a cysteine 86-dependent manner to block necroptosis. (A) Recombinant NTD or NTD-C86S (1 μM) was incubated at 37 °C with or without NSA, and 50 ng protein from each treatment was loaded for SDD-AGE. (B and C) NTD-DmrB or NTD-C86S-DmrB cells were treated as indicated. Cell lysates were subjected to nonreducing or reducing SDS/PAGE (B) or SDD-AGE (C). (D) NTD-DmrB or NTD-C86S-DmrB cells were subjected to the indicated treatments, and cell survival was determined by CellTiter-Glo assay. Data are presented as mean ± SD. IB, immunoblot.
Next, we tested if NSA affected polymer formation in vivo. Compound necrostatin-1 (Nec-1) blocks RIPK1 activity (38), which is not required for necroptosis in the NTD-DmrB cells. As shown in Fig. 6B, neither Nec-1 nor NSA could block dimerizer-induced tetramer formation in NTD-DmrB cells (lanes 3 and 4). However, NSA blocked polymer formation (Fig. 6C, lane 4). When cysteine 86 was mutated to serine, NSA was no longer able to block polymer formation (Fig. 6C, lane 8). Consequently, NSA blocked necroptosis in NTD-DmrB cells, but not in NTD-C86S-DmrB cells (Fig. 6D). These results suggest that NSA conjugates cysteine 86 of human MLKL to block MLKL polymer formation and necroptosis.
NSA Blocks MLKL Polymer Formation and Necroptosis in Human Cells but Not in Mouse Cells.
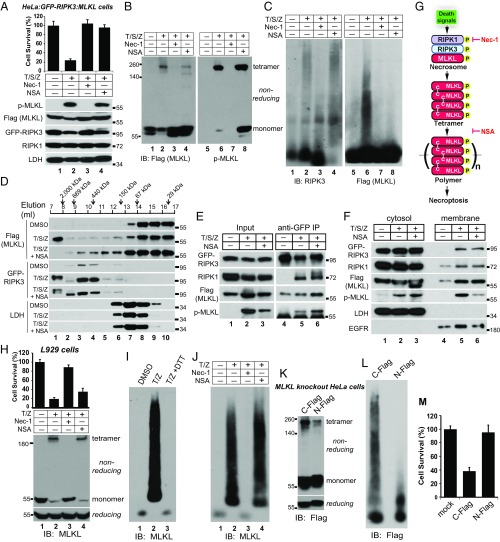
In HeLa:GFP-RIPK3:MLKL cells, both Nec-1 and NSA blocked necroptosis (Fig. 7A). By inhibiting RIPK1 activity, Nec-1 blocked MLKL phosphorylation (Fig. 7A, lane 3) and tetramer formation (Fig. 7B, lane 3). On the other hand, NSA modestly reduced MLKL phosphorylation and tetramer formation (Fig. 7 A and B, lane 4). Moreover, phosphorylated MLKL was still present in the tetramers (Fig. 7B, lane 8). The RIPK3 polymers did not form in Nec-1–treated samples but still formed in NSA-treated samples (Fig. 7C, lanes 3 and 4). Importantly, MLKL polymers did not form in NSA-treated samples (lane 8), indicating that NSA conjugation to cysteine 86 of MLKL blocks MLKL polymer formation. This again suggests that MLKL polymers and RIPK3 polymers are distinct entities. On the gel filtration column, in both T/S/Z- and T/S/Z plus NSA-treated samples, RIPK3 migrated to the void volume, consistent with the SDD-AGE result. However, in the T/S/Z plus NSA-treated sample, MLKL did not migrate to the void volume, confirming that NSA blocked MLKL polymer formation (Fig. 7D, lane 1). NSA also blocked MLKL polymer formation in HT-29 cells (Fig. S2). It has been shown that NSA enhances necrosome formation (14, 16). We examined its effect on phosphorylated MLKL. Consistent with previous results, NSA increased RIPK3 interaction with RIPK1 and MLKL. Interestingly, it also enhanced the presence of phosphorylated MLKL in the necrosome (Fig. 7E, lane 6). With T/S/Z plus NSA treatment, RIPK3, RIPK1, and MLKL still translocated to the membrane fraction (Fig. 7F, lane 6). However, less phosphorylated MLKL was present in the membrane fraction compared with T/S/Z treatment alone (Fig. 7F, lanes 5 and 6). These results suggest that NSA enhances necrosome formation but blocks the polymerization of MLKL tetramers and subsequent cell death (Fig. 7G).
Fig. 7.
NSA blocks MLKL polymer formation in human cells, and N-terminal–tagged MLKL fails to form polymers to promote necroptosis. (A, Upper) HeLa:GFP-RIPK3:MLKL cells were subjected to the indicated treatments, and cell survival was determined by CellTiter-Glo assay. Data are presented as mean ± SD. (Lower) Cell lysates were subjected to Western blotting with the indicated antibodies. (B) Cells were treated as indicated, and cell lysates were subjected to nonreducing SDS/PAGE and detected with Flag or p-MLKL antibody. (C) Cells lysates were subjected to SDD-AGE and detected with RIPK3 or Flag antibody. (D) Cells were treated with DMSO, T/S/Z, or T/S/Z plus NSA, and cell lysates were separated by gel filtration. The fractions were subjected to Western blotting with the indicated antibodies. (E) Cell lysates were subjected to immunoprecipitation with an anti-GFP antibody and were analyzed by Western blotting with the indicated antibodies. (F) Cells were separated into cytosol and crude membrane fractions and were analyzed by Western blotting with the indicated antibodies. LDH is a cytosol marker, and EGF receptor (EGFR) is a membrane marker. (G) Working model. NSA does not block necrosome formation. Instead, it blocks the polymerization of the tetramers to inhibit necroptosis. (H, Upper) L929 cells were subjected to the indicated treatments, and cell survival was determined by CellTiter-Glo assay. Data are presented as mean ± SD. (Lower) Cell lysates were subjected to nonreducing or reducing SDS/PAGE and were detected with mouse MLKL antibody. (I) DMSO- or T/Z-treated lysates or T/Z-treated lysates incubated with 10 mM DTT were separated by SDD-AGE and detected with mouse MLKL antibody. (J) L929 cells were treated as indicated, and cell lysates were subjected to SDD-AGE and detected with mouse MLKL antibody. (K and L) C-terminal Flag-tagged MLKL or N-terminal Flag-tagged MLKL was overexpressed in MLKL-knockout HeLa cells. Cell lysates were subjected to nonreducing or reducing SDS/PAGE (K) or SDD-AGE (L). (M) C-Flag or N-Flag MLKL was overexpressed in MLKL-knockout HeLa cells, and cell survival was determined by CellTiter-Glo assay. Data are presented as mean ± SD. IB, immunoblot.
Fig. S2.
NSA blocks MLKL polymer formation in HT-29 cells. (A) HT-29 cells were treated as indicated, and cell lysates were subjected to nonreducing or reducing SDS/PAGE and detected with the indicated antibodies. (B) HT-29 cells were treated as indicated, and cell lysates were subjected to SDD-AGE and detected with MLKL antibody. IB, immunoblot.
NSA cannot conjugate mouse MLKL because mouse MLKL lacks the cysteine corresponding to cysteine 86 of human MLKL (14). It did not block necroptosis efficiently in mouse L929 cells (Fig. 7H). It also did not block tetramer formation (Fig. 7H, lane 4). During necroptosis mouse MLKL formed polymers that could be dissociated with DTT incubation, suggesting that they are also disulfide bond-dependent (Fig. 7I, lane 3). Importantly, Nec-1 totally blocked polymer formation, while NSA did not (Fig. 7J), correlating with their ability to block cell death.
N-Terminal–Tagged MLKL Fails to Form Polymers to Promote Necroptosis.
It was reported that N-terminal–tagged MLKL could not induce necroptosis (20, 26), but the reason is not well understood. C-terminal Flag-tagged or N-terminal Flag-tagged MLKL was overexpressed in MLKL-knockout HeLa cells. Both formed tetramers (Fig. 7K). However, unlike MLKL-C-Flag, N-Flag-MLKL failed to form large polymers (Fig. 7L). This correlated with their ability to induce cell death (Fig. 7M), again suggesting that polymer formation is essential for MLKL to induce necroptosis.
Discussion
The following experiments lead us to believe that disulfide bond-dependent MLKL fibers are necessary and sufficient for necroptosis induction. (i) MLKL forms SDS-resistant polymers on SDD-AGE. (ii) Large MLKL polymers elute in the void volume of gel filtration, with a size more than 2 million Da. (iii) MLKL polymers are proteinase K-resistant. (iv) MLKL polymers are fibers under EM. (v) MLKL polymers are independent of RIPK1 and RIPK3 fibers. (vi) MLKL polymers are DTT-sensitive. (vii) Recombinant NTD of MLKL forms polymers in vitro. (viii) NTD polymers bind the amyloid dye Congo red. (ix) The NTD with all cysteines mutated fails to form polymers in vitro and in vivo. Moreover, it cannot induce necroptosis efficiently. (x) NSA conjugation to cysteine 86 of human MLKL blocks MLKL polymer formation and subsequent necroptosis. (xi) N-terminal–tagged MLKL cannot form polymers, providing a possible explanation for its inability to induce necroptosis, shown previously (19, 26).
The MLKL polymers have some unique features. First, they are disulfide bond-dependent, gain-of-function fibers. Although some misfolded neurodegenerative proteins such as PrP and SOD1 form loss-of function fibers that contain disulfide bonds (39), here we show that disulfide bonds are required for the gain-of-function activity of amyloid-like MLKL fibers. There is also disulfide bond formation in the MAVS polymers, but it is not required for MAVS activity (32). Second, the MLKL polymerization process has a relatively stable tetramer step, which is separable from polymers. For example, NSA could preserve the tetramers and block polymer formation, and N-Flag-MLKL could form tetramers but could not form polymers. These results suggest that polymerization from tetramers to polymers is a tightly regulated process. Third, unlike the β-sheet structure of RIPK1 and RIPK3 amyloids (27), MLKL polymers possibly contain mainly helical structures, because the entire NTD is comprised of helixes (21, 40, 41). This is similar to the MAVS and ASC polymers which are also comprised of α-helical bundles and do not bind thioflavin T (32–34). It has been suggested that amyloid fibrils be defined as “fibrillar polypeptide aggregates with cross-β conformation” (42). We consider MLKL polymers amyloid-like because they are fibrillar polypeptide aggregates that bind the amyloid dye Congo red but possibly do not form cross-β conformation.
MLKL localizes in a reducing environment of the cytosol, but it still forms disulfide bonds upon necroptosis induction. One possibility is that within the necrosome, the RIPK1/3 polymers serve as a nucleating factor to concentrate and phosphorylate MLKL, which induces MLKL conformation changes to a state in which cysteines are in close proximity to form intra- and intermolecular disulfide bonds. Disulfide bond formation would further stabilize the adopted conformation and facilitate downstream polymerization. A piece of supporting evidence is that purified NTD spontaneously forms dimers and tetramers in vitro (Fig. 4J). Moreover, in cultured cells, after the addition of dimerizer, NTD-DmrB forms polymers instead of only dimers despite the reducing cytosolic environment (Fig. 5C). These results suggest that NTD could undergo conformation shifts to form disulfide bonds when brought together. A second possibility is that the necroptotic signal generates a local burst of reactive oxygen species (ROS), which could induce oxidation of MLKL. A recent report demonstrated that upon necroptosis induction mitochondria ROS promoted RIPK1 autophosphorylation and RIPK3 recruitment (43). It is possible that the same mitochondria ROS could regulate MLKL disulfide bond formation and polymerization. There is also a possibility that MLKL tetramers could be transported to an oxidizing environment, such as the lumen of the endoplasmic reticulum or lysosome.
NSA conjugates cysteine 86 of human MLKL and blocks MLKL polymer formation. However, the C86S mutant and mouse MLKL without the corresponding cysteine still form tetramers and polymers, suggesting that cysteine 86 itself is not essential for polymer formation. One logical explanation is that conjugated NSA occupies the space between the tetramer building blocks and prevents them from coming close enough to polymerize. This is supported by the facts that NSA does not totally block tetramer formation and that the C86S mutant forms polymers in the presence of NSA (Fig. 6).
N-terminal–tagged MLKL fails to form large polymers and cannot induce necroptosis (19, 26). These results suggest that the exact N-terminal sequences are important for polymer formation in the cells. One possibility is that the bulky N-terminal tag, like NSA conjugation, creates steric hindrance for polymerization. Another possibility is that the exact N-terminal sequences are involved in binding to other proteins or lipids on the membranes, which are important for polymer formation. Supporting evidence for membrane involvement in MLKL polymerization is that recombinant NTD oligomerizes after exposure to liposome (21). Furthermore, N-Flag-MLKL could translocate to the plasma membrane (26), but it could not form polymers (Fig. 7L), suggesting that polymerization occurs after membrane translocation. NTD-4CS-DmrB also could translocate to membrane fractions but could not form polymers (Fig. 5), again suggesting that polymerization might occur on the membranes.
Multiple models have been proposed for how MLKL executes necroptosis. Our result that MLKL forms polymers does not contradict the proposed models. It is possible that the reported octamer (26) is another stable intermediate in the process of forming polymers. It is also conceivable that the MLKL polymers would generate a large insult on the membranes through exposed positive charges. Alternatively, the MLKL polymers could recruit ion channels or others to dysregulate their function. Furthermore, the MLKL polymers could potentially serve as platforms to recruit and nucleate other downstream signaling molecules. This sequential polymerization model has been demonstrated for the ASC inflammasome, in which the sensors form amyloid fibers to induce ASC fiber formation, which in turn induces caspase 1 fiber formation, eventually leading to caspase 1 activation (33, 34). It is worth mentioning that these potential polymer functions are not mutually exclusive. Future experiments are needed to elucidate the exact mechanism.
An interesting note is that fungi also use a pore-forming domain, termed the “HeLo-like” (HELL) domain, to induce cell death (44). The HELL domain is an amyloid-controlled, cell membrane-targeting, cell death-inducing domain. It also targets a lipase to the membrane to help dismantle cell membrane. Most interestingly, the HELL domain is homologous to the NTD of MLKL. It seems that evolution has selected the MLKL-like proteins for executing programmed necrosis. This raises many interesting questions: How is MLKL polymerization regulated? How is MLKL membrane targeting regulated? Are there other proteins associated with MLKL at the membranes? Is a lipase involved? Answering these questions would provide a clearer picture of how MLKL induces necroptosis.
Materials and Methods
General Reagents and Stable Cell Lines.
The antibodies and other reagents as well as the methods for generating the stable cell lines are listed in Supporting Information.
Nonreducing SDS/PAGE.
Nonreducing SDS/PAGE is carried out as is regular SDS/PAGE, except that cell lysates were mixed with SDS sampling buffer without the reducing agent 2-Mercaptoethanol.
SDD-AGE and 2D SDD-AGE.
A detailed protocol is described in ref. 36. Briefly, 1% agarose was boiled in 1× Tris-acetate-EDTA (TAE), and SDS solution was added to a final of 0.1% to cast the gel. Cell lysates were loaded with sample buffer (0.5× TAE, 5% glycerol, 2% SDS, and 0.02% bromophenol blue). The gel was run at 4 V/cm gel length in 1× TAE with 0.1% SDS. The proteins were transferred to a PVDF membrane with TBS buffer [20 mM Tris (pH 7.4) and 150 mM NaCl] using capillary transfer. The membrane was then processed by Western blotting. For 2D analysis, an entire lane of the 1D gel was cut out and laid in the gel-casting tray. The second gel was poured around the gel slice and run as the regular SDD-AGE.
Cell Lysates and Immunoprecipitation.
Cell pellets were lysed with five volumes of lysis buffer [50 mM Tris (pH 7.4), 137 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 10% glycerol, supplemented with protease inhibitors] and incubated on ice for 30 min. Cell lysates were spun down at 20,000 × g for 15 min, and the supernatant was collected. Lysates (1 mg) were incubated with 20 µL anti-Flag agarose at 4 °C overnight. The beads were washed five times with lysis buffer and eluted with 60 µL elution buffer (0.2 M glycine, pH 2.8) for 5 min. The eluates were immediately neutralized with 6 µL of 1 M Tris, pH 7.4.
For crude membrane fractionation, cell pellets were resuspended in five volumes of buffer A [20 mM Tris (pH 7.4), 10 mM KCl, and 1 mM MgCl2] and incubated on ice for 20 min. The cells were passed through a 22-gauge needle 30 times and centrifuged at 500 × g for 10 min. The supernatant was centrifuged again at 20,000 × g for 10 min and saved as cytosol fraction. The pellet was extracted with lysis buffer, centrifuged at 20,000 × g for 10 min, and saved as the crude membrane fraction.
Negative Staining.
Two hundred mesh carbon/formvar-coated copper grids were rendered hydrophilic by glow-discharge in air. Protein sample (5 µL) was applied to the grid and incubated for 30 s. After wicking, the samples were stained with 5 µL of 1% uranyl acetate for 1 min, wicked, and air dried for a minimum of 15 min. Images were obtained on a FEI Tecnai G2 Spirit electron microscope.
Cell Death Assays.
(i) The CellTiter-Glo assay (Promega) was performed according to the manufacturer’s instructions. Luminescence was recorded with a Synergy 2 machine (BioTek). (ii) For PI staining, 2,000 cells were plated in each well of a 96-well glass-bottomed plate. After cell death induction, 1 µM PI and 10 µg/mL DAPI were added to cells for 10 min at room temperature. Images were taken using a Cytation 3 machine (BioTek). Percentage of cell death was calculated by dividing the number of PI+ cells by the number of DAPI+ cells. (iii) SYTOX Green staining was performed as for PI staining except that 1 µM SYTOX Green was used. Three wells were used for each treatment.
Recombinant Protein Purification and Polymerization.
cDNA encoding amino acids 2–178 of human MLKL was cloned into pGEX-4T vector. Cysteine mutants were generated by site-directed mutagenesis. GST fusion proteins were purified from BL21 Escherichia coli cells with glutathione-Sepharose beads (GE) according to the standard protocol. The GST tag was cleaved from beads with thrombin. The proteins were further purified through gel filtration and Q-Sepharose columns. (i) Purified NTD was dialyzed against PBS buffer and incubated at 37 °C in PBS buffer containing 0.1% Triton X-100 for polymerization. (ii) Aβ42 peptide (AnaSpec) was dissolved in double-distilled H2O at 350 µM. It was diluted in PBS containing 0.1% Triton X-100 to a final concentration of 10 µM and incubated at 37 °C for polymerization.
Congo Red Binding.
Congo red was dissolved in PBS to 100 µM and passed through a 0.22-µm filter. Polymers were incubated with 50 µM of Congo red at room temperature for 10 min, and the absorbance was measured with a wavelength scan from 400–600 nm using a Synergy 2 machine (BioTek).
SI Materials and Methods
General Reagents and Methods.
Recombinant TNF-α, Smac-mimetic, and anti-human RIPK3 were prepared as described previously (12). The following reagents and antibodies were used: Z-VAD-FMK (ApexBio), Necrostain-1 (Calbiochem), Necrosulfonamide (Millipore), dimerizer (635058; Clontech), proteinase K (Sigma), Congo red (Sigma), Aβ42 peptide (AS-24224; AnaSpec), anti–β-amyloid (AS-56074; AnaSpec), anti-Flag M2 antibody and affinity gel (Sigma), anti-human MLKL (GTX107538; Genetex), anti-mouse MLKL (OAAB10571; Aviva Systems Biology), anti–phospho-MLKL S358 (ab187091; Abcam), anti-RIPK1 (551042; BD), anti-LDH (ab53292; Abcam), anti-Actin (Sigma), anti-LAMP1 (sc-17768; Santa Cruz), anti-EGFR (no. 2646; Cell Signaling), and anti-GFP (A11122; Life Technologies).
For human cells, 20 ng/mL TNF, 100 nM Smac-mimetic, and 20 µM Z-VAD-FMK were used. For L929 cells, 2 ng/mL TNF and 20 µM Z-VAD-FMK were used. For DmrB cells, 20 nM dimerizer and 20 µM Z-VAD-FMK were used. For compound treatment, 10 µM Nec-1 and 5 µM NSA were used. Generally, cells were treated for 20 h for cell death analysis. For cell lysates used for Western blotting, SDD-AGE, or immunoprecipitation, cells were treated for 6 h before harvesting.
Cell Culture and Stable Cell Lines.
HT-29, L929, and HeLa cells were cultured in DMEM (high glucose) supplemented with 10% FBS. All the HeLa stable lines were generated in the background of previously reported HeLa-TetR cells that expressed the Tet repressor (TetR) (16). (i) For the MLKL-knockout HeLa line, MLKL knockout in the HeLa-TetR background was generated according to the protocol described in ref. 45. Briefly, oligo targeting human MLKL with the sequence GCTGCCCTGGAGGAGGCTAATGG was cloned into the gRNA vector. It was cotransfected with a Cas9-expressing vector into HeLa-TetR cells. MLKL knockout was confirmed by Western blotting and sequencing. (ii) For the HeLa:GFP-RIPK3:MLKL line, Tet-inducible GFP-RIPK3 and Tet-inducible MLKL-C-HA-3xFlag were stably expressed in the MLKL-knockout HeLa cells. (iii) For the NTD-DmrB, NTD-4CS-DmrB, and NTD-C86S-DmrB lines, amino acids 1–190 of human MLKL fused to the DmrB domain with a C-terminal 3xFlag were driven by a Tet-inducible promoter to construct the NTD-DmrB vector. This construct was stably expressed in the MLKL-knockout HeLa line to establish the NTD-DmrB cell line. All the cysteine mutants were generated by site-directed mutagenesis. The NTD-4CS-DmrB and NTD-C86S-DmrB stable lines were established using the same strategy. For all doxycycline (Dox)-inducible stable lines, 50 ng/mL Dox was added 24 h before necroptosis induction to induce transgene expression.
Transfection.
For the CellTiter-Glo assay, 2,000 cells were plated in each well of a 96-well plate, and 25 ng of DNA was transfected with Lipofectamine 2000 (Invitrogen). For Western blotting and SDD-AGE, 1 million cells were plated in a 10-cm plate, and 5 µg of DNA was transfected with Lipofectamine 2000.
Statistical Analysis.
Statistical analysis was performed with Excel. At least three different samples were used. Data are presented as mean ± SD.
Data Availability.
All relevant data are available from the authors.
Acknowledgments
We thank Hong Yu, Dr. Kate Phelps, Dr. Anza Darehshouri, and the staff at the University of Texas Southwestern Electron Microscopy Core for excellent technical assistance and Dr. Randal Halfmann, Dr. James Chen, and Dr. Peter Douglas for critical suggestions. This work is supported by the Welch Foundation Grant I1827 and Fellowships 2T32GM008203-26A1 (to A.J.), TL1TR1104 (to S.H.-A.), and GM111049-01A1 (to E.R.). Z.W. is the Virginia Murchison Linthicum Scholar in Medical Research, and a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar (Grant R1222).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707531114/-/DCSupplemental.
References
- 1.Kroemer G, et al. Nomenclature Committee on Cell Death 2009 Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2010;22:263–268. doi: 10.1016/j.ceb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan FK. Fueling the flames: Mammalian programmed necrosis in inflammatory diseases. Cold Spring Harb Perspect Biol. 2012;4:a008805. doi: 10.1101/cshperspect.a008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linkermann A, et al. Necroptosis in immunity and ischemia-reperfusion injury. Am J Transplant. 2013;13:2797–2804. doi: 10.1111/ajt.12448. [DOI] [PubMed] [Google Scholar]
- 5.Sun L, Wang X. A new kind of cell suicide: Mechanisms and functions of programmed necrosis. Trends Biochem Sci. 2014;39:587–593. doi: 10.1016/j.tibs.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 7.Mocarski ES, Guo H, Kaiser WJ. Necroptosis: The Trojan horse in cell autonomous antiviral host defense. Virology. 2015;479-480:160–166. doi: 10.1016/j.virol.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinlich R, Oberst A, Beere HM, Green DR. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017;18:127–136. doi: 10.1038/nrm.2016.149. [DOI] [PubMed] [Google Scholar]
- 9.Holler N, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 10.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez DA, et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2016;23:76–88. doi: 10.1038/cdd.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Z, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su L, et al. A plug release mechanism for membrane permeation by MLKL. Structure. 2014;22:1489–1500. doi: 10.1016/j.str.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dondelinger Y, et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Quarato G, et al. Sequential engagement of distinct MLKL phosphatidylinositol-binding sites executes necroptosis. Mol Cell. 2016;61:589–601. doi: 10.1016/j.molcel.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand JM, et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci USA. 2014;111:15072–15077. doi: 10.1073/pnas.1408987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia B, et al. MLKL forms cation channels. Cell Res. 2016;26:517–528. doi: 10.1038/cr.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D, et al. The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol Cell Biol. 2017;37:e00497-16. doi: 10.1128/MCB.00497-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 29.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 30.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel RM, et al. Death-effector filaments: Novel cytoplasmic structures that recruit caspases and trigger apoptosis. J Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dick MS, Sborgi L, Rühl S, Hiller S, Broz P. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat Commun. 2016;7:11929. doi: 10.1038/ncomms11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. 2008;17:838. doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 38.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 39.Mossuto MF. Disulfide bonding in neurodegenerative misfolding diseases. Int J Cell Biol. 2013;2013:318319. doi: 10.1155/2013/318319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 41.Xie T, et al. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 2013;5:70–78. doi: 10.1016/j.celrep.2013.08.044. [DOI] [PubMed] [Google Scholar]
- 42.Fändrich M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell Mol Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. RIP1 autophosphorylation is promoted by mitochondrial ROS and is essential for RIP3 recruitment into necrosome. Nat Commun. 2017;8:14329. doi: 10.1038/ncomms14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daskalov A, et al. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci USA. 2016;113:2720–2725. doi: 10.1073/pnas.1522361113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available from the authors.