Significance
Sociobiological theory proposed that similarities between human and animal societies reflect similar evolutionary origins. We used comparative genomics to test this controversial idea by determining whether superficial behavioral similarities between humans and honey bees reflect shared molecular mechanisms. We found unique and significant enrichment for autism spectrum disorder-related genes in the neurogenomic signatures of a high-level integration center of the insect brain in bees unresponsive to two different salient social stimuli. These results demonstrate deep conservation for genes implicated in autism spectrum disorder in humans and genes associated with social responsiveness in honey bees. Comparative genomics thus provides a means to test theory on the biology of social behavior.
Keywords: autism, evolution, honey bee, social behavior, transcriptomics
Abstract
E. O. Wilson proposed in Sociobiology that similarities between human and animal societies reflect common mechanistic and evolutionary roots. When introduced in 1975, this controversial hypothesis was beyond science’s ability to test. We used genomic analyses to determine whether superficial behavioral similarities in humans and the highly social honey bee reflect common molecular mechanisms. Here, we report that gene expression signatures for individual bees unresponsive to various salient social stimuli are significantly enriched for autism spectrum disorder-related genes. These signatures occur in the mushroom bodies, a high-level integration center of the insect brain. Furthermore, our finding of enrichment was unique to autism spectrum disorders; brain gene expression signatures from other honey bee behaviors do not show this enrichment, nor do datasets from other human behavioral and health conditions. These results demonstrate deep conservation for genes associated with a human social pathology and individual differences in insect social behavior, thus providing an example of how comparative genomics can be used to test sociobiological theory.
To accomplish what is necessary to survive and reproduce—such as obtaining food and shelter, finding mates, and protecting and caring for offspring—animals must often engage in diverse interactions with members of their species. Although some animal species live solitary lifestyles with minimal social interactions, others like humans and honey bees live in sophisticated societies marked by regular and extensive social interactions. In addition, individuals within the same species differ in social responsiveness, with well-documented variation among humans (1), primates (2), and other mammals (3).
Individual variations in social interactions and the biological factors driving them represent major constituents of human mental health. For example, autism spectrum disorders (ASDs) are primarily characterized by a lack of the capacity to engage in reciprocal social interactions (4). ASDs are a key focus in contemporary psychiatric genomics, and studies have revealed the complexity of these disorders: genetic factors, epigenetics, developmental deficits, and environmental factors are all thought to contribute to ASDs (5, 6).
E. O. Wilson proposed in Sociobiology (7) that similarities between the social structures of human and nonhuman animal societies reflect common evolutionary origins. When introduced in 1975, this controversial hypothesis was beyond science’s ability to test (8). In addition, specific questions about whether these similarities might have arisen from common ancestry or from convergent evolution were not explicitly considered in the development of early sociobiological theory. However, this type of information is particularly important to our understanding of the relationship between animal behaviors and human mental health disorders. Emerging evidence for conserved molecular mechanisms for social behaviors between invertebrates and vertebrates (9–14) suggests that there are similar connections to mental health disorders.
Honey bees have highly sophisticated and well-described social behaviors, including symbolic communication (15), alloparental care (16), and kin recognition (17). They have been used extensively in sociogenomic analyses of normal behavior (18), but never before for explorations of social deficits. Because honey bees show strong individual differences in social behaviors, we used them to explore molecular mechanisms underlying differences in social responsiveness.
Results and Discussion
We first hypothesized there are individual bee differences in responsiveness to various social stimuli, associated with unique neurogenomic signatures. To test this hypothesis, we exposed groups of 10 7-d-old honey bee nestmates to a social responsiveness assay in the laboratory, which involved exposures to two different social stimuli. One stimulus provided a social challenge: an unrelated bee as a territorial threat, which provokes an aggressive response (10, 17). The other stimulus provided a social opportunity: a queen larva, which provokes alloparental care (19). We tested 246 groups from seven genetically distinct source colonies. To rigorously test for stable individual differences in social responsiveness, each group was given an assay involving serial administration of both stimuli in a randomized order followed by a second exposure to both stimuli 1 h later. Because each individual was thus tested four times, different levels of responsiveness to social stimuli were identified with high confidence.
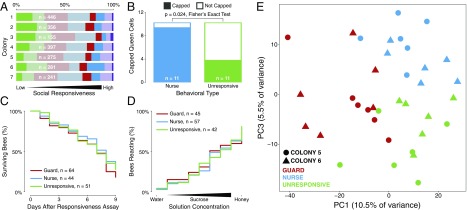
We observed a full range of social responsiveness, from individuals consistently unresponsive to either social stimulus to individuals consistently responding strongly to both stimuli (Fig. 1A). We focused on the following three behavioral types: 9.3% responded consistently only to the social opportunity (“nurses”), 7.7% responded consistently only to the social challenge (“guards”), and 14% never responded to either stimulus (“unresponsive”). We did not consider further the bees that responded to both stimuli because they accounted for only a small proportion (∼1%), consistent with the fact that the tendency to defend the hive and nurse brood at the same time is not commonly observed due to the division of labor that operates in a honey bee colony.
Fig. 1.
Behavioral responses to social stimuli vary among individual bees. (A) Proportion of individuals of different behavioral types. Unresponsive individual honey bees (deep green) did not display a response to social stimuli. Guards (deep red) responded with aggression to two social challenge trials. Nurses (deep blue) responded with nursing to two social opportunity trials. Highly responsive individuals (deep purple) responded to all four social stimulus trials. Other categories include bees that showed weak responses to social stimuli (light green), single responses to a social challenge (light red) or social opportunity (light blue), or another mixture of behavioral responses (light purple). (B) Exposure to a strong social stimulus did not change the behavior of unresponsive individuals. There was (C) no difference in survival or (D) sucrose perception of individuals with different levels of social responsiveness. Survival: log-rank test, χ2(2) = 1.56, P = 0.46; proboscis extension response: Kruskal–Wallis test, χ2(2) = 0.093, P = 0.95. (E) Principal-component analysis (PCA) of genes differentially expressed in the MBs at FDR of <0.10 revealed two PCs that together reliably separate guards, nurses, and unresponsive bees (n = 12 per group).
The occurrence of unresponsive bees was not an artifact of exposure to a subthreshold stimulus. When exposed individually to a queen larva, unresponsive bees also underperformed, even though this test provided each individual with a stronger social stimulus than it would receive in a group. Only 4 out of 11 unresponsive bees showed alloparental care, whereas 10 out of 11 nurse bees did (Fisher’s exact test, P = 0.024; Fig. 1B). In addition, unresponsive bees showed no differences in survival or the ability to detect sensory stimuli relative to other behavioral types (Fig. 1C), indicating that they were not obviously sick or deficient in sensory abilities.
To explore the molecular basis of this social unresponsiveness, we generated profiles of gene expression for nurses, guards, and unresponsive bees. We used RNA sequencing (RNA-seq) to profile the mushroom bodies (MBs), a region of the insect brain involved in multimodal sensory integration (20) and recently implicated in honey bee social behavior (10).
There were a large number (1,057) of differently expressed genes (DEGs) between the three behavioral types [edgeR general linear model (GLM) blocked for colony, false-discovery rates (FDR) of <0.10, Dataset S1; in Dataset S1, part S1.2 shows that smaller numbers of DEGs also were detected with pairwise analyses between the behavioral groups]. The magnitude of this difference was unexpected as the bees tested were highly related to one another due to haplodiploidy and artificial insemination; they also were laboratory reared until testing and exposed to the same stimuli simultaneously. It is not possible to know to what extent differences in MB gene expression reflect baseline neurogenomic states or responses to stimulus exposure, but given that all bees selected for transcriptomic analysis either showed consistent responsiveness or nonresponsiveness to repeated stimulus exposure, we think it likely that baseline neurogenomic states figured prominently.
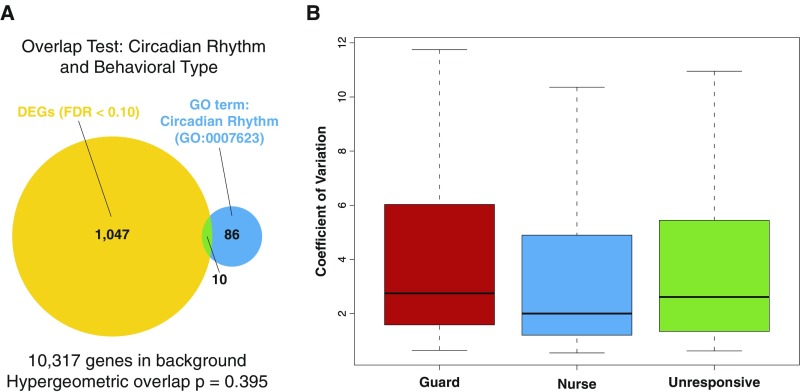
Gene ontology analysis of the “bee DEG list” revealed enrichment for molecular functions related to chaperones and ATP binding (Dataset S2). Circadian rhythm-related genes were not enriched, indicating that the observed differences in responsiveness were not due to acute differences in arousal (Fig. S1).
Fig. S1.
No evidence for differences in MB expression of circadian rhythm genes among the three different behavioral types suggests that differences in circadian-related arousal do not explain the observed differences in social responsiveness. (A) The gene ontology (GO) term for circadian rhythm is not enriched in the bee DEG list. (B) The observed coefficients of variation for the 96 genes classified in the circadian rhythm GO term are similar between the three different behavioral types (n = 12 per group).
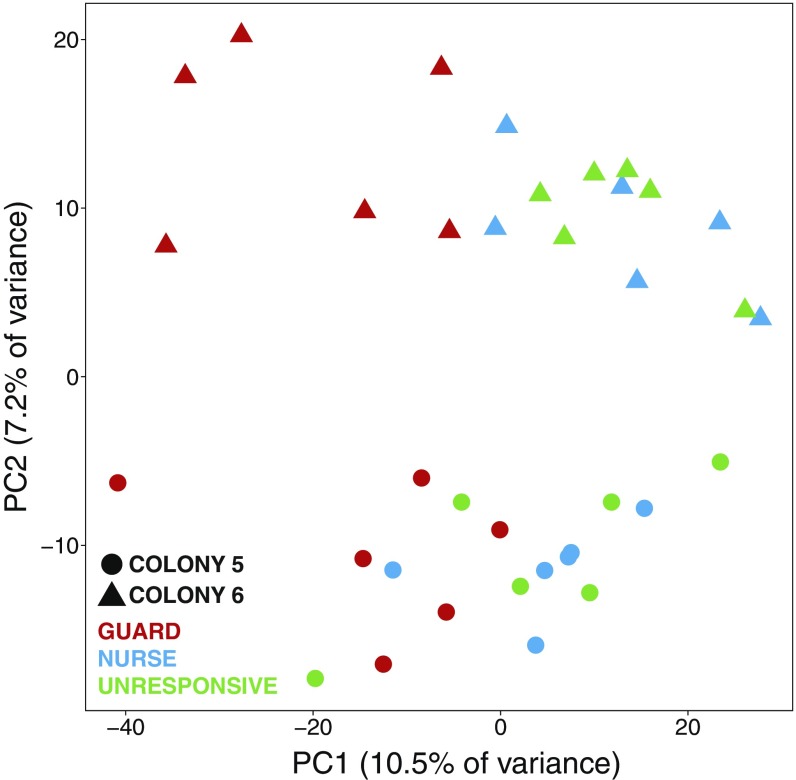
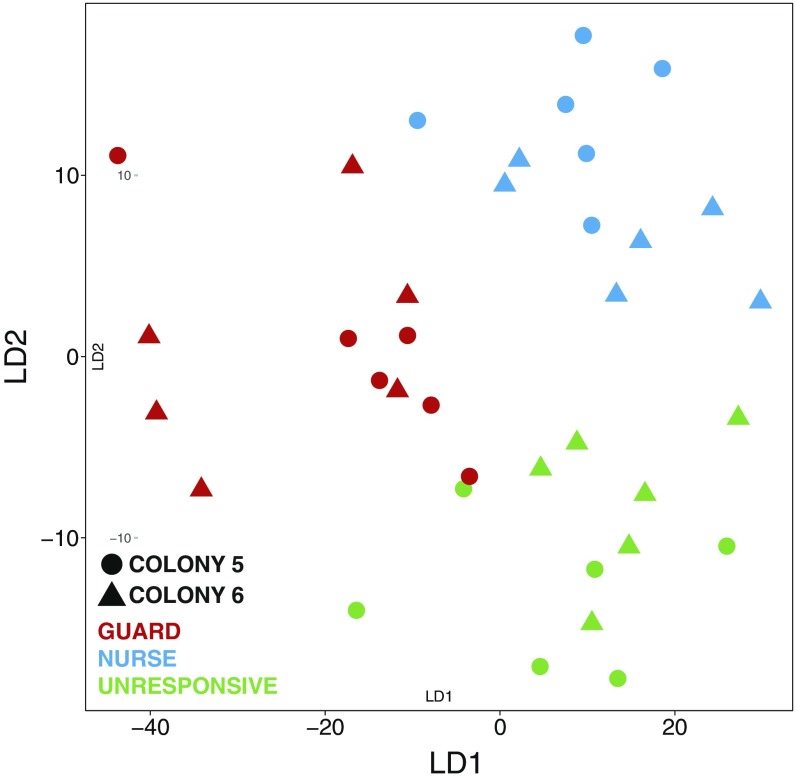
The distinctiveness of the unresponsive individuals was further revealed by principal-component analysis (PCA) of the bee DEG list. PCA clearly separated the three behavioral types (Fig. 1E). PC1, PC2, and PC3 accounted for 10.5%, 7.2%, and 5.5% of the variance in MB gene expression, respectively. PC2 separated bees by the two independent colonies we sampled (Fig. S2), and PC1 and PC3 together separated the three behavioral types with great precision (Fig. 1E). The PCA results were confirmed by penalized linear discriminant analysis (Fig. S3). These findings demonstrate that unresponsive individuals have a unique MB transcriptomic profile that is a function of multiple molecular factors.
Fig. S2.
PC2 highlights the differences in MB gene expression between the two source colonies that produced the bees used in this study (n = 18 per colony).
Fig. S3.
Penalized LDA confirms the PCA results, showing a clear separation in brain gene expression between the three behavioral types independent of colony origin (n = 12 per group).
We examined the 50 genes with the highest loadings in PC1 and PC3 to further explicate the bee DEGs associated with these social responsiveness axes. The genes highly loaded in PC1 include a number of ion channels and ion channel modulators as well as the nuclear receptor ftz-f1, a transcriptional regulator associated with social responsiveness in honey bees (21) and mice (22). The genes highly loaded in PC3 include chaperone proteins generally related to the HSP90 complex, the nucleoporin complex, and hormone signaling (23) (Dataset S3).
If social responsiveness in bees is functionally related in some way to ASD, sociobiological theory would predict that the bee DEG list described above should be enriched for ASD-related genes. To test this hypothesis, we examined the overlap between the bee DEG list and several previously published human ASD gene sets. We improved existing methods for assessing cross-species gene list overlap, developing a test called Orthoverlap for the purpose of comparative analyses. This test facilitates comparison of gene lists across long evolutionary distances by weighting for the strength of evidence for overlap within orthogroups of genes.
We used this methodology to test for similarities between the bee DEG set and DEGs from humans ASD patients. We found significant overlaps between the bee DEG list and two independent DEG lists from expression studies of postmortem brain tissue (24, 25) (Table 1, DEG lists). Because transcriptomic analyses are correlative, we also compared the bee DEG list to curated sets of gene variants associated with ASD from the Simons Foundation Autism Research Initiative (SFARI), at least some of which are assumed to have causal effects on ASD phenotypes (26). We found significant overlap between the bee DEG list and the SFARI gene list. The SFARI database is divided into seven sets of genes that vary in the confidence level with which they are related to ASD. When comparing the bee DEG list to the highest confidence SFARI sets, the overlap was statistically significant. When weaker confidence SFARI sets were included, statistical significance weakened proportionately (Table 1, SFARI lists).
Table 1.
Evolutionary conservation of autism-related genes in honey bees
| Human list | No. human genes | Overlapping orthogroups | P value | FDR |
| DEG lists* | ||||
| Ref. 24, INITIAL COHORT DEGs | 928 | 76 | 0.0026 | 0.0150 |
| Ref. 25, Cortex DEGs | 1,087 | 108 | 0.0310 | 0.0521 |
| SFARI lists† | ||||
| Tier 1 (syndromic genes only) | 83 | 16 | 0.0066 | 0.0150 |
| Tier 2 (high confidence + tier 1 genes) | 89 | 17 | 0.0046 | 0.0150 |
| Tier 3 (strong candidate + tier 2 genes) | 120 | 22 | 0.0067 | 0.0150 |
| Tier 4 (suggestive evidence + tier 3 genes) | 250 | 31 | 0.0347 | 0.0521 |
| Tier 5 (minimal evidence + tier 4 genes) | 474 | 44 | 0.2391 | 0.3074 |
| Tier 6 (hypothesized + tier 5 genes) | 609 | 49 | 0.4202 | 0.4202 |
| Tier 7 (not supported + tier 6 genes) | 628 | 51 | 0.4143 | 0.4202 |
The top significant enrichment clusters of functional terms enriched in SFARI (evidence tier 3) and the two human DEG lists overlapping with the bee DEG list were generally related to ion channels and included GABA receptor and voltage-gated ion channel genes. The genes overlapping with ref. 25 suggested an additional enrichment for heat shock proteins (Dataset S4, parts B–D). These findings suggest a deep conservation in the functional genomic systems related to social behavior in the animal kingdom.
To explore whether these results might be false positives, we performed two sets of control analyses using the same analytical methods. First, we compared the bee DEG list with gene lists related to several other human disease phenotypes. We observed no significant enrichment for any of them, including schizophrenia-related (unadjusted P = 0.82) or depression-related genetic variant lists (unadjusted P = 0.84). Second, we compared the three ASD-related gene list sources described above with three other honey bee MB DEG lists unrelated to social responsiveness. We again observed no significant enrichment (Dataset S4). The significant overlap thus appears to be unique to the honey bee social responsiveness and human ASD gene sets. This finding provides additional evidence for functional conservation of genes involved in the coordination of social behaviors across phyla (9), and demonstrates that this conservation also extends to deficits in social behavior.
Disorders of human cognition and social behavior have been associated with human-specific genomic features (27). In addition, it is highly likely that social insects and humans evolved their distinct repertoire of complex social behaviors independently of each other (28), with the forces of natural selection acting differently on social behaviors in insects and humans. In humans, social interactions are presumed to represent a strong selective force: individuals who, for whatever reason, cannot engage in normal social interactions lose access to common resources and should have diminished fitness. In social insects, the fitness of an individual is dependent on the performance of the whole colony, and perhaps it is less costly for a large insect society to tolerate unresponsive individuals than to actively exclude them. Supporting this speculation, inactive individuals have been reported in several social insect species (29) and are usually interpreted as providing colonies with the advantages of a “reserve” labor force that will act when the colony faces a stressful situation (30). However, inactive honey bees do not always respond to changes in colony needs (31), and here we have identified individuals at the extremes of social responsiveness spectra who may not have any adaptive value to their colony.
Conclusions
Social insects may be particularly fruitful sources of behavioral phenotypes relevant to the study of human psychiatric diseases. Their large societies appear to tolerate a broad range of behavioral phenotypes, including highly repetitive behavior (32, 33), unusually high levels of activity (34), and as reported here, unresponsiveness to various social stimuli.
Despite profound differences between honey bee and human societies, we have documented strong similarities in the genes associated with social responsiveness. It is not possible to discern whether these similarities arose from either common ancestry or convergent evolution, but our findings provide further support for conserved genetic “toolkits” that are used in independent evolutions of social behavior (9). Comparative genomics thus can determine in a rigorous and unbiased manner whether behavioral similarities between humans and distantly related species reflect common mechanisms, thus providing a means to further explore sociobiological theory.
Materials and Methods
Bees.
Adult worker honey bees (Apis mellifera) were obtained from colonies maintained in apiaries according to standard commercial methods at the University of Illinois Bee Research Facility (Urbana, IL) from June to July 2015. We used workers derived from two different colony types: colonies headed by queens who mated naturally and colonies headed by single drone-inseminated (SDI) queens, who were instrumentally inseminated with semen from a single drone. For the proboscis extension reflex (PER), capping, and survival experiments, we used four different colonies headed by naturally mated queens. For the social responsiveness and transcriptomics experiments, we used bees derived from two SDI queens. Because of haplodiploidy, SDI worker offspring are highly related to each other (average coefficient of relatedness, r = 0.75), thus decreasing within-trial genetic variation.
One-day-old adult bees were obtained by removing frames of honeycomb containing pupae from colonies, placing them in an incubator (34 °C, 50 ± 5% relative humidity), and then monitoring their emergence every 24 h. One-day-old bees were individually marked on their thoraces with a spot of paint (Testors PLA) and placed in groups. Bees were kept in the laboratory in vertically oriented petri dishes (100 × 20 mm) with a beeswax foundation sheet placed on the “wall” of the dish to mimic in-hive conditions. Dishes were supplied with one tube of honey (∼1.4 mL), 30% sucrose solution (2 mL), and a mixture of fresh frozen pollen and 30% sugar solution (∼10-mm-diameter ball).
Social Responsiveness Assay.
Groups of 10 individually marked 7-d-old adult bees were held in petri dishes inside an incubator room maintained to mimic the hive environment (34 ± 1 °C, 50 ± 10% relative humidity). The bees were held in the dish from adult emergence with ad libitum food supply and were not exposed to outside stimuli until the beginning of the experiment. All behavioral observations were performed in the incubator room under white light.
To search for unresponsive bees, we developed an assay using two established behavioral assays: the resident–intruder assay (17) and the nursing assay (19). The resident–intruder assay is a 5-min survey of all aggressive interactions shown by resident bees toward an unrelated bee following its introduction to the group; some individuals (typically two to four per group) react with highly aggressive behaviors, attacking the intruder with biting and stinging. The nursing assay is a 5-min survey of all nurturing interactions between adult bees and a 4-d-old queen larva in a waxen queen cell that is introduced to the group. Some individuals (typically two to three) respond by entering the queen cell to inspect (short visits of less than 10 s) or feed (long visits of 11 s or more).
Both assays were performed on the same group of bees in succession and in random order according to a coin flip, and then both assays were repeated in random order 1 h after the completion of the first set. Individuals exhibiting highly aggressive behavior (biting and stinging) in both intruder assays but no response to the queen larva were classified as guards; individuals exhibiting alloparenting in both nursing assays but no response in the resident–intruder assay were classified as nurses. Individuals that did not respond at all in the four trials (two trials of each assay) were defined as unresponsive. The rest of the individuals in the group were defined by their behavior, which included those that showed weak responses (e.g., antennation) toward a social stimulus (Fig. 1A, light green), those that responded to only one of the stimuli (guard once: light red; nurse once: light blue), those that showed mixed responses (light purple), or those that showed strong responses to all social stimuli (deep purple). Only guards, nurses, and unresponsive individuals were used for further analysis. Individual observers conducted each behavioral assay blind to the results of previous assays. The behavioral experiment was repeated with bees from seven different unrelated colonies.
Survival.
The survival of all of the individuals in each group was monitored for 9 d after the conclusion of the behavioral experiment. The observer monitoring survival was blind to the behavioral type of each individual; matching survival and behavioral type was done after all of the data were collected. We measured the survival of the individuals from 28 groups including 64 nurses, 44 guards, and 51 unresponsive bees.
Response to Sugar Solution.
The PER assay (35) was used to measure the response to sugar solution for individuals belonging to each of the three behavioral types. Bees were collected from the group after the end of the assay by anesthesia with CO2. Each individual was harnessed to a stand where its body was fixed, its head was free to move, and its proboscis was free to extend. Each individual was left on the stand for 2 h to ensure that it was hungry. Increasing concentrations of sugar solution (water, 0.1%, 0.3%, 1%, 3%, 10%, 30%, 50%, honey) were used to test responsiveness to sugar. The sugar stimulus was presented by a gentle touch of both antennae using a 20-µL micropipette. The responses of each individual—extension or no extension of the proboscis—were recorded for each sugar concentration. Unlimited water was provided for the bees at the beginning of the test and before each presentation of the sugar solution to help eliminate responses based on thirst. Bees from 42 groups, including 57 nurses, 45 guards, and 42 unresponsive individuals, were tested. The observer was blind to the behavioral type of the individuals while performing PER testing.
Queen Pupal Cell Capping Test.
One unresponsive bee and one nurse each from 11 groups were moved into new petri dishes as described above. One 4-d-old queen larva in a queen cell was introduced into each dish for 24 h. At this age, bees typically add wax to the queen cell and enclose the developing queen by capping the cell. Capping of the cell was assessed on the day after the introduction. A Fisher’s exact test was used to compare the rate of cell capping in each behavioral type. This assay extends the bee’s exposure to a social signal for a longer period, allowing insight into the stability of the social responsiveness phenotype. This assay was designed to test how increasing the strength of a social stimulus affected unresponsive bees’ responses.
MB Gene Expression Analysis.
Only bees tested in the social responsiveness behavioral assay were used for MB gene expression analysis. Nurses, guards, and unresponsive individuals were collected and frozen immediately after all behavioral testing, which was ≈70 min after the presentation of the first social stimulus. They were flash-frozen in liquid nitrogen and then transferred into marked 1.5-mL microcentrifuge tubes in a dry-ice bucket. The bees were stored at −80 °C until tissue dissection. Thirty-six individuals (three behavioral types and two colonies; six bees per group per colony) were included in the gene expression analysis.
Heads were separated from bodies on dry ice and placed in a dissection dish with 200-proof ethanol and dry ice to prevent RNA degradation during dissection. The cuticle of the head capsule was removed, and each head was placed in RNAlaterICE (Life Technologies) at −20 °C for 14–18 h. The heads were then opened, hypopharyngeal glands were removed, and the whole brain was removed. The optic lobes were removed, and a horizontal incision was made across the midbrain through the posterior protocerebral lobe using a fine scalpel. The lower part of the midbrain, containing the antennal lobes and the subesophageal ganglion, was removed. The upper part of the midbrain containing the MBs and some surrounding tissue (∼10%) was used for gene expression analysis. This MB preparation has been described in detail in previous work (10). The MB of each individual were placed in a new 1.5-mL microcentrifuge tube and kept frozen at −80 °C until RNA extraction. RNA extraction was performed with a PicoPure RNA Isolation Kit (lot no. 1210063; Applied Biosystems) according to the manufacturer’s specifications, including a DNase treatment (Qiagen) to remove genomic DNA contamination. A total of 550 ng of total RNA from each sample was used for whole-transcriptome expression analysis. RNA integrity was ensured with a Bioanalyzer 2100 (Agilent). Bioanalyzer RNA integrity numbers were not calculated, because honey bee 28S rRNA contains an AU-rich “hidden break” region, which causes it to split into two during heat denaturing and migrate with the 18S rRNA peak (36, 37). Consequently, we used qualitative assessment as a quality control for RNA integrity, ensuring that the electropherograms within this experiment appeared to be internally consistent and comparable to previously observed electropherograms for high-quality RNA from honey bees (Fig. S4).
Fig. S4.
Electropherograms of input total RNA showing consistent quality between samples. (A) Electropherograms for colony R22 total RNA samples. (B) Electropherograms for colony R23 samples.
RNA-Seq, Data Processing, and Analysis.
RNA-seq libraries were constructed with the TruSeq Stranded mRNA HT (high-throughput kit, catalog no. RS-122-2103; Illumina) using an ePMotion 5075 robot (Eppendorf). The libraries were uniquely barcoded (36 barcodes total), quantified by quantitative PCR, and pooled into a single pool of equimolar concentration as per instructions. Single-end sequencing (read length, 100 nt) was performed on the pooled libraries across five lanes of an Illumina HiSeq 2500 sequencer using a TruSeq SBS sequencing kit with v4 chemistry. FASTQ files were generated with CASAVA 1.8.2. Library preparation and RNA-seq were performed at the W. M. Keck Center for Comparative and Functional Genomics at the Roy J. Carver Biotechnology Center (University of Illinois). Demultiplexed RNA-seq libraries produced a median of 33.6 million reads (range, 28.5 million to 49.3 million reads) per sample. Raw FASTQ files have been deposited in the Sequence Read Archive under accession number SRP089994, raw counts have been deposited in the Gene Expression Omnibus under accession number GSE87001, and TMM-normalized counts per million values are available in Dataset S1, part S1.3.
Sequencing reads were aligned to the A. mellifera 4.5 reference genome (38) using TopHat2 with Bowtie2. Numbers of reads per gene were counted with HTSeq-count, for 15,314 genes (OGs 3.2). A total of 10,317 genes had more than one count per million (cpm) in six or more samples and were included in the analysis. Gene expression levels were compared between the three behavioral types using an ANOVA-like implementation of the GLM, with colony used as blocking factor, in edgeR (39). P-value correction for multiple testing was done using the FDR method (40) across all factors simultaneously; DEGs were defined as those with an FDR of <0.10. PCA was performed in R using the prcomp function, and the confirmatory penalized linear discriminant analysis was performed using the PenalizedLDA function in the R package penalizedLDA (41). DAVID analysis was performed using version 6.8 of the knowledgebase, and significant annotation clusters were defined as those with enrichment scores of ≥3 using medium classification stringency. Phenotype data used for analysis of cpm data are available in Dataset S1, part S1.4.
Orthoverlap Analysis.
We developed a statistical test for enrichment between gene sets that come from different species. This test, which we call Orthoverlap, assumes that, in each species, the number of genes in an orthogroup that belong to that species’ gene set of interest follows a hypergeometric distribution, with parameters determined by the size of the orthogroup, the gene set size, and the number of genes in the background for the species (where the background is all genes from a species that are in the orthogroups common to both species). Under the null hypothesis, these hypergeometric random variables are assumed to be independent across the two species. Orthoverlap converts the hypergeometric variables to Z scores and then measures their relatedness across species using the Pearson correlation coefficient. Finally, it obtains a permutation P value for this correlation coefficient by randomly permuting the orthology relationships between the species, similar to a test that has been described previously (42).
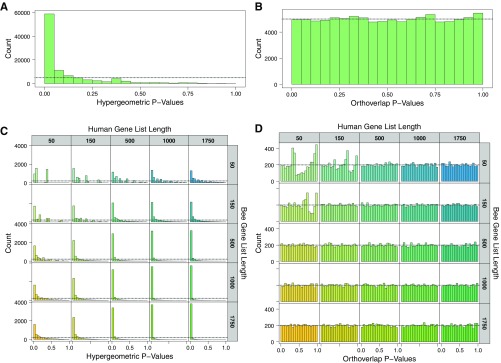
This procedure identifies enrichment at the level of gene orthogroups and is able to correctly account for species-specific orthogroup sizes. By using a Z score, it down-weights large orthogroups if they contain few genes of interest, and up-weights smaller orthogroups if most of their constituent genes are of interest. Furthermore, the procedure negatively weights evidence of mismatch, that is, when large orthogroups in one species contain no or very few genes in one gene set while the corresponding orthogroups in the other species contain many genes in the other gene set. Our test assumes that more genes of interest within the same orthogroup across both species constitutes stronger evidence of overlap. We ran computational simulations with multiple gene list sizes and confirmed that this method adequately controls false positives compared with a simpler and more established hypergeometric method for testing orthogroups with DEGs in each species (Fig. S5).
Fig. S5.
Simulations using random gene lists from honey bee and human demonstrate that Orthoverlap controls for false positives. (A) Hypergeometric tests on orthogroup overlap lead to many false positives as shown by a spike in P values less than 0.05. This result is likely due to the nonindependence introduced by a few large orthogroups. (B) Orthoverlap on simulated data shows an expected uniform distribution of P values. (C) Hypergeometric test results are further skewed at different list sizes with the most false-positive results in large lists. (D) Orthoverlap shows a uniform distribution of the P values, although in very small list sizes (50/50 and 50/150) there is some deviation from a uniform distribution. Nevertheless, random gene lists at all sizes have ≈5% false positives as expected. Orthoverlap was used in the current study on a gene list of 1,057, where there is a clear uniform distribution.
The code used to run Orthoverlap is released under the GNU GPL 3.0 and can be found in the R package msaul (https://github.com/msaul/msaul). We ran Orthoverlap using a million permutations on all orthogroups common to Homo sapiens and Apis mellifera in OrthoDB, version 9 (43).
Human autism gene sets of interest were derived from the Gene Scoring Module set from the September 2016 revision of the SFARI database (24) and genes identified from microarray (25) and RNA-seq (26) gene expression experiments on postmortem samples of brain tissue from autism patients. The SFARI genes were divided into seven tiers of evidence, beginning with including only the genes with the strongest evidence of a relationship to autism (Syndromic genes) in tier 1; the genes with the strongest and second strongest evidence of a relationship to autism (Syndromic and High Confidence genes) in tier 2 and adding in new and weaker evidence into each tier until tier 7, which includes the genes from all classes of evidence of a relationship with autism (Syndromic, High Confidence, Strong Candidate, Suggestive Evidence, Minimal Evidence, Hypothesized, and Not Supported genes). From the expression studies, we used the lists of all genes found differentially expressed across multiple conditions (from ref. 25, supplementary data file INITIAL COHORT table; from ref. 26, supplementary table 2).
To validate the method and results, we ran two sets of control analyses. Honey bee control gene sets were derived from two RNA-seq expression sets from honey bee MB: (i) DEGs comparing active and inactive male honey bees (44) and (ii) DEGs comparing worker bees trained to forage at specific times of day (45). Human control gene sets were derived as follows. One set of human control gene sets from genome-wide association study results was derived from HuGE Phenopedia (44), keeping only genes that had been replicated in at least two studies. Another set of control human gene sets was derived from differential expression sets for human whole-transcriptome brain postmortem tissue studies from patients with schizophrenia (46) and Alzheimer’s disorder (47) diagnoses (Dataset S4).
FDR correction was performed in two different ways: a pooled method where all P values from all Orthoverlap tests were corrected together, and an experiment-wise method where FDR correction was performed separately for controls and for human autism vs. bee responsiveness lists. We report the experiment-wise results to avoid skewing positive results of interest by adding in many expected negative results, but we note that the only tests reported as significant at a pooled FDR of <0.10 were four of the tests from the human autism vs. bee responsiveness lists.
Supplementary Material
Acknowledgments
We are grateful to A. M. Bell for inspiring the research question. We thank L. J. Stubbs, A. M. Bell, S. Sinha, A. B. Barron, and S. A. Ament for critical reading of the manuscript, and C. D. Nye for bee management. This research was supported by Simons Foundation Grant SFLife 291812 (to G.E.R. and L. J. Stubbs, principal investigators), NSF Grant DMS-1613005 (to S.D.Z.), United States–Israel Binational Agricultural Research and Development Postdoctoral Fellowship Award FI-462-2012 (to H.Y.S.), and a Carl R. Woese Institute for Genomic Biology Postdoctoral Fellowship (to M.C.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-seq files reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE87001). Raw FASTQ files have been deposited in the Sequence Read Archive (accession no. SRP089994).
See Commentary on page 9502.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708127114/-/DCSupplemental.
References
- 1.Bölte S, Poustka F, Constantino JN. Assessing autistic traits: Cross-cultural validation of the social responsiveness scale (SRS) Autism Res. 2008;1:354–363. doi: 10.1002/aur.49. [DOI] [PubMed] [Google Scholar]
- 2.Faughn C, et al. Brief report: Chimpanzee social responsiveness scale (CSRS) detects individual variation in social responsiveness for captive chimpanzees. J Autism Dev Disord. 2015;45:1483–1488. doi: 10.1007/s10803-014-2273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer SF, van der Vegt BJ, Koolhaas JM. Individual variation in aggression of feral rodent strains: A standard for the genetics of aggression and violence? Behav Genet. 2003;33:485–501. doi: 10.1023/a:1025766415159. [DOI] [PubMed] [Google Scholar]
- 4.American Psychiatric Association DSM-5 Task Force . Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th Ed American Psychiatric Association; Arlington, VA: 2013. [Google Scholar]
- 5.Abrahams BS, Geschwind DH. Advances in autism genetics: On the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355, and erratum (2008) 9:493. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson EO. Sociobiology: The New Synthesis. Belknap Press of Harvard Univ Press; Cambridge, MA: 1975. [Google Scholar]
- 8.Segerstråle UCO. Defenders of the Truth: The Battle for Science in the Sociobiology Debate and Beyond. Oxford Univ Press; Oxford: 2000. [Google Scholar]
- 9.Rittschof CC, et al. Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci USA. 2014;111:17929–17934. doi: 10.1073/pnas.1420369111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shpigler HY, et al. Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav. 2017;16:579–591. doi: 10.1111/gbb.12379. [DOI] [PubMed] [Google Scholar]
- 11.Saul MC, et al. Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 2017;27:959–972. doi: 10.1101/gr.214221.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell P, Reep RL, Stoll ML, Ophir AG, Phelps SM. Conservation and diversity of Foxp2 expression in muroid rodents: Functional implications. J Comp Neurol. 2009;512:84–100. doi: 10.1002/cne.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malki K, et al. Transcriptome analysis of genes and gene networks involved in aggressive behavior in mouse and zebrafish. Am J Med Genet B Neuropsychiatr Genet. 2016;171:827–838. doi: 10.1002/ajmg.b.32451. [DOI] [PubMed] [Google Scholar]
- 14.Bukhari SA, et al. Temporal dynamics of neurogenomic plasticity in response to social interactions in male threespined sticklebacks. PLoS Genet. doi: 10.1371/journal.pgen.1006840. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frisch KV. The Dance Language and Orientation of Bees. Harvard Univ Press; Cambridge, MA: 1967. [Google Scholar]
- 16.Winston ML. The Biology of the Honey Bee. Harvard Univ Press; Cambridge, MA: 1987. [Google Scholar]
- 17.Breed MD. Nestmate recognition in honey bees. Anim Behav. 1983;31:86–91. doi: 10.1006/anbe.1997.0581. [DOI] [PubMed] [Google Scholar]
- 18.Zayed A, Robinson GE. Understanding the relationship between brain gene expression and social behavior: Lessons from the honey bee. Annu Rev Genet. 2012;46:591–615. doi: 10.1146/annurev-genet-110711-155517. [DOI] [PubMed] [Google Scholar]
- 19.Shpigler HY, Robinson GE. Laboratory assay of brood care for quantitative analyses of individual differences in honey bee (Apis mellifera) affiliative behavior. PLoS One. 2015;10:e0143183. doi: 10.1371/journal.pone.0143183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zars T. Behavioral functions of the insect mushroom bodies. Curr Opin Neurobiol. 2000;10:790–795. doi: 10.1016/s0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 21.Chandrasekaran S, et al. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci USA. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grgurevic N, Büdefeld T, Rissman EF, Tobet SA, Majdic G. Aggressive behaviors in adult SF-1 knockout mice that are not exposed to gonadal steroids during development. Behav Neurosci. 2008;122:876–884. doi: 10.1037/0735-7044.122.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Zhang Z, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 2007;282:37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrahams BS, et al. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doan RN, et al. Homozygosity Mapping Consortium for Autism Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 2016;167:341–354.e12. doi: 10.1016/j.cell.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Robinson GE, Jakobsson E. Conservation in mammals of genes associated with aggression-related behavioral phenotypes in honey bees. PLoS Comput Biol. 2016;12:e1004921. doi: 10.1371/journal.pcbi.1004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charbonneau D, Dornhaus A. When doing nothing is something. How task allocation strategies compromise between flexibility, efficiency, and inactive agents. J Bioeconomics. 2015;17:217–242. [Google Scholar]
- 30.Hasegawa E, Ishii Y, Tada K, Kobayashi K, Yoshimura J. Lazy workers are necessary for long-term sustainability in insect societies. Sci Rep. 2016;6:20846. doi: 10.1038/srep20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson GE, Page RE., Jr Genotypic constraints on plasticity for corpse removal in honey-bee colonies. Anim Behav. 1995;49:867–876. [Google Scholar]
- 32.Robinson GE, Underwood BA, Henderson CE. A highly specialized water-collecting honey bee. Apidologie. 1984;15:355–358. [Google Scholar]
- 33.Moore AJ, Breed MD, Moor MJ. The guard honey bee: Ontogeny and behavioural variability of workers performing a specialized task. Anim Behav. 1987;35:1159–1167. [Google Scholar]
- 34.Tenczar P, Lutz CC, Rao VD, Goldenfeld N, Robinson GE. Automated monitoring reveals extreme interindividual variation and plasticity in honeybee foraging activity levels. Anim Behav. 2014;95:41–48. [Google Scholar]
- 35.Scheiner R, Page RE, Erber J. Sucrose responsiveness and behavioral plasticity in honey bees (Apis mellifera) Apidologie. 2004;35:133–142. [Google Scholar]
- 36.Winnebeck EC, Millar CD, Warman GR. Why does insect RNA look degraded? J Insect Sci. 2010;10:159. doi: 10.1673/031.010.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara H, Ishikawa H. Molecular mechanism of introduction of the hidden break into the 28S rRNA of insects: Implication based on structural studies. Nucleic Acids Res. 1986;14:6393–6401. doi: 10.1093/nar/14.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elsik CG, et al. HGSC production teams Honey Bee Genome Sequencing Consortium Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. [Google Scholar]
- 41.Witten DM, Tibshirani R. Penalized classification using Fisher’s linear discriminant. J R Stat Soc Series B Stat Methodol. 2011;73:753–772. doi: 10.1111/j.1467-9868.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisinger BE, Saul MC, Driessen TM, Gammie SC. Development of a versatile enrichment analysis tool reveals associations between the maternal brain and mental health disorders, including autism. BMC Neurosci. 2013;14:147. doi: 10.1186/1471-2202-14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriventseva EV, et al. OrthoDB v8: Update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naeger NL, Robinson GE. Transcriptomic analysis of instinctive and learned reward-related behaviors in honey bees. J Exp Biol. 2016;219:3554–3561. doi: 10.1242/jeb.144311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and genopedia: Disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maycox PR, et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14:1083–1094, and erratum (2010) 15:442–443. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- 47.Hokama M, et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: The Hisayama study. Cereb Cortex. 2014;24:2476–2488. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.