Significance
Development of the blood–brain barrier (BBB) requires spatiotemporal coordination of cerebrovascular endothelial cells (ECs) and pericytes. Until now, the molecular mechanism(s) coordinating the pericyte–EC behaviors during this process have been incompletely understood. In this study, combining the analysis of EC-/pericyte-specific Cd146-KO mice and in vitro BBB models, we report CD146 as a dynamic coordinator regulating the communication between ECs and pericytes within the neurovascular unit during BBB development. Our study demonstrates that a single cell-adhesion receptor, CD146, acts as an essential regulator to coordinate pericyte–EC communication and BBB formation during embryogenesis. Furthermore, it identifies CD146 as a potential key therapeutic target for neurological diseases related to cerebrovascular disorders.
Keywords: CD146, claudin-5, PDGFRβ, spatiotemporal expression, blood–brain barrier
Abstract
The blood–brain barrier (BBB) establishes a protective interface between the central neuronal system and peripheral blood circulation and is crucial for homeostasis of the CNS. BBB formation starts when the endothelial cells (ECs) invade the CNS and pericytes are recruited to the nascent vessels during embryogenesis. Despite the essential function of pericyte–EC interaction during BBB development, the molecular mechanisms coordinating the pericyte–EC behavior and communication remain incompletely understood. Here, we report a single cell receptor, CD146, that presents dynamic expression patterns in the cerebrovasculature at the stages of BBB induction and maturation, coordinates the interplay of ECs and pericytes, and orchestrates BBB development spatiotemporally. In mouse brain, CD146 is first expressed in the cerebrovascular ECs of immature capillaries without pericyte coverage; with increased coverage of pericytes, CD146 could only be detected in pericytes, but not in cerebrovascular ECs. Specific deletion of Cd146 in mice ECs resulted in reduced brain endothelial claudin-5 expression and BBB breakdown. By analyzing mice with specific deletion of Cd146 in pericytes, which have defects in pericyte coverage and BBB integrity, we demonstrate that CD146 functions as a coreceptor of PDGF receptor-β to mediate pericyte recruitment to cerebrovascular ECs. Moreover, we found that the attached pericytes in turn down-regulate endothelial CD146 by secreting TGF-β1 to promote further BBB maturation. These results reveal that the dynamic expression of CD146 controls the behavior of ECs and pericytes, thereby coordinating the formation of a mature and stable BBB.
Blood–brain barrier (BBB) development is a sequential and well-orchestrated process that commences when brain endothelial cells (ECs; BECs) invade the embryonic neuroectoderm from the surrounding vascular plexus and induce BBB properties by establishing paracellular tight junctions (TJs) (1). Endothelial TJs are formed by a complex of transmembrane proteins, including claudins and occludin, as well as cytoplasmic adaptors such as zonula occludens protein 1 (ZO-1), thus creating a high-resistance paracellular barrier to molecules and ions (2). Compelling evidence shows that claudin-5 plays a key role in the induction of BBB properties, and specific loss of claudin-5 in mice results in a more leaky BBB (3–5). Following the establishment of the TJs, the BECs of nascent vessels recruit pericytes to the endothelial walls, which improve the barrier function of BECs by stabilizing TJs and decreasing transcytosis, and are crucial for maturation of the BBB (6). Importantly, pericytes suppress the expression of leukocyte adhesion molecules (LAMs) in BECs to reduce the invasion of immune cells into the CNS, therefore regulating CNS immune surveillance, a critical feature of the mature BBB (6, 7). Thus, as a dynamic interface with a range of interrelated functions, the BBB results from extremely effective TJs, pericyte recruitment, and regulation of leukocyte extravasation, thereby generating the mature physical and immune regulatory functions of the BBB (8).
Recently, extensive efforts have been made to investigate the underlying molecular mechanisms that regulate the sequential formation of the BBB. The activation of the VEGFR2 and Wnt–β-catenin pathways in BECs have been shown to induce angiogenesis and mature vessel morphology in the developing CNS (9–12). Subsequently, TGF-β/TGF-βR and Ang-1/Tie-2 signaling promote further BBB maturation (6). During angiogenesis, the BECs of nascent vessels recruit pericytes to the endothelial surface by releasing PDGF-B (13, 14). Disruption of this attachment and interaction may cause BBB dysfunction and neuroinflammation in CNS disease (15). Despite the importance of pericyte–EC interactions in the regulation of BBB development, little is known about the molecular mechanisms that spatiotemporally modulate their communication during the gradual process of BBB development (16, 17).
CD146 (also known as MCAM, S-endo-1, P1H12, and MUC18) was originally identified as a novel endothelial biomarker for angiogenesis in the tumor progression of several malignancies, including melanoma, prostate cancer, and breast cancer (18). In the CNS, CD146 is also involved in multiple sclerosis (19, 20) and Alzheimer’s disease (www.malacards.org/). Recent studies have shown that CD146 is constitutively expressed in the pericytes of several organs and functions as a component of endothelial junctions to reduce the paracellular permeability of peripheral ECs (21–24). However, in the CNS, the expression pattern of CD146 in the cerebrovasculature, and its role during BBB development remain largely unknown.
The aim of the present study was to explore the specific role of CD146 in BBB formation, especially its involvement in pericyte–EC interactions during this gradual process. We investigated the expression patterns of CD146 in the mouse cerebrovasculature at a series of developmental stages and further explored the roles of CD146 in pericyte/EC communication during BBB development by generating pericyte- and EC-specific Cd146-KO mice, respectively. In vivo vascular permeability analysis in mice combined with in vitro BBB models using mouse and human cerebrovascular ECs and pericytes was performed to examine the function and integrity of the BBB. Our results revealed that CD146 is required for BBB development by dynamically coordinating pericyte–EC communication during embryogenesis. As BBB breakdown is involved in many neurological disorders, including brain tumors, stroke, CNS infections, and neurodegenerative diseases (25, 26), the present study could provide new insights into therapeutic strategies to modulate the BBB in neurological disorders associated with BBB dysfunction.
Results
Dynamic CD146 Expression Correlates with BBB Development.
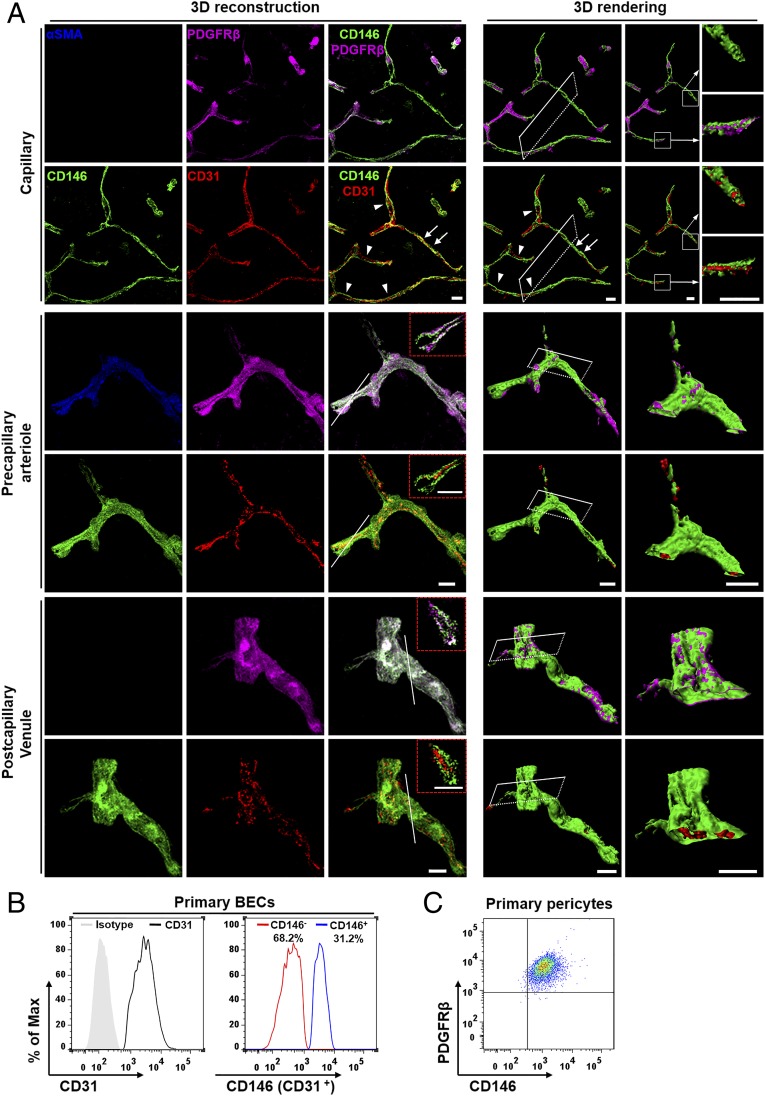
The BBB is highly developed at the level of the brain microvascular network comprising capillaries, arterioles, and venules (27). To determine the involvement of CD146 in the development of the BBB, we first investigated the expression of CD146 in the capillaries, precapillary arterioles, and postcapillary venules of the mouse brain at 4–6 wk. As shown in Fig. 1A, costaining of the brain sections from cortex with antibodies against CD146, CD31 (a marker of ECs), PDGF receptor-β (PDGFRβ; a marker of pericytes), or α-smooth muscle actin (αSMA; a marker of arterioles) revealed two distinct CD146 expression patterns in the CNS vessels. In immature brain microvessels without pericyte coverage, CD146 was expressed in the BECs. However, in the microvessels covered with pericytes, CD146 expression was exclusively observed in pericytes and not in BECs (Fig. 1A). Similar expression patterns of CD146 were also observed in the hippocampus, cerebellum, olfactory bulb, and corpus callosum, where CD146 was expressed in the BECs of immature brain microvessels without pericyte coverage, and was expressed only in pericytes of the microvessels but not in pericyte-covered BECs (Fig. S1). Flow cytometry showed that BECs isolated from the mouse brain exhibited heterogeneity in CD146 expression (Fig. 1B), whereas pericytes isolated from the mouse brain constitutively expressed CD146 (Fig. 1C). The analysis of CD146 expression suggests a heterogeneity between vascular segments with and without pericyte coverage within the vascular bed (arterioles/capillaries/venules).
Fig. 1.
Expression of CD146 in BECs and pericytes is dynamic in mice. (A) Brain sections (40 μm thick) from the cortex of mice at 4–6 wk were stained for CD146 (green), CD31 (EC marker; red), PDGFRβ (pericyte marker; purple), and αSMA (artery marker; blue) to indicate CD146 expression in capillaries, precapillary arterioles, and postcapillary venules. Shown are 3D reconstructions of confocal image z-stacks of brain vessels and the 3D surface rendering of epifluorescence images. Cut-open images were then created from the 3D surface rendering of vessels to reveal the inner vessel wall. In capillaries, segments without pericyte coverage showed CD146 expression in the BECs (arrows). However, in the capillaries covered with pericytes (arrowheads), CD146 was expressed in pericytes but not in BECs. White squares indicate areas shown on the right of each panel. In brain precapillary arterioles and postcapillary venules, CD146 expression was exclusively observed in pericytes and not in BECs. The dashed red insets indicate a confocal z-slice located at the white line of each panel, confirming that the expression of CD146 was exclusively observed in pericytes and not in BECs of precapillary arterioles and postcapillary venules. (Scale bars: 10 μm.) At least 20 capillaries, 20 precapillary arterioles, and 20 postcapillary venules from the cortex were analyzed. (B) Flow-cytometry analysis of CD146 expression in murine BECs (stained with antibodies against CD146 and CD31) showed heterogeneity in CD146 expression. The ratio between CD146− and CD146+ BECs was approximately 2:1. (C) Flow-cytometry analysis of CD146 expression in murine brain pericytes (stained with antibodies against CD146 and PDGFRβ). Pericytes constitutively expressed CD146. Data are from one experiment representative of three independent experiments with five mice (A) or six to eight mice per group (B and C).
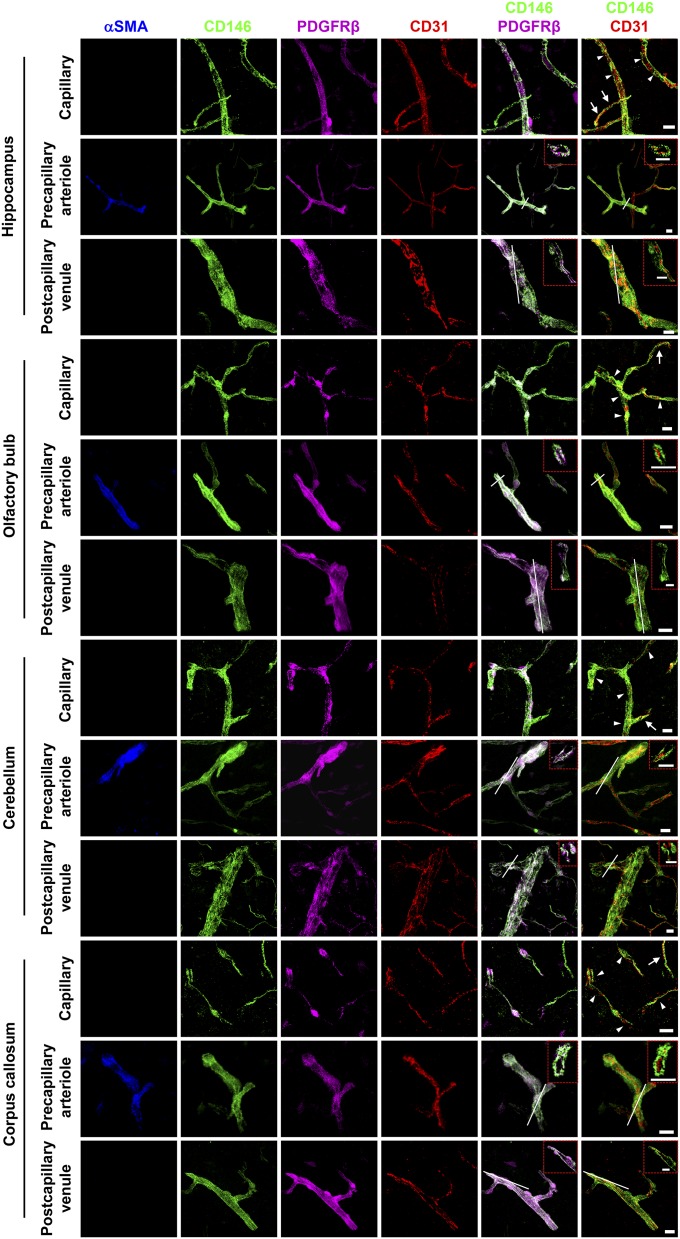
Fig. S1.
The expression of CD146 in brain vessels at 4–6 wk. Brain sections (40-μm thickness) from the hippocampus, olfactory bulb, cerebellum, and corpus callosum (white matter) of mice at 4–6 wk were stained for CD146 (green), CD31 (EC marker; red), PDGFRβ (pericyte marker; purple), and αSMA (artery marker; blue). Shown are 3D reconstructions of confocal image z-stacks of brain capillaries, precapillary arterioles, and postcapillary venules. The same expression pattern of CD146 was observed in different brain regions where CD146 was expressed in the BECs of immature capillaries without pericyte coverage (arrows) and was only expressed in pericytes of brain capillaries covered with pericytes (arrowheads), precapillary arterioles, and postcapillary venules. The dashed red rectangles indicate a confocal z-slice located at the white line of each panel, confirming that the expression of CD146 was exclusively observed in pericytes but not in BECs of precapillary arterioles and postcapillary venules. At least 20 capillaries, 20 precapillary arterioles, and 20 postcapillary venules from the hippocampus, olfactory bulb, cerebellum, and corpus callosum were analyzed. (Scale bars: 10 μm.) Data are from one experiment representative of three independent experiments with five mice.
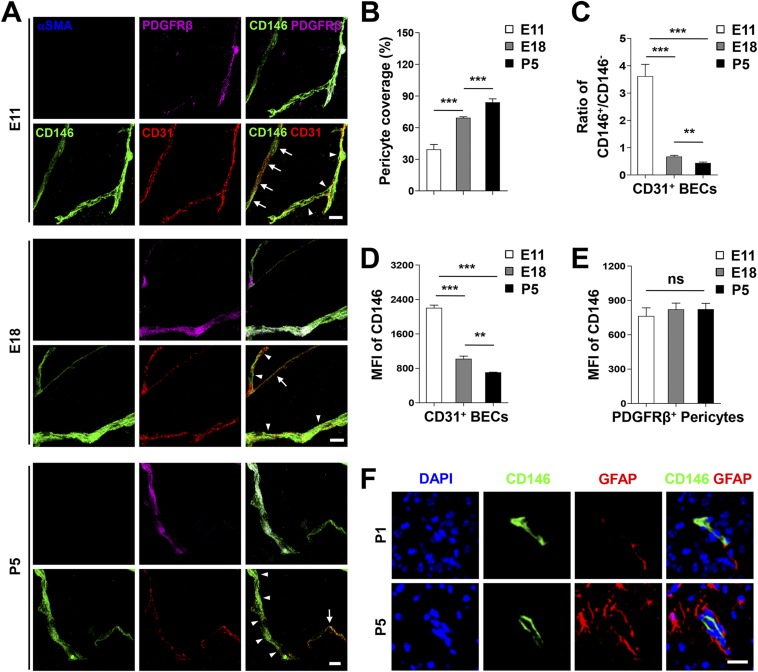
Capillaries constitute the largest and tightest fraction of the microvasculature (27, 28). During early development of the BBB, the angiogenesis of mice brain capillary begins at embryonic day (E) 10, when the BECs enter the cortex and vascularize the CNS (1). The functions of the BBB gradually develop with the coverage and recruitment of pericytes to cerebrovascular elements at approximately E15.5 (29). To further examine the correlation of the dynamic expression of CD146 and the recruitment of pericytes at different BBB developmental stages, we next investigated the expression patterns of CD146 in mouse cerebral capillaries at E11, E18, and postnatal day (P) 5, respectively. At E11, CD146 was expressed in the BECs of the nascent vessels and attached pericytes (Fig. S2A). At E18 and P5, along with increased pericyte coverage, CD146 was expressed mainly in the pericytes but not in BECs covered with pericytes, whereas BECs without pericyte coverage still expressed CD146 (Fig. S2 A and B). These observations are consistent with the finding that the percentage of CD146-positive BECs as well as the expression level of CD146 in BECs decreased dramatically during the course of BBB maturation (Fig. S2 C and D). By contrast, CD146 expression in pericytes was relatively stable during BBB development (Fig. S2E). Additionally, there was no detectable expression of CD146 in mouse astrocytes at P1 and P5 (Fig. S2F). These results demonstrate that, even though CD146 is constitutively expressed in pericytes, its expression in BECs is dynamic during BBB development. The dynamic expression of CD146 in ECs and pericytes may indicate progressive functional changes at different stages of BBB formation.
Fig. S2.
Dynamic CD146 expression correlates with pericyte recruitment during BBB development. (A) Three-dimensional reconstructions of confocal-image z-stacks of brain capillaries stained for CD146 (green), PDGFRβ (pericyte marker; purple), CD31 (EC marker; red), and αSMA (artery marker; blue) in cortex from mice at E11, E18, and P5. The dynamic expression pattern of CD146 was observed at different BBB developmental stages. Arrows indicate capillaries without pericyte coverage; arrowheads indicate capillaries with pericyte coverage. (Scale bars: 10 μm.) (B) Quantification of pericyte coverage by analyzing percent length of CD31+ capillaries as opposed to PDGFRβ+ pericytes as shown in A. (C) Quantification of the ratio of CD146+/CD146− BECs from mice was analyzed by flow cytometry (stained with antibodies against CD146 and CD31). The percentage of CD146+ BECs decreased with BBB development. (D) MFI of CD146 in CD31+ murine BECs showed a significant decrease from E11 to P5. (E) MFI of CD146 in PDGFRβ+ murine brain pericytes was relatively stable during BBB development. (F) CD146 is not expressed in astrocytes in mice. Brain sections of mice at ages P1 and P5 were stained for CD146 (green) and GFAP (astrocyte marker; red). Nuclei were stained with DAPI (blue). (Scale bar: 20 μm.) Data are from one experiment representative of three independent experiments with five mice (A, B, and F) or six to eight mice per group (C–E) (**P < 0.01 and ***P < 0.001).
Loss of Cd146 Results in BBB Breakdown.
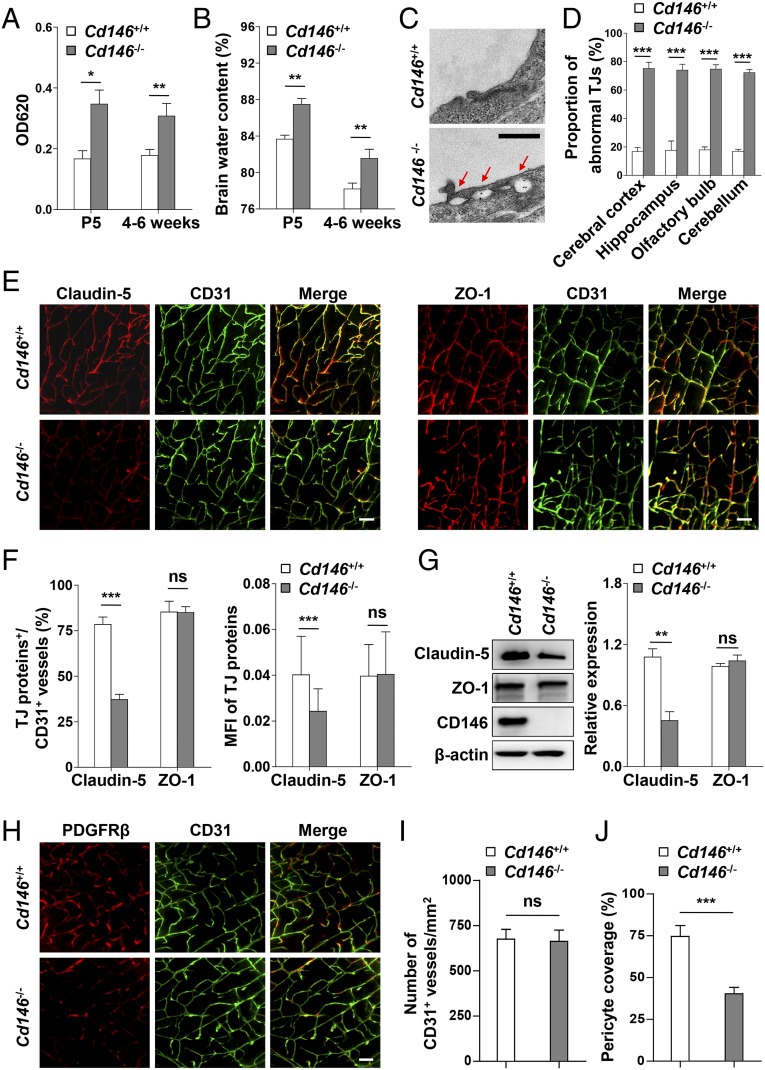
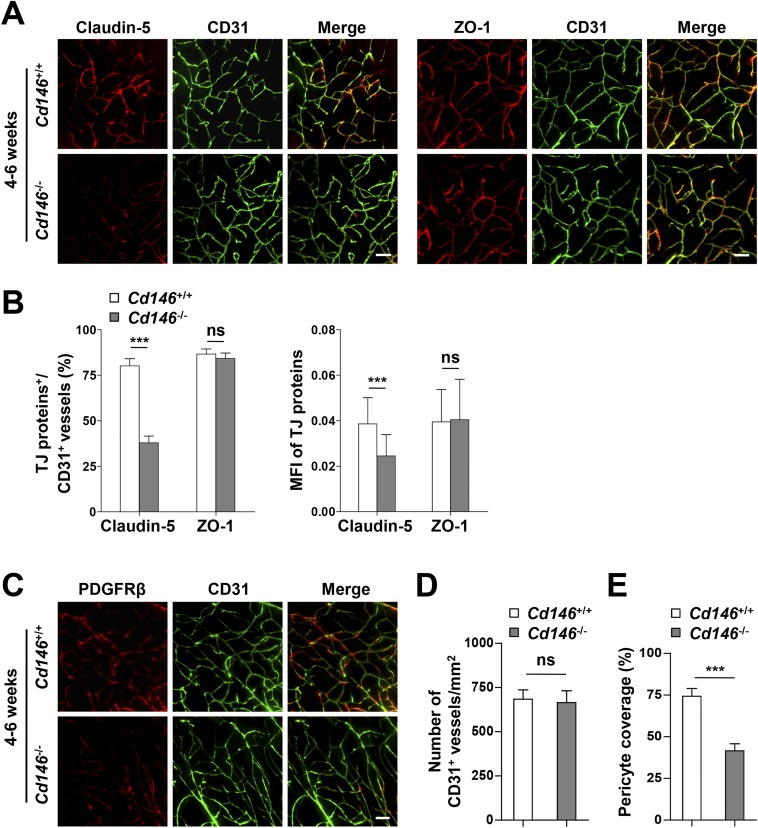
To explore the role of CD146 during BBB development, we first evaluated BBB function in Cd146-KO mice. It was found that the permeability of the BBB to Evans blue dye (which labels albumin) and brain water content were significantly higher in the neonatal and 4–6-wk-old KO mice than in the WT littermate control mice (Fig. 2 A and B). As BBB permeability is highly dependent on cerebrovascular endothelial TJs (4), we next checked the integrity of endothelial TJs in the brain of Cd146-KO mice. Electron microscopy (EM) analysis demonstrated that abnormal paracellular gaps along the apical side of BECs were present in Cd146-KO mice (Fig. 2 C and D). The expression of TJ proteins in the cerebrovasculature of Cd146-KO mice was further investigated. Compared with WT mice, Cd146-KO mice expressed less claudin-5, but showed comparable levels of ZO-1 (Fig. 2 E–G and Fig. S3 A and B). In addition, it was found that deletion of Cd146 caused a reduction of pericyte coverage without affecting the number of vessels (Fig. 2 H–J and Fig. S3 C–E). Together, these data suggest that CD146 is essential for BBB integrity.
Fig. 2.
Cd146 deficiency results in impaired BBB integrity in mice. (A) Cd146+/+ and Cd146−/− mice at P5 or 4–6 wk were given an i.p. injection or i.v. injection of Evans blue dye, respectively, and the absorption of Evans blue extracted from the mouse brain was measured by a microplate spectrophotometer at 620 nm. (B) Brain water content of Cd146+/+ and Cd146−/− mice (P5 or 4–6 wk). (C) EM images of TJs of brain capillaries in cortex from Cd146+/+ and Cd146−/− mice (P5). Red arrows indicate altered junctional strands. (Scale bar: 500 nm.) (D) Quantification of the abnormal endothelial TJ structure of brain capillaries in cerebral cortex, hippocampus, olfactory bulb, and cerebellum from Cd146+/+ and Cd146−/− mice (P5; at least 50 TJs were analyzed per group). (E) Brain sections from cortex of mice at P5 were costained for CD31 (green) and claudin-5 or ZO-1 (red) and analyzed by light-sheet fluorescence microscopy (LSFM) after being optically cleared by using organic solvents. Representative maximum-intensity projections (MIPs) of 40 virtual single slices from Cd146+/+ and Cd146−/− mice are shown. (Scale bars: 50 μm.) (F) Quantification of the number of CD31+ capillaries expressing claudin-5 or ZO-1 and the mean fluorescence intensity (MFI) of claudin-5 or ZO-1 in CD31+ capillaries demonstrate reduction of claudin-5 in capillaries from Cd146−/− mice compared with those from Cd146+/+ mice. (G) Western blot analysis of the expression of claudin-5 and ZO-1 in murine BECs purified from Cd146+/+ and Cd146−/− mice. (H) Brain sections from cortex of mice at P5 were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146+/+ and Cd146−/− mice are shown. (Scale bar: 50 μm.) (I) Quantification of the number of CD31+ capillaries in cortex from Cd146+/+ and Cd146−/− mice. No difference was detected. (J) Pericyte coverage was quantified by analyzing percent length of CD31+ capillaries opposed to PDGFRβ+ pericytes. The capillaries of cortex from Cd146−/− mice showed a decrease in pericyte coverage (*P < 0.05, **P < 0.01, and ***P < 0.001). Data are from one experiment representative of three independent experiments with eight mice per genotype (A, B, D and G) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (F, I and J).
Fig. S3.
Cd146 deficiency results in reduced claudin-5 expression and impaired pericyte recruitment. (A) Brain sections from cortex of mice at 4–6 wk were costained for CD31 (green) and claudin-5 (red) or ZO-1 (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146+/+ and Cd146−/− mice are shown. (Scale bars: 50 μm.) (B) Quantification of the number of CD31+ capillaries expressing claudin-5 or ZO-1 and the MFI of claudin-5 or ZO-1 in CD31+ capillaries showed reduction of claudin-5 in capillaries of cortex from Cd146−/− mice compared with those from Cd146+/+ mice at P5 and 4–6 wk. (C) Brain sections from cortex of mice at 4–6 wk were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146+/+ and Cd146−/− mice are shown. (Scale bar: 50 μm.) (D) Quantification of the number of CD31+ capillaries of brain cortex from Cd146+/+ and Cd146−/− mice. No difference was detected. (E) Pericyte coverage was quantified by analyzing percent length of CD31+ capillaries as opposed to PDGFRβ+pericytes. The capillaries of brain cortex from Cd146−/− mice showed a decrease in pericyte coverage (***P < 0.001). Data are from one experiment representative of three independent experiments with five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (B, D, and E).
EC Cd146 Deletion Results in Reduced Claudin-5 Expression and BBB Breakdown.
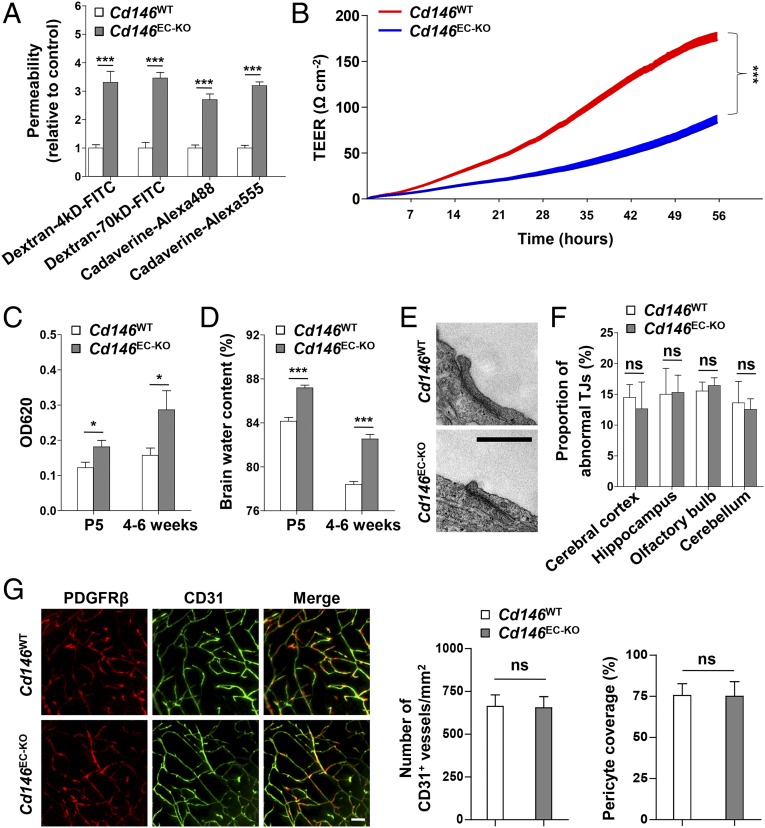
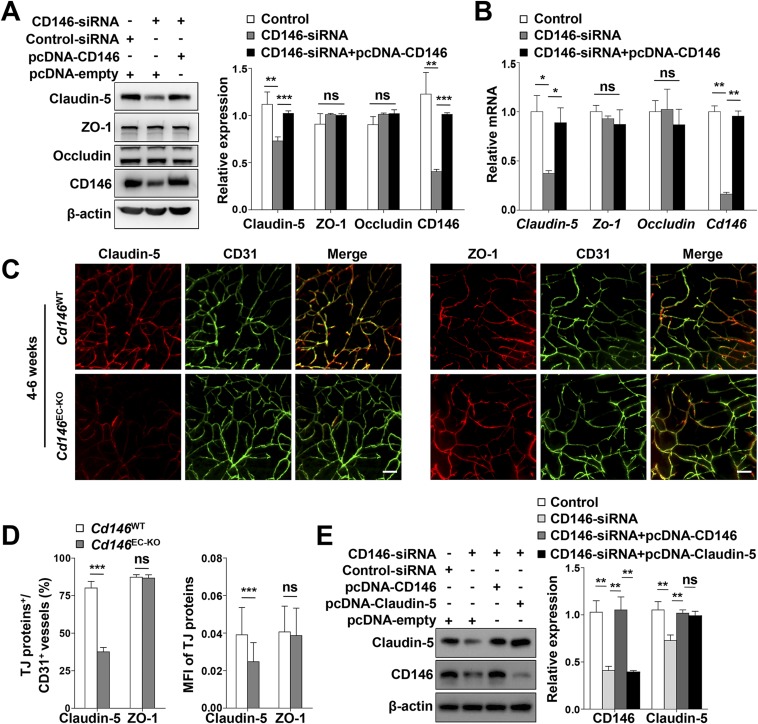
The aforementioned results indicated that CD146 expression was down-regulated from pericyte-free ECs to pericyte-covered ECs, whereas it was consistent in pericytes during BBB development. We therefore analyzed the effects of endothelial CD146 on BBB permeability first. To this end, primary BECs of the EC-specific Cd146-KO mice (Cd146EC-KO) and their WT littermates were first isolated and identified. The cerebral capillaries isolated from these mice still express TJ proteins (claudin-5 and ZO-1), and BECs obtained from these fragments retain key features of the BBB. By performing diffusion assays by using tracers of different sizes (fluorescent dextrans, 70 kDa and 4 kDa; and cadaverine, 640 Da and 950 Da), the paracellular permeability of the primary mouse BEC monolayer was shown to be increased after Cd146 deletion (Fig. 3A). The reduction of transendothelial electrical resistance (TEER) of monolayers of primary mouse BECs confirmed the decrease of paracellular tightness after KO of endothelial Cd146 (Fig. 3B). This suggested an essential role of endothelial CD146 in the regulation of paracellular tightness of the endothelial barrier. Furthermore, we investigated the role of endothelial CD146 in the regulation of paracellular tightness in humans by using the human BEC line hCMEC/D3 (30). Consistent with the effects on primary mouse BECs, the paracellular tightness of the hCMEC/D3 monolayer was decreased by CD146 knockdown and could be rescued after the restoration of CD146 expression, as measured by the diffusion assay using dextrans and cadaverine (Fig. S4A). Collectively, these data suggested that endothelial CD146 is indeed required for the paracellular tightness of BECs in mice and humans.
Fig. 3.
Endothelial Cd146 deletion leads to reduced claudin-5 expression and BBB breakdown without affecting TJ structures and pericyte coverage. (A) The paracellular permeability of the brain capillary ECs isolated from Cd146WT and Cd146EC-KO mice was assessed by tracers of different sizes, fluorescent dextrans (4 kDa and 70 kDa), and cadaverine (640 Da and 950 Da; n = 10 per group). (B) Measurements of paracellular tightness of the brain capillary ECs isolated from Cd146WT and Cd146EC-KO mice (n = 8 per group). The paracellular tightness was examined by measuring the TEER using an RTCA-SP instrument. (C) Cd146WT and Cd146EC-KO mice at P5 or 4–6 wk were given an i.p. injection or i.v. injection of Evans blue dye, respectively. The absorption of Evans blue was measured by microplate spectrophotometer at 620 nm. (D) Brain water content of Cd146WT and Cd146EC-KO mice (P5 or 4–6 wk). (E) EM images of the endothelial TJs of brain capillaries in cortex from Cd146WT and Cd146EC-KO mice (P5). (Scale bar: 500 nm.) (F) Quantification of the abnormal TJ structure of brain capillaries in cerebral cortex, hippocampus, olfactory bulb, and cerebellum from Cd146WT and Cd146EC-KO mice (P5; at least 50 TJs were analyzed per group). (G) Brain sections from cortex of mice at P5 were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146EC-KO mice are shown. The number of CD31+ capillaries and pericyte coverage in Cd146WT and Cd146EC-KO mice was quantified. No difference was detected (*P < 0.05 and ***P < 0.001). (Scale bar: 50 μm.) Data are from one experiment representative of three independent experiments with eight mice per genotype (C, D, and F) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (G).
Fig. S4.
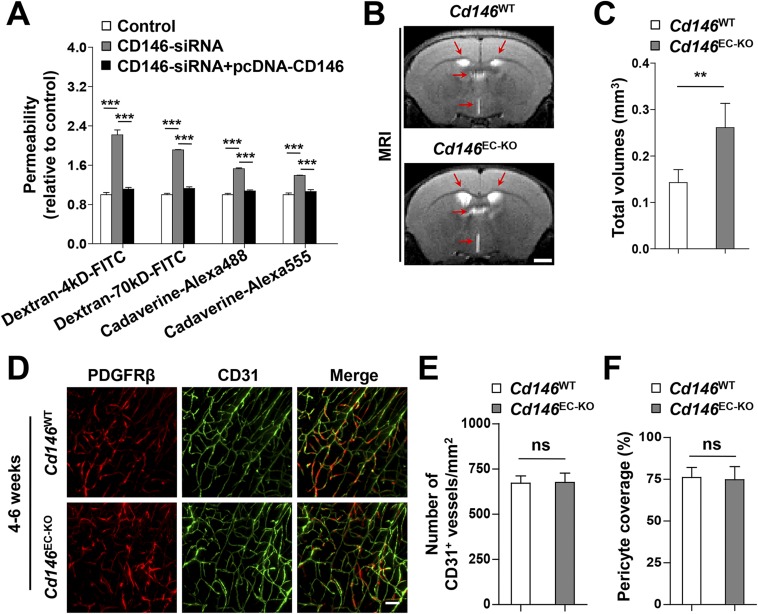
Deletion of endothelial Cd146 leads to BBB breakdown and enlargement of brain ventricles. (A) hCMEC/D3 cells were transfected with CD146 siRNA or cotransfected with CD146 siRNA and CD146-expressing plasmid. The paracellular permeability was assessed by using fluorescent tracers of different sizes of dextrans (4 kDa and 70 kDa) and cadaverine (640 Da and 950 Da; n = 10 per group). (B) Representative images at the same brain level were chosen from the volumes aligned to the Waxholm space. T2-weighted coronal MRI scans showing enlargement of the ventricles (red arrows) in Cd146EC-KO mice compared with that in Cd146WT mice at 4–6 wk. (Scale bar: 1,000 μm.) (C) Quantification of the total volumes of ventricles, including left lateral ventricle, right lateral ventricle, third ventricle, and fourth ventricle in Cd146EC-KO mice and Cd146WT mice at 4–6 wk. (D) Brain sections from cortex of mice at 4–6 wk were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146EC-KO mice are shown. (Scale bar: 50 μm.) (E) Quantification of the number of CD31+ capillaries of brain cortex from Cd146WT and Cd146EC-KO mice. No difference was detected. (F) Pericyte coverage was quantified by analyzing percent length of CD31+ capillaries opposed to PDGFRβ+ pericytes. No difference was detected (**P < 0.01 and ***P < 0.001). Data are from one experiment representative of three independent experiments with eight mice per genotype (C) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (E and F).
To further explore the role of endothelial CD146 in BBB permeability in vivo, we next examined BBB integrity in Cd146EC-KO mice using the Tg (Tek-cre) system (31). Compared with littermate controls, Cd146EC-KO mice showed impaired BBB integrity to Evans blue (i.e., albumin; Fig. 3C). Additionally, in the brains of P5 or 4–6-wk-old Cd146EC-KO mice, an increased ventricle size, as well as water content, was observed compared with their WT littermates (Fig. S4 B and C and Fig. 3D). However, there were no visible changes in the morphology of TJs, vessel number, or pericyte coverage (Fig. 3 E–G and Fig. S4 D–F). These observations indicate that Cd146 deletion in mouse BECs results in impairment of BBB function, leading to brain edema.
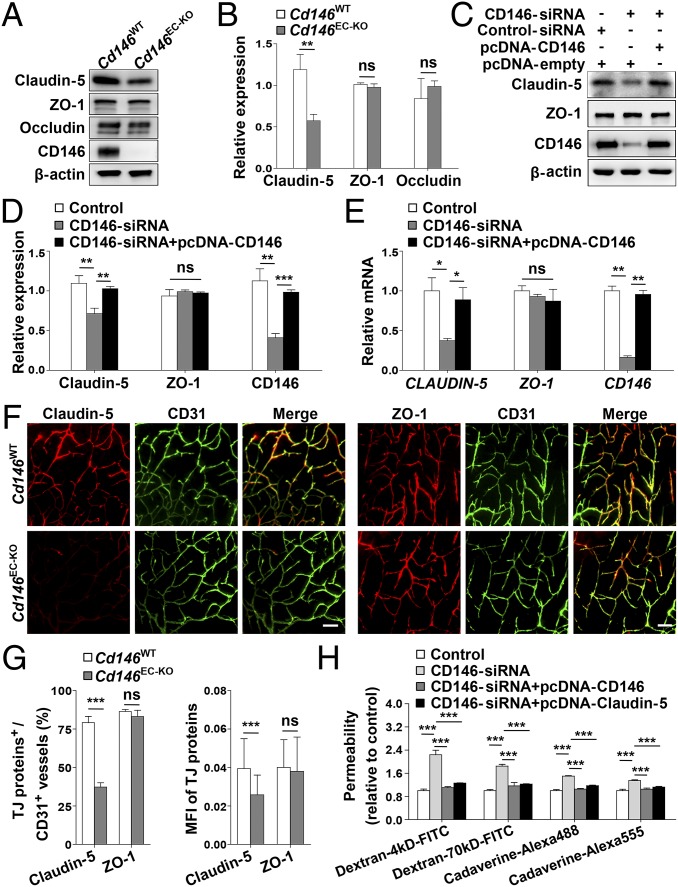
To explore the regulatory mechanisms of CD146 on the endothelial barriers during BBB development, we first used a tandem mass tag-based quantitative proteomics approach to map the relative proteome changes in bEND.3 cells following CD146 knockdown. It was found that knockdown of CD146 resulted in down-regulated expression of claudin-5, without affecting the expression of other important endothelial barrier-related proteins, including other TJ family members, transporters, and LAMs (Table S1). We next tested these findings in different cell lines and found that, in comparison with WT controls, loss or knockdown of CD146 down-regulated the expression of claudin-5, but not ZO-1 or occludin, in primary BECs (Fig. 4 A and B), hCMEC/D3 cells (Fig. 4 C–E), and bEND.3 cells (Fig. S5 A and B), respectively. To further confirm the regulatory role of endothelial CD146 on claudin-5 in vivo, we next compared the expression of claudin-5 in BECs of the Cd146EC-KO mice and their WT littermates. In the cerebral microvessels of the WT littermates, claudin-5 was strongly expressed in the cortical vascular trees of P5 or 4–6-wk-old mice, whereas expression was lower in the BECs of the Cd146EC-KO mice (Fig. 4 F and G and Fig. S5 C and D). These in vitro and in vivo results indicate an essential role of endothelial CD146 in vascular permeability and suggest claudin-5 as a downstream regulator of CD146 mediating BBB permeability. To find further evidence for this hypothesis, in vitro rescue experiments were performed, by expressing exogenous human claudin-5 combined with the dextran and cadaverine diffusion assay. It was found that the reduced paracellular tightness caused by knockdown of CD146 could be rescued by restoring claudin-5 expression (Fig. 4H and Fig. S5E). Together, these observations suggest that CD146 is necessary for the induction of claudin-5 expression in BECs, thereby regulating BBB integrity.
Table S1.
Selected proteins expressed in bEND.3 cells transfected with siRNA specifically targeting Cd146
| Protein name | Gene symbol | siRNA/Ctrl* | SD |
| CD146 (MCAM, MUC18) | Mcam | −0.66 | 0.03 |
| TJ | |||
| Claudin-5 | Cldn5 | −0.81 | 0.03 |
| TJ protein ZO-1 | Tjp1 | 0.98 | 0.02 |
| TJ protein ZO-2 | Tjp2 | 1.04 | 0.01 |
| Junctional adhesion molecule A | F11r | 1.03 | 0.03 |
| Occludin | Ocln | 1.08 | 0.06 |
| Angiogenesis | |||
| VEGF receptor 1 | Flt1 | 1.11 | 0.03 |
| VEGF receptor 3 | Flt4 | 0.88 | 0.06 |
| Cell division control protein 42 homolog | Cdc42 | 1.02 | 0.02 |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase γ-2 | Plcg2 | 0.96 | 0.03 |
| Nonspecific protein-tyrosine kinase | Src | 1.01 | 0.01 |
| Catenin β-1 | Ctnnb1 | 1.07 | 0.02 |
| Low-density lipoprotein receptor-related protein 5 | Lrp5 | 0.98 | 0.1 |
| Pan-endothelial | |||
| Cadherin-2 | Cdh 2 | 1.07 | 0.05 |
| Cadherin-5 | Cdh 5 | 1.09 | 0.05 |
| Cadherin-6 | Cdh 6 | 1.11 | 0.15 |
| Tyrosine-protein kinase receptor Tie-1 | Tie1 | 0.99 | 0.05 |
| Influx transporters | |||
| Solute carrier family 2, member 1 | Slc2a1 | 1.06 | 0.06 |
| Solute carrier family 12, member 2 | Slc12a2 | 1.01 | 0.03 |
| Solute carrier family 12, member 4 | Slc12a4 | 1.02 | 0.04 |
| Solute carrier family 23, member 2 | Slc23a2 | 1.04 | 0.1 |
| Solute carrier family 25, member 40 | Slc25a40 | 1.03 | 0.14 |
| Efflux transporters | |||
| ATP-binding cassette, subfamily A, member 1 | Abca1 | 1.12 | 0.04 |
| ATP-binding cassette, subfamily B, member 6 | Abcb6 | 1.04 | 0.04 |
| ATP-binding cassette, subfamily D, member 1 | Abcd1 | 1.06 | 0.04 |
| ATP-binding cassette, subfamily G, member 2 | Abcg2 | 1.01 | 0.02 |
| Permeability modulators | |||
| Plasmalemma vesicle-associated protein | Plvap | 1.17 | 0.07 |
| Leukocyte adhesion | |||
| Galectin-3 | Lgals3 | 1.16 | 0.08 |
| Intercellular adhesion molecule | Icam1 | 0.92 | 0.02 |
| Activated leukocyte cell-adhesion molecule | Alcam | 1.11 | 0.03 |
Selected proteins are listed with comparative protein expression data from bEND.3 cells transfected with siRNA specifically targeting Cd146 and controls. Direction of fold change is given if ≥ 1.18 or ≤ 0.84.
Fig. 4.
CD146 in BECs up-regulates the expression of claudin-5. (A) Western blot analysis of the expression of claudin-5, ZO-1, and occludin in murine BECs purified from Cd146WT and Cd146EC-KO mice. (B) Quantification of the expression level of claudin-5, ZO-1, and occludin in murine BECs purified from Cd146WT and Cd146EC-KO mice. (C–E) hCMEC/D3 cells were transfected with CD146 siRNA or cotransfected with CD146 siRNA and CD146-expressing plasmids. The expression of claudin-5 and ZO-1 was analyzed by Western blotting (C and D) or real-time PCR (E). (F) Brain sections from cortex of mice at P5 were costained for CD31 (green) and claudin-5 or ZO-1 (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146EC-KO mice are shown. (Scale bars: 50 μm.) (G) Quantification of the number of CD31+ capillaries expressing claudin-5 or ZO-1 and the MFI of claudin-5 or ZO-1 in CD31+ capillaries in cortex showed a reduction of claudin-5 in Cd146EC-KO mice compared with Cd146WT mice. (H) hCMEC/D3 cells were transfected with CD146 siRNA or cotransfected with CD146 siRNA and CD146-expressing plasmid or with CD146 siRNA and claudin-5–expressing plasmid, respectively. The paracellular permeability was assessed by tracers of different sizes of fluorescent dextrans (4 kDa and 70 kDa) and cadaverine (640 Da and 950 Da; n = 10 per group; *P < 0.05, **P < 0.01, and ***P < 0.001). Data are from one experiment representative of three independent experiments with eight mice per genotype (B) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (G).
Fig. S5.
Down-regulation or deletion of endothelial Cd146 results in reduced claudin-5 expression. (A and B) bEND.3 cells were transfected with CD146 siRNA or cotransfected with CD146 siRNA and CD146-expressing plasmid. The expression of claudin-5, ZO-1, and occludin was analyzed by Western blotting (A) or real-time PCR (B). (C) Brain sections from cortex of mice at 4–6 wk were costained for CD31 (green) and claudin-5 (red) or ZO-1 (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146EC-KO mice are shown. (Scale bars: 50 μm.) (D) Quantification of the number of CD31+ capillaries expressing claudin-5 or ZO-1 and the MFI of claudin-5 or ZO-1 in CD31+ capillaries showed reduction of claudin-5 in capillaries of brain cortex from Cd146EC-KO mice compared with those from Cd146WT mice. (E) hCMEC/D3 cells were transfected with CD146 siRNA, cotransfected with CD146 siRNA and CD146-expressing plasmid, or cotransfected with CD146 siRNA and claudin-5–expressing plasmid. The expression of claudin-5 and CD146 was analyzed by Western blotting (*P < 0.05, **P < 0.01, and ***P < 0.001). Data are from one experiment representative of three independent experiments with five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (D).
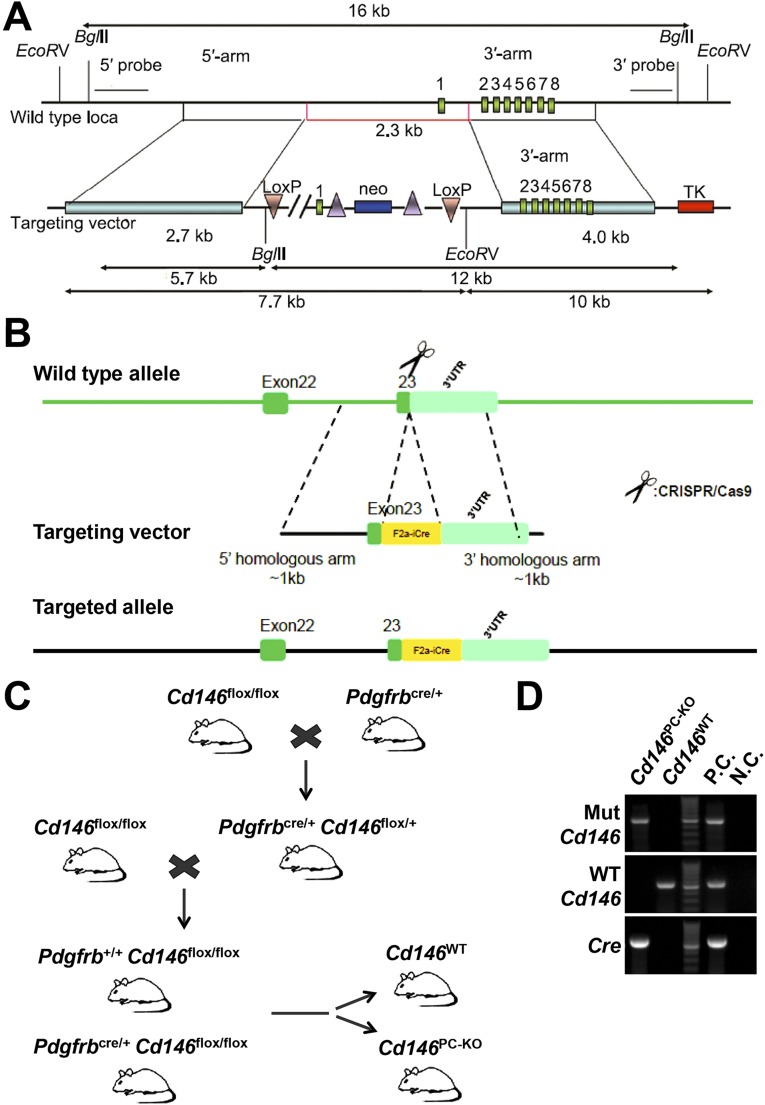
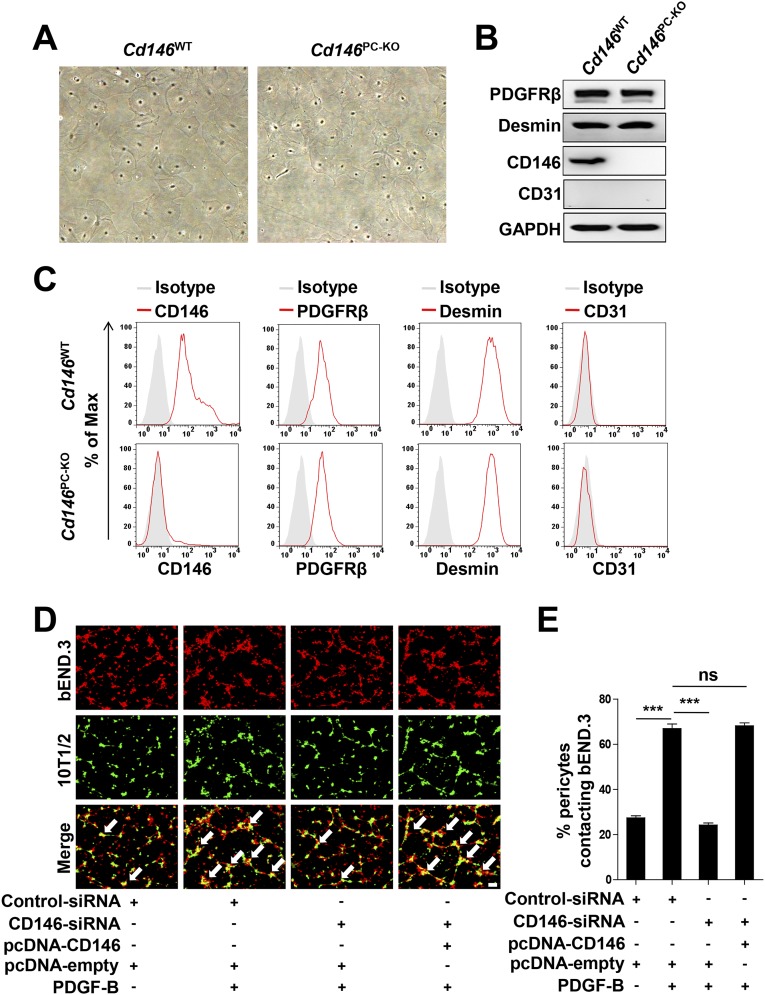
Pericyte Cd146 Deletion Results in Impaired Pericyte Recruitment and Breakdown of the BBB.
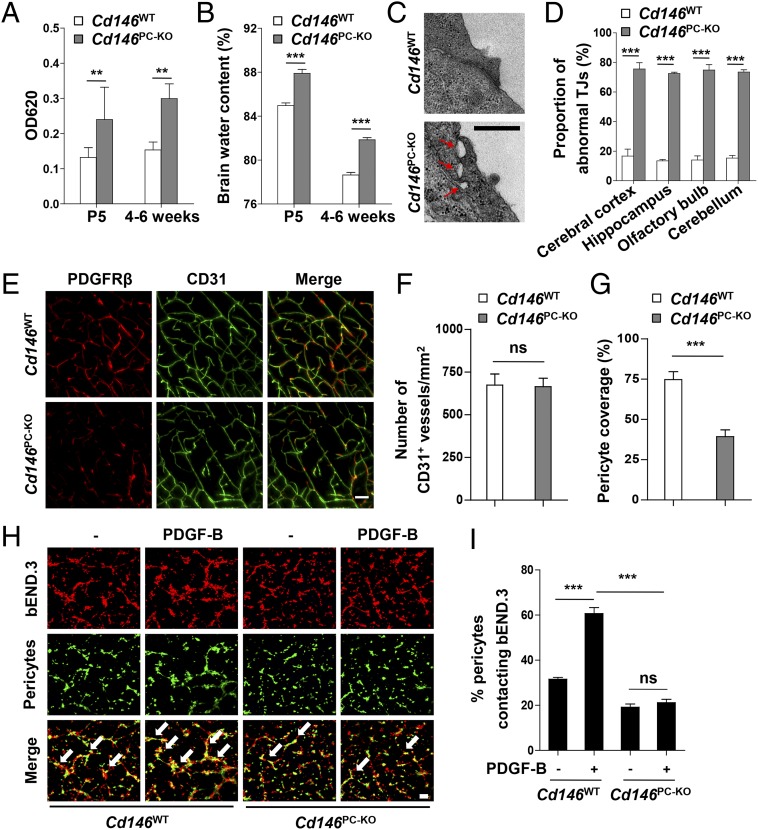
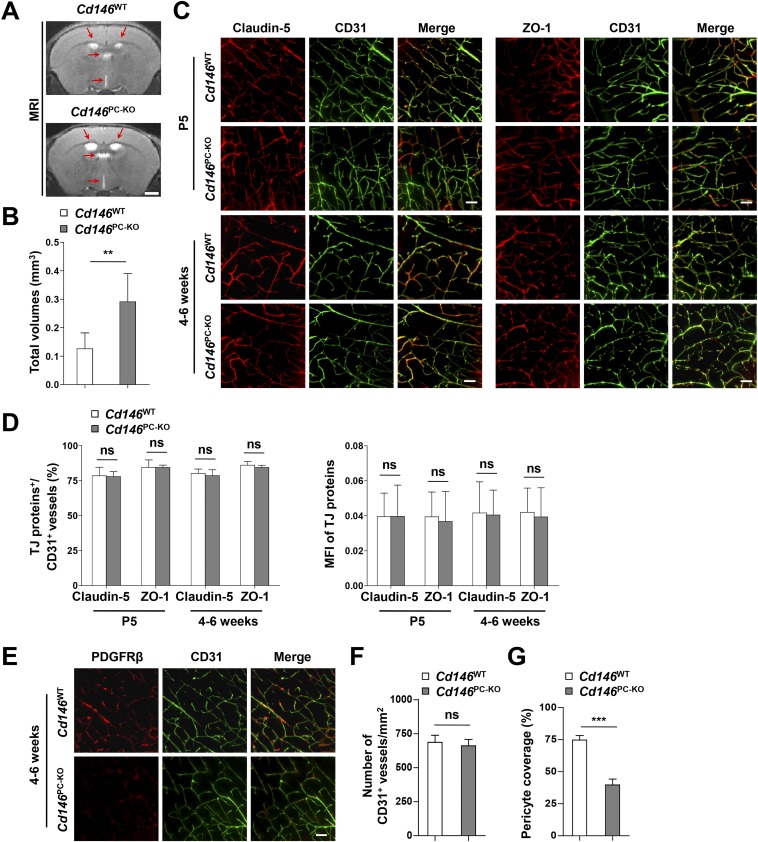
Pericyte coverage is essential for BBB integrity during embryogenesis (15, 32). To determine whether pericyte-expressed CD146 regulates BBB development, we generated pericyte-specific Cd146-KO mice (Cd146PC-KO) by using the Cre/LoxP system (Fig. S6). The BBB permeability and ventricle size in Cd146PC-KO mice at the age of P5 or 4–6 wk were increased in comparison with the littermate controls (Fig. 5 A and B and Fig. S7 A and B). Furthermore, abnormal cerebral endothelial TJ structure was observed in Cd146PC-KO mice, whereas the expression of TJ proteins was not affected (Fig. 5 C and D and Fig. S7 C and D). Moreover, pericyte-specific Cd146 deletion resulted in reduced pericyte coverage of the CNS microvessels, but did not affect the vessel number (Fig. 5 E–G and Fig. S7 E–G), indicating that the impairment of the BBB could be the result of insufficient pericyte coverage and recruitment.
Fig. S6.
Generation and characterization of pericyte-specific Cd146-KO mice. (A) Targeting strategy for generation of Cd146floxed/floxed mice; shown are the WT locus of mouse Cd146 gene (Top) and the targeting construct (Bottom). A LoxP site (3′loxp) was cloned upstream of the promoter, and the frt-Neo-frt-loxp cassette was cloned downstream of exon 1. (B) Targeting strategy for generation of Pdgfrbcre/+ mice. (C) Mating scheme to generate pericyte-specific Cd146-KO mice (Pdgfrbcre/+Cd146flox/flox, i.e., Cd146PC-KO) and control WT littermates (Pdgfrb+/+Cd146flox/flox, i.e., Cd146WT). (D) Genotyping of Cd146PC-KO and Cd146WT mice by PCR analysis of genomic DNA. A 595-bp fragment from WT Cre gene, a 550-bp fragment from WT Cd146 gene (WT Cd146), and a 502-bp fragment from floxed Cd146 gene (Mut Cd146) were PCR-amplified with specific primers. Genomic DNA from Pdgfrbcre/+ mice was used as positive control (P.C.) for Cre analysis; genomic DNA from Cd146floxed/floxed mice were used as positive control for Mut Cd146 analysis; genomic DNA from C57BL/6 mice were used as positive control for WT Cd146 analysis. H2O was used as negative control (N.C.) for all three PCR analyses. Data represent three independent experiments.
Fig. 5.
Pericyte Cd146 deletion results in impaired pericyte recruitment and BBB breakdown. (A) Cd146WT and Cd146PC-KO mice at P5 or 4–6 wk were given an i.p. injection or i.v. injection of Evans blue dye, respectively, and the absorption of Evans blue extracted from the mouse brain was measured by a microplate spectrophotometer at 620 nm. (B) Brain water content of Cd146WT and Cd146PC-KO mice (P5 or 4–6 wk). (C) EM images of TJs of brain capillaries in the cortex from Cd146WT and Cd146PC-KO mice (P5). Red arrows indicate altered TJ alignment. (Scale bar: 500 nm.) (D) Quantification of the abnormal TJ structure of brain capillaries in cerebral cortex, hippocampus, olfactory bulb, and cerebellum from Cd146WT and Cd146PC-KO mice (P5; at least 50 TJs were analyzed per group). (E) Brain sections from the cortex of mice at P5 were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146PC-KO mice are shown. (Scale bar: 50 μm.) (F) Quantification of the number of CD31+ capillaries in cortex from Cd146WT and Cd146PC-KO mice. No difference was detected. (G) Pericyte coverage was quantified by analyzing percent length of CD31+ capillaries opposed to PDGFRβ+ pericytes. Decreased pericyte coverage in capillaries was observed in the cortex of Cd146PC-KO mice. (H) bEND.3 cells and brain microvessel pericytes isolated from Cd146WT and Cd146PC-KO mice were labeled by PKH26-red and CFSE, respectively, and were cocultured in Matrigel-coated culture slides for 6 h. White arrows indicate pericytes contacting bEND.3 cells. (Scale bar: 100 μm.) (I) Quantification was performed by measuring the merged cells from 15 fields of 3 independent experiments (**P < 0.01 and ***P < 0.001). Data are from one experiment representative of three independent experiments with eight mice per genotype (A–D) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (F and G).
Fig. S7.
Pericyte Cd146 deletion results in the enlargement of brain ventricles and impaired pericyte recruitment. (A) The representative images at the same brain level were chosen from the volumes aligned to the Waxholm space. T2-weighted coronal MRI scans showing enlargement of the ventricles (red arrows) in Cd146PC-KO mice compared with that in Cd146WT mice at 4–6 wk. (Scale bar: 1,000 μm.) (B) Quantification of the total volumes of ventricles, including left lateral ventricle, right lateral ventricle, third ventricle, and fourth ventricle in Cd146PC-KO mice and Cd146WT mice at 4–6 wk. (C) Brain sections from cortex of mice at 4–6 wk were costained for CD31 (green) and claudin-5 (red) or ZO-1(red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146PC-KO mice are shown. (Scale bars: 50 μm.) (D) Quantification of the number of CD31+ capillaries expressing claudin-5 or ZO-1 and the MFI of claudin-5 or ZO-1 in CD31+ capillaries. No difference was detected. (E) Brain sections from cortex of mice at age 4–6 wk were costained for CD31 (green) and PDGFRβ (red) and analyzed by LSFM after being optically cleared by using organic solvents. Representative MIPs of 40 virtual single slices from Cd146WT and Cd146PC-KO mice are shown. (Scale bar: 50 μm.) (F) Quantification of the number of CD31+ capillaries in brain cortex from Cd146WT and Cd146PC-KO mice. No difference was detected. (G) Pericyte coverage was quantified by analyzing percent length of CD31+ capillaries as opposed to PDGFRβ+ pericytes. The capillaries of brain cortex from Cd146PC-KO mice showed a decrease in pericyte coverage (**P < 0.01 and ***P < 0.001). Data are from one experiment representative of three independent experiments with eight mice per genotype (B) or five mice per genotype, at least eight MIPs per mouse, and five random fields per MIP (D, F, and G).
To confirm the role of CD146 in pericyte recruitment, an in vitro cell recruitment assay was applied to mimic pericyte recruitment and attachment to BECs. First, primary pericytes from the brains of Cd146WT and Cd146PC-KO mice were isolated and identified. As shown in Fig. S8 A–C, these cells expressed the pericyte markers PDGFRβ and desmin, but were negative for the EC marker CD31. The cell recruitment results indicated that pericyte attachment to bEND.3 cells was significantly impaired by using Cd146-deficient pericytes (Fig. 5 H and I). Similar results were obtained by using the pericyte progenitor cell line 10T1/2 and bEND.3 cells (Fig. S8 D and E). These findings suggest that CD146 plays an important role in pericyte recruitment to BECs.
Fig. S8.
Pericyte Cd146 deletion impairs pericyte recruitment. (A) Morphology of primary murine CNS pericytes isolated from Cd146WT and Cd146PC-KO mice. (B and C) Characterization of primary murine CNS pericytes isolated from Cd146WT and Cd146PC-KO mice by Western blotting (B) or flow-cytometry analysis (C) using specific antibodies as indicated. (D) bEND.3 cells and 10T1/2 cells were labeled by PKH26-red and CFSE, respectively, and were cocultured in Matrigel-coated culture slides for 6 h. White arrows indicate pericytes contacting bEND.3 cells. (Scale bar: 100 μm.) (E) Quantification was performed by measuring the merged cells from 15 fields of three independent experiments (***P < 0.001). Data represent three independent experiments.
CD146 Mediates Pericyte Recruitment by Acting as a Coreceptor for PDGFRβ.
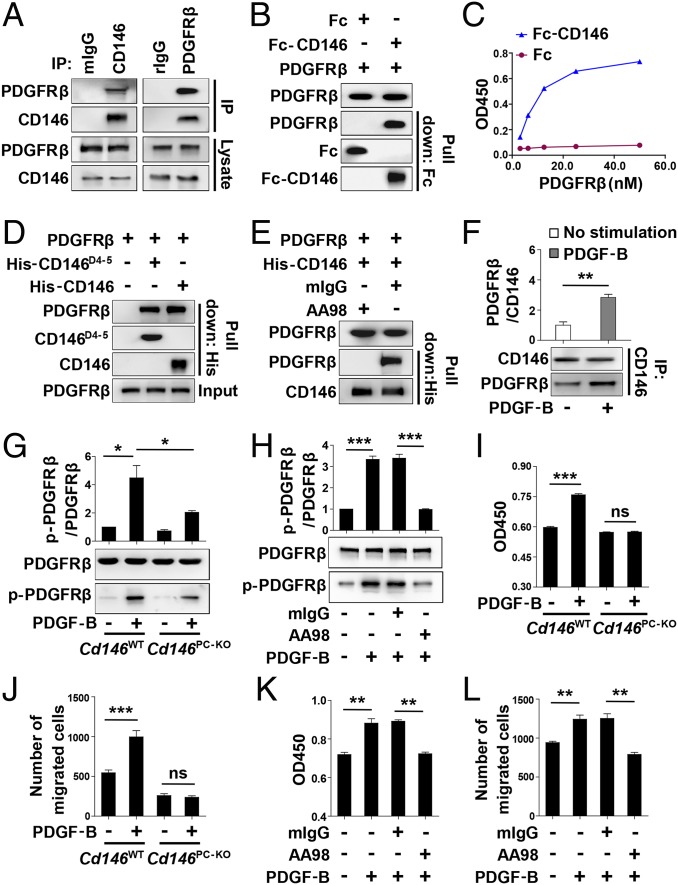
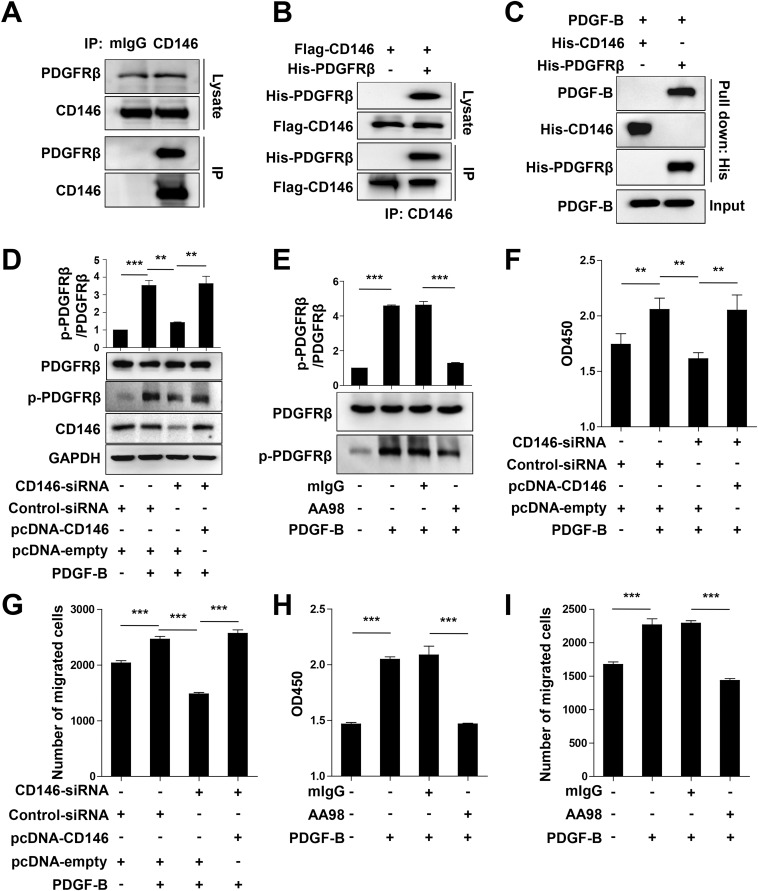
As we showed that pericyte-specific Cd146 deletion impaired pericyte recruitment, we next explored the molecular mechanisms that govern CD146-mediated pericyte recruitment to ECs. It has been reported that the major pathway regulating pericyte recruitment is the PDGF-B/PDGFRβ pathway (6), and the signaling molecules associated with CD146, such as Akt and PI3-kinase, are also present downstream of PDGFRβ. Moreover, CD146 has been shown to induce cell migration and proliferation that converge with responses elicited by PDGFRβ (14, 33). Based on these findings, we hypothesized that CD146 is involved in mediating pericyte recruitment to the BECs by participating in PDGFRβ activation. To prove this, we first examined the physical association of CD146 with PDGFRβ. As shown in Fig. 6A and Fig. S9A, the coimmunoprecipitation assay confirmed the binding between CD146 and PDGFRβ in 10T1/2 and human smooth muscle cells that express endogenous CD146 and PDGFRβ. This binding was also confirmed in CD146- and PDGFRβ-transfected HEK293 cells (Fig. S9B). The direct binding between CD146 and PDGFRβ was further proved by using Fc pull-down assay and ELISA (Fig. 6 B and C). The pull-down assay also revealed that the structural epitope of CD146 involved in binding to PDGFRβ is located in the domain 4–5 region of CD146 (CD146D4-5; Fig. 6D). This finding was confirmed by using the anti-CD146 antibody AA98, which recognizes CD146D4-5 and abrogates the interaction between CD146 and PDGFRβ (Fig. 6E). Moreover, PDGF-B stimulation enhanced the interaction of CD146 and PDGFRβ, indicating that this interaction is ligand-dependent (Fig. 6F). By contrast, CD146 did not show any direct interaction with PDGF-B (Fig. S9C). Taken together, these results suggest a direct physical interaction between CD146 and PDGFRβ.
Fig. 6.
CD146 activates PDGFRβ and promotes pericyte proliferation and migration. (A) Coimmunoprecipitation of endogenous CD146 and PDGFRβ in 10T1/2 cells. (B) Direct interaction between CD146 and PDGFRβ detected by Fc pull-down assay. (C) Fc or Fc-CD146 was added to wells coated with different concentrations of PDGFRβ, and ELISA was performed. (D) PDGFRβ directly binds CD146D4-5. PDGFRβ was first incubated with His-tagged CD146-ECD or CD146D4-5, respectively. The complex was pulled down by anti-His antibody. (E) Anti-CD146 AA98 abrogates CD146 and PDGFRβ interaction. His-CD146-ECD was incubated with PDGFRβ in the presence of mIgG or AA98 (50 μg/mL). (F) PDGF-B stimulation enhances the interaction of CD146 and PDGFRβ. Pericytes were stimulated without or with PDGF-B (20 ng/mL). (G) Cd146 deletion leads to impaired PDGFRβ activation. CNS pericytes isolated from Cd146WT and Cd146PC-KO mice were incubated with PDGF-B (20 ng/mL). The level of p-PDGFRβ was detected by Western blotting. (H) AA98 impairs PDGF-B–induced PDGFRβ activation in CNS pericytes isolated from Cd146WT mice. (I and J) The proliferation and migration of CNS pericytes isolated from Cd146WT and Cd146PC-KO mice in the presence of PDGF-B (20 ng/mL) were determined by CCK-8 assay (I) and transwell Boyden chamber assay (J). (K and L) Proliferation and migration of CNS pericytes from Cd146WT mice during PDGF-B (20 ng/mL) stimulation without or with anti-CD146 AA98 (50 μg/mL) were determined by CCK-8 assay (K) and transwell Boyden chamber assay (L). Data represent three independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001).
Fig. S9.
Targeting CD146 results in impaired PDGFRβ activation and cell proliferation and migration in 10T1/2 cells. (A) Coimmunoprecipitation (IP) assay of endogenous CD146 and PDGFRβ in human smooth muscle cells. CD146 from cell lysates was immunoprecipitated with anti-CD146 AA1. (B) HEK293 cells were transfected with Flag-CD146 and His-PDGFRβ. Cell lysates were immunoprecipitated with anti-CD146 AA1. (C) PDGF-B was incubated with His-PDGFRβ or His-CD146, and then anti-His antibody was used to precipitate the complex. (D) 10T1/2 cells were transfected with CD146-siRNA or cotransfected with CD146-siRNA and CD146-expressing plasmid, and then stimulated with PDGF-B (20 ng/mL). PDGFRβ activation was detected by Western blotting. Quantification of the relative p-PDGFRβ/PDGFRβ index is shown. (E) 10T1/2 cells were stimulated with PDGF-B (20 ng/mL) in the presence of mIgG or AA98 (50 μg/mL). PDGFRβ activation was detected by Western blotting. Quantification of the relative p-PDGFRβ/PDGFRβ index is shown. (F and G) 10T1/2 cells were treated as described in D. Cell proliferation and migration were detected by using a CCK-8 assay (F) and transwell Boyden chamber assay (G), respectively. (H and I) 10T1/2 cells were treated as described in E. The cell proliferation and migration were detected by CCK-8 assay (H) and transwell Boyden chamber assay (I), respectively (**P < 0.01 and ***P < 0.001). Data represent three independent experiments.
We next investigated whether CD146 engages in PDGFRβ activation in response to PDGF-B. As shown in Fig. 6 G and H, PDGF-B–induced PDGFRβ phosphorylation was impaired in Cd146-deficient pericytes as well as in CD146+ pericytes treated with AA98. Similar results were also obtained in 10T1/2 cells (Fig. S9 D and E). These findings strongly suggest that CD146 is a coreceptor of PDGFRβ, and is required for PDGF-B–induced PDGFRβ activation.
Finally, two in vitro pericyte models (primary brain pericytes and 10T1/2 cells) were chosen to evaluate the function of CD146 in PDGF-B–promoted pericyte proliferation and migration, two essential steps in pericyte recruitment to ECs. PDGF-B–induced pericyte proliferation and migration were abrogated in Cd146-deficient pericytes and in AA98-treated CD146+ pericytes (Fig. 6 I–L and Fig. S9 F–I), confirming the essential role of CD146 in PDGF-B–induced pericyte recruitment.
Pericyte-Derived TGF-β1 Down-Regulates Endothelial CD146 Expression.
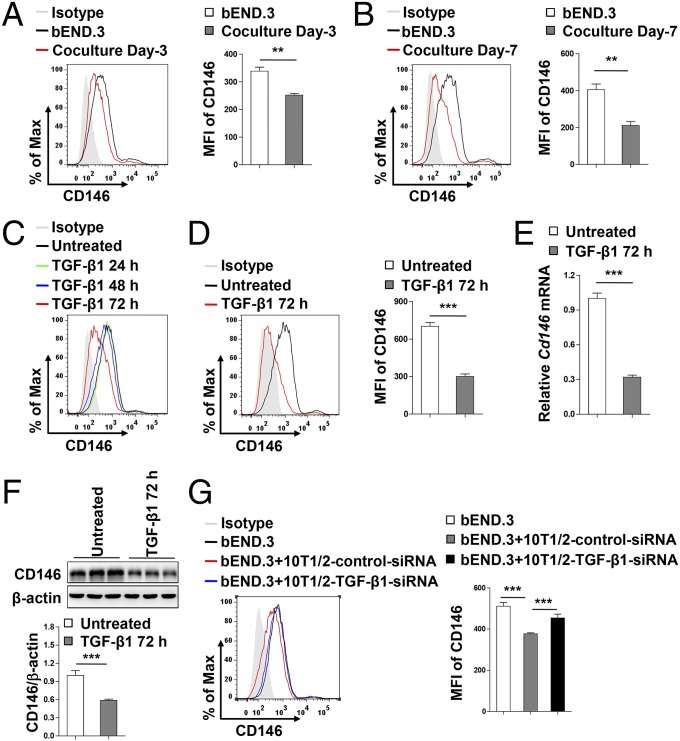
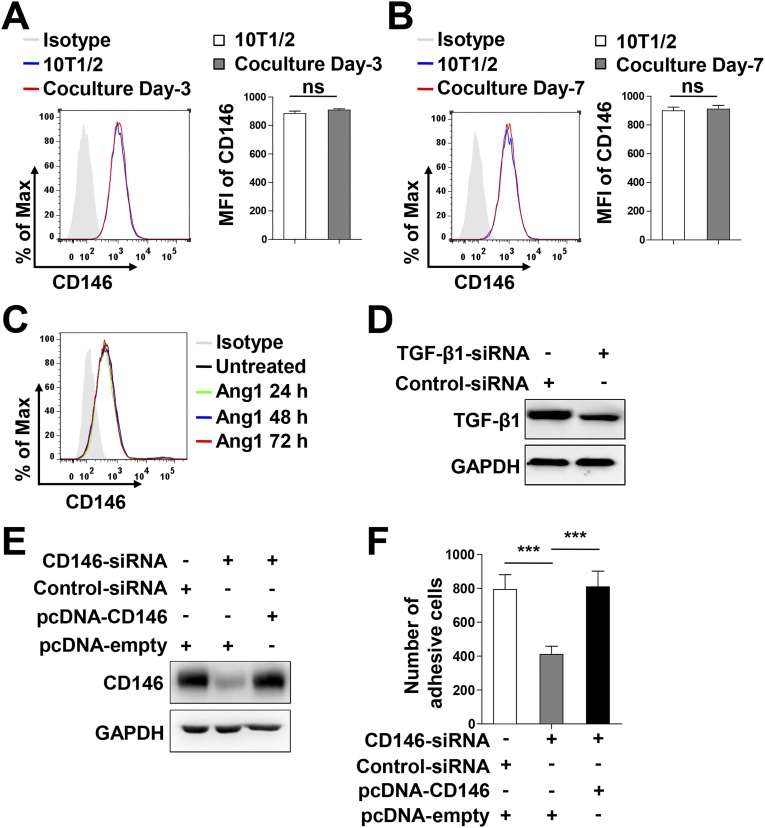
Given the observation that, after recruitment of pericytes to vessels, CD146 expression was dramatically decreased in BECs covered with pericytes during BBB development, we hypothesized that the consistent down-regulation of endothelial CD146 might be the result of paracrine signals from pericytes. To test the potential effects of surrounding pericytes on endothelial CD146 expression, coculture experiments were performed. As shown in Fig. 7 A and B and Fig. S10 A and B, coculturing bEND.3 cells with pericytes for 3 or 7 d resulted in the gradually reduced expression of endothelial, but not pericytic, CD146. A previous study showed that the interaction between BECs and pericytes mainly occurred through paracrine TGF-β1 and Ang1 signals (6). We observed that, after treatment of bEND.3 cells with TGF-β1, but not with Ang1, the expression of CD146 was down-regulated in a time-dependent manner (Fig. 7 C and D and Fig. S10C). The TGF-β1–mediated down-regulation of endothelial CD146 was also confirmed at the mRNA and protein levels (Fig. 7 E and F). Conversely, knockdown of TGF-β1 in pericytes inhibited the pericyte-mediated down-regulation of endothelial CD146 (Fig. 7G and Fig. S10D). It is well known that pericytes suppress the expression of LAMs in BECs to reduce the movement of immune cells into the CNS, thereby regulating CNS immune surveillance (7, 15). We found that the down-regulation of CD146 in hCMEC/D3 cells by pericytes was also associated with a reduction in the adhesive properties of immune cells to hCMEC/D3 (Fig. S10 E and F). Taken together, our results indicate that, during the later BBB maturation phase, pericytes down-regulate endothelial CD146 expression through TGF-β1.
Fig. 7.
Pericytes down-regulate endothelial CD146 expression through TGF-β1. (A and B) Flow-cytometry analysis of CD146 expression in bEND.3 cells cocultured without or with pericytes for 3 d (A) or 7 d (B). (C) Flow-cytometry analysis of CD146 expression in bEND.3 cells stimulated without or with TGF-β1 (10 ng/mL) for 24 h, 48 h, and 72 h. (D–F) bEND.3 cells were stimulated without or with TGF-β1 (10 ng/mL) for 72 h. Expression of CD146 in bEND.3 was determined by flow cytometry (D), real-time PCR (E), or Western blotting (F). (G) 10T1/2 cells transfected with control siRNA or TGF-β1 siRNA were cocultured with bEND.3 cells. The expression of CD146 in bEND.3 was determined by flow cytometry. (Right) Quantification of the MFI of CD146 (**P < 0.01 and ***P < 0.001). Data represent three independent experiments.
Fig. S10.
Pericytes down-regulate endothelial CD146 expression through TGF-β1 signal, but not Ang1. (A and B) Flow-cytometry analysis of CD146 expression in 10T1/2 cell coculture without or with bEND.3 cells for 3 d (A) or 7 d (B). (C) Flow-cytometry analysis of CD146 expression in bEND.3 cells stimulated without or with Ang1 (10 ng/mL) for 24 h, 48 h, and 72 h. (D) TGF-β1 expression in 10T1/2 cells transfected with control siRNA or TGF-β1 siRNA was detected by Western blotting. (E) hCMEC/D3 cells were transfected with CD146 siRNA or cotransfected with CD146 siRNA and CD146-expressing plasmid. CD146 expression levels were measured by using Western blotting. (F) hCMEC/D3 cells were transfected as described in E. Adhesion of human leukocytes to hCMEC/D3 cell monolayer was detected by flow cytometry analysis (***P < 0.001). Data represent three independent experiments.
Discussion
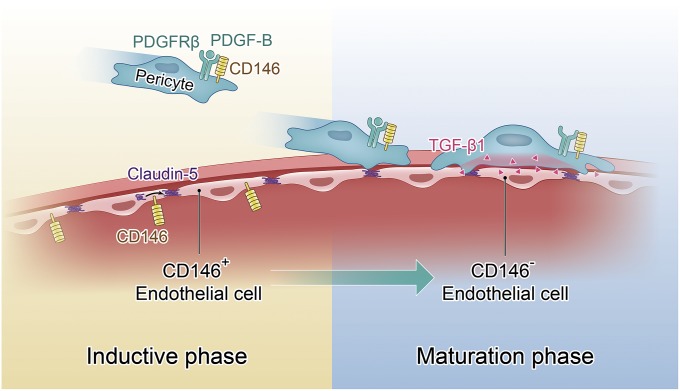
BBB development is a progressive process controlled by close coordination of BECs and pericytes (7). However, the mechanisms that govern this process have thus far remained elusive (16, 17). Here, we show that CD146 functions as a key spatiotemporal molecule to orchestrate the development of the BBB. Initially, CD146 is expressed in the BECs of nascent vessels and is a critical regulator of claudin-5 expression. Subsequently, CD146 expressed in pericytes promotes pericyte recruitment to the BECs by directly regulating PDGF-B/PDGFRβ signaling. Meanwhile, CD146 expression in BECs is rapidly down-regulated by pericyte-secreted TGF-β1, which may contribute to CNS immune quiescence (Fig. 8). Thus, CD146 regulates BBB development through temporal and cell type-specific effects.
Fig. 8.
Dynamic coordination of BEC–pericyte communication by CD146 controls BBB formation during embryogenesis. At the initial stage of BBB development, CD146 expression in the cerebrovascular ECs promotes barrier induction by up-regulating claudin-5. Subsequently, pericyte-expressed CD146 functions as a coreceptor of PDGFRβ to recruit pericytes to BECs. Following the pericyte recruitment and attachment, endothelial CD146 is finally down-regulated by pericyte-derived TGF-β1, contributing to further BBB maturation.
In the initial stages of BBB development, CD146 is expressed in the BECs and pericytes. By investigating the detailed expression pattern of CD146 in different brain regions during BBB development, we identified that (i) the expression of CD146 in brain microvessels was mainly in the pericytes but not in BECs covered with pericytes, whereas BECs without pericyte coverage still expressed CD146; (ii) the dynamic expression pattern of CD146 in the cerebral microvasculature was observed in different regions of the brain, including olfactory bulb, cerebral cortex, hippocampus, cerebellum, and corpus callosum, indicating its universality; (iii) endothelial CD146 showed an identical expression pattern among different vascular segments, including capillaries, precapillary arterioles, and postcapillary venules, being strongly expressed in capillary ECs without pericyte coverage but extremely weak in the ECs of precapillary arterioles and postcapillary venules as a result of near-complete pericyte ensheathment; and (iv) this expression pattern of CD146 could be observed in CNS microvessels at different stages during BBB development, including E11, E18, in P5, and in 4–6-wk-old mice.
As the expression profiles could be indicative of function, we hypothesized that CD146 played distinct roles in the stages of BBB development. Previous studies demonstrated that CD146 is a component of the endothelial junctions responsible for the integrity of peripheral ECs in vitro (22–24, 34, 35). The present findings confirmed that CD146 is critical for BBB induction and promotes BEC integrity in vitro and in vivo. Furthermore, we proved that endothelial expression of CD146 could up-regulate claudin-5 as a downstream signaling mediator of this process at the stage of BBB induction.
Compared with the Cd146EC-KO mice, Cd146-KO mice displayed a reduction in pericyte coverage and abnormal TJ structures, suggesting that the role of CD146 in BBB development is not limited to BBB induction by regulating claudin-5 expression in BECs, but is also responsible for sufficient pericyte recruitment and BBB maturation. Consistent with a recent report showing the requirement of pericytes for TJ formation (15), we observed that genetic deletion of Cd146 in pericytes results in insufficient pericyte coverage, abnormal TJs, and impaired BBB function. This was further supported by the results of the in vitro cell recruitment assay, demonstrating an important role of CD146 in pericyte recruitment to BECs.
The present study also revealed that CD146 participates in pericyte recruitment through functioning as a coreceptor of PDGFRβ. PDGF-B/PDGFRβ signaling is known as the most important pathway for promoting CNS pericyte recruitment to BECs (14). Homozygous deletion of Pdgfb or Pdgfrb results in a lack of CNS pericytes and BBB dysfunction (32). Here, we demonstrated that CD146 knockdown or disruption of the CD146/PDGFRβ interaction impaired PDGF-B–induced PDGFRβ activation, highlighting the critical role of CD146 as a coreceptor for PDGFRβ in pericyte recruitment.
It is now well established that the activation of receptor tyrosine kinase (RTK) is a complex process whereby receptors are generally associated with several proteins to fulfill efficient receptor activation initiated by ligand engagement (36). Several studies have shown that the cooperation of PDGFRβ with other membrane proteins such as CD44, low-density lipoprotein receptor-related protein, and cell surface tissue transglutaminase facilitates receptor activation by modulating kinase activity (37–39). Our observation that CD146 acts as a coreceptor for PDGFRβ not only sheds light on the mechanism of PDGFRβ activation but also provides evidence that cell adhesion molecules function as coreceptors for RTKs (36).
Furthermore, the present results demonstrate that EC-specific CD146 expression is correlated with BBB development. Following the successful pericyte recruitment and attachment at the stage of BBB maturation, the endothelial expression of CD146 was found to be down-regulated by pericyte-derived TGF-β1 in BECs covered with pericytes. Moreover, the ratio between CD146− and CD146+ BECs was approximately 2:1 in the mature BBB, which is in agreement with previous studies showing that the ratio between BECs and pericytes in the brain vasculature is 1:1–3:1 (40). By using a coculture model, we found that pericytes gradually decreased the expression level of CD146 in bEND.3 cells along with pericyte recruitment. In the absence of CD146 in BECs covered with pericytes, pericytes recruited by the CD146/PDGFRβ axis during BBB maturation might be able to maintain the structure of TJs. Our findings also provided evidence that CD146 expression in bEND.3 cells is regulated by TGF-β1 secreted by adjacent pericytes. This result further supports earlier findings that the interaction between BECs and pericytes is mainly mediated by TGF-β1 in the BBB maturation phase (32).
The present findings have important implications for understanding the roles of CD146 in BBB maturation, during which pericytes suppress expression of LAMs in BECs to reduce extravasation of inflammatory cells to the CNS (7). In the present study, we found that adhesion of immune cells to hCMEC/D3 cells was reduced after knockdown of endothelial CD146, suggesting that the down-regulation of endothelial CD146 might be important for maintaining the CNS immune system in a quiescent state during BBB development, a critical feature of the mature BBB. Consistent with this, previous studies from our laboratory and others demonstrated that CD146 is significantly elevated in BECs to promote neuroinflammation under several pathological conditions (19, 41). The expression variations of CD146 in the BBB of the normal cerebrovasculature and neurological disorders further indicate that CD146 is a key regulator of CNS immune surveillance. It could act as an important clinical diagnostic factor and as a potential therapeutic target in multiple sclerosis and other neuroinflammatory diseases.
In conclusion, the present work demonstrates that a single cell adhesion receptor, CD146, acts as an essential regulator to coordinate BBB formation during embryogenesis. This finding not only provides important insights into the molecular mechanisms governing the interaction between BECs and pericytes to modulate BBB development, but also highlights the possibility for targeting CD146 as a feasible therapeutic strategy for dampening neuroinflammation associated with BBB breakdown.
Materials and Methods
Antibodies and Reagents.
Anti-CD146 antibodies, including mouse anti-CD146 AA98 and AA4, have been described previously (42). Other antibodies used in this study are listed in Table S2.
Table S2.
Antibodies used in this study
| Antibodies | Cat. no. |
| From Abcam | |
| Anti-GAPDH | ab8245 |
| Anti-PDGFRβ | ab32570 |
| Anti-desmin | ab15200 |
| Anti-αSMA | ab5694 |
| Anti-CD31 | ab56299 |
| Anti-GFAP | ab7260 |
| Anti–β-actin | ab8226 |
| Anti-His | ab18184 |
| From Cell Signaling Technology | |
| Anti–p-PDGFRβ | 3161 |
| From Invitrogen | |
| Anti-ZO-1 | 40–2200 |
| Anti-claudin-5 | 35–2500 |
| Anti-occludin | 71–1500 |
| Alexa Fluor 488 goat anti-rat IgG | A11006 |
| Alexa Fluor 555 goat anti-mouse IgG | A21422 |
| Alexa Fluor 647 goat anti-rabbit IgG | A21244 |
| From R&D Systems | |
| Anti–TGF-β1 | MAB240 |
| From Biolegend | |
| PE-conjugated anti-mouse CD31 | 102407 |
| PE/Cy7 anti-mouse CD146 | 134713 |
Recombinant Fc (cat. no. 10702-HNAH), CD146-Fc (cat. no. 10115-H02H), and PDGFRβ (cat. no. 10514-H08H) were obtained from Sino Biological. Recombined TGF-β1 (cat. no. 100–21) and PDGF-B (cat. no. 500-P47) were from Peprotech. Angiopoietin-1 (cat. no. 923-AN-025) was from R&D Systems.
Mice.
Constitutive Cd146-KO mice (Cd146−/−), EC-specific Cd146-KO mice (Cd146EC-KO), and pericyte-specific Cd146-KO mice (i.e., Cd146PC-KO) were used in this study. All KO mice were generated by using a Cre/loxP recombination system as described in SI Materials and Methods. All mice were maintained in a pathogen-free facility. All animal experiments were performed in compliance with the guidelines for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of the Institute of Biophysics of the Chinese Academy of Sciences.
Immunofluorescence Staining of Mouse Brains.
Mouse brains were fixed in 4% PFA overnight, dehydrated, and embedded in optimal cutting temperature compound. Tissue slices (40 µm thick) were blocked in PBS solution with 1% BSA and 1% Triton X-100 for 4–6 h, then with primary antibody at room temperature overnight with agitation, and then for 8 h with secondary antibody. Between each incubation step, slides were washed with PBS solution three times for 5 min. Confocal images were obtained with a Zeiss LSM 880 imaging system under a 63× water-immersion objective.
Cerebral capillaries, precapillary arterioles, and postcapillary venules were identified according to features established in earlier publications (5, 43, 44) and briefly as follows: (i) αSMA-negative vessels with a vascular lumen diameter < 10 μm were identified as capillaries; (ii) αSMA-positive vessels terminating before αSMA-negative capillaries were identified as precapillary arterioles; and (iii) αSMA-negative vessels with a vascular lumen diameter 10–20 μm were identified as postcapillary venules.
Immunolabeling and Clearing of Mouse Brains.
Immunolabeling was performed as previously described (45). For the tissue clearing before imaging, immunolabeled brain samples were dehydrated in methanol ranging from 25% to 100% (in PBS solution) for 3 h with a step of 25% at room temperature. Samples were then cleared with BABB solution (benzyl alcohol and benzyl benzoate in a 1:2 volume ratio) overnight at room temperature (46). Images were taken with light-sheet fluorescence microscopy (LSFM; LaVision BioTec).
Image Acquisition and Analysis of LSFM Data.
The immunolabeled and cleared brains were imaged on a commercial light-sheet fluorescence microscope with a 5.0× magnification, a 2× objective lens (Mv PLAPO2VC; Olympus) and a 6-mm working distance dipping cap. This configuration led to a pixel size of 0.65 μm. We used a super continuum white light laser (SuperKEXTREME 80 mHz VIS; NKT Photonics) with a wavelength spectrum ranging from 400 to 2,400 nm. For the detection of brain vessels, the filters were set as follows: Alexa Fluor 488, excitation 500/20 nm; emission 535/30 nm; Alexa Fluor 555, excitation 577/25 nm; emission 632/60 nm. The step size was set to 2 μm for z-stacks scanning and a total scan range of the brain sample up to 1 mm. The measurements were performed with exposure times of 385 ms per slice, resulting in a total acquisition time of ∼2 min per brain resection sample, i.e., cortex, hippocampus, cerebellum, and olfactory bulb. The images in TIFF format were further postprocessed in the ImageJ package FIJI, version1.51 (fiji.sc/Fiji). Maximum-intensity projection (MIP) of 40 virtual single slices was selected for analysis.
In Vitro Pull-Down Assays.
The pull-down assays were performed as previously described (further details provided in SI Materials and Methods ) (47).
Coimmunoprecipitation.
Coimmunoprecipitation was performed as previously described (further details provided in SI Materials and Methods) (18).
Cell Recruitment Assay.
The cell recruitment assay was performed as previously described (48). bEND.3 cells (3 × 105 cells per milliliter) and pericytes (1.5 × 105 cells per milliliter), labeled with PKH26 and CFSE, respectively, were cocultured in Matrigel-coated culture slides. At 6 h after stimulation with PDGF-B (20 ng/mL), images were taken on a confocal laser scanning microscope (Fluoview FV 1000; Olympus) with an IX81 digital camera (Olympus). Quantification was made by counting the number of bEND.3-pericyte attachments from fifteen fields from three independent experiments.
Isolation and Culture of Mouse Brain Pericytes and Capillary ECs.
The isolation and culture of mouse brain pericytes and capillary ECs are described in SI Materials and Methods.
EM.
P5 mice were euthanized, and the left ventricle was perfused with 2.5% glutaraldehyde and 4% PFA. Subsequently, brains were extracted and cut into small pieces (< 1 mm3). Tissues were fixed in 2.5% glutaraldehyde overnight at 4 °C and then processed for EM.
Evans Blue (Albumin) Permeability.
Mice at P5 or 4–6 wk were given an i.p. injection or i.v. injection of Evans blue dye, respectively. The mice were euthanized and perfused with PBS solution through the left ventricle. Brains were then extracted and dissolved in formamide. After extraction in formamide for 48 h at 55 °C, the absorption (at 620 nm) of the Evans blue extracted from the brain was measured by using a microplate spectrophotometer.
Brain Water Content.
Brain water content was evaluated to assess the development of edema as described previously (49) and detailed in SI Materials and Methods.
Assessment of TEER.
An xCELLigence System RTCA-SP instrument (ACEA Biosciences) was used to measure the paracellular tightness of the BEC monolayer. A total of 20,000 BECs were grown in the 96-well E-plate (ACEA Biosciences) and the paracellular tightness was examined by using the RTCA-SP instrument. The TEER, presented in units of ohms per square centimeter, was taken automatically every 2 min and analyzed as paracellular tightness.
Real-Time PCR.
Real-time PCR was performed as previously described (detailed in SI Materials and Methods) (50).
Cell Proliferation and Migration Assays.
Cell proliferation and migration assays are described in SI Materials and Methods.
Statistical Analysis.
All experiments were performed independently at least three times. The results are shown as the mean ± SEM. One or two-way ANOVA was used to compare differences between groups in the different experiments. Differences with a P value < 0.05 were considered statistically significant.
SI Materials and Methods
Mice.
WT C57BL/6J mice were obtained from the Department of Laboratory Animal Science of Peking University Health Science Center. All KO mice were generated by using a Cre/loxP recombination system. Cd146EC-KO mice were generated through the Tg (Tek-cre) system as described previously (31). To generate Cd146−/− mice, EIIacre/+ mice [B6; FVB-TgN (EIIa-Cre) C5379Lmgd], which carry a Cre transgene under the control of the adenovirus EIIa promoter that targets expression of Cre recombinase to the early mouse embryo (51, 52), were obtained from the Animal Model Research Center at Nanjing University. These mice were then crossed with Cd146floxed/floxed mice (obtained from the Animal Model Research Center at Nanjing University, backcrossed onto a C57BL/6J background for a minimum of nine generations) to obtain EIIaCre/+Cd146+/− mice. The EIIaCre/+Cd146+/− mice were crossed with C57BL/6J mice to obtain the Cd146+/− mice. The Cd146+/− mice were intercrossed to obtain Cd146−/− mice.
For the generation of Cd146PC-KO (Pdgfrbcre/+Cd146floxed/floxed) mice, Pdgfrbcre/+Cd146+/+ mice (obtained from Beijing Biocytogen) were first crossed with Pdgfrb+/+Cd146floxed/floxed mice. The Pdgfrbcre/+Cd146floxed/+ mice were then backcrossed with Pdgfrb+/+Cd146floxed/floxed mice to obtain Pdgfrbcre/+Cd146floxed/floxed mice, which were annotated Cd146PC-KO mice. Pdgfrb+/+Cd146floxed/floxed mice (annotated as Cd146WT mice here) were used as controls. All genotypes were confirmed by PCR analysis and identified by sequencing. Cd146 deficiency in brain pericytes was confirmed by using Western blotting and flow cytometry analyses (Fig. S6).
In Vitro Pull-Down Assays.
A total of 200 ng Fc or Fc-CD146 was incubated with 200 ng PDGFRβ for 2 h at 4 °C in Hepes buffer. The beads were then incubated with proteins for 1 h at 4 °C. The bound proteins were washed three times with Hepes buffer and analyzed by Western blotting.
Coimmunoprecipitation.
Cells were washed three times with ice-cold PBS solution before being lysed in 500 µL ice-cold RIPA buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM phenylmethanesulfonyl fluoride, and protease inhibitor mixtures) for 40 min. Supernatants were then incubated with antibodies at 4 °C overnight. The complexes were precipitated with protein G-Sepharose (Santa Cruz Biotechnology). The immunoprecipitates were extensively washed with washing buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 0.1% SDS, 0.5% deoxycholate, 0.1% Nonidet P-40, 1 mM phenylmethanesulfonyl fluoride, and protease inhibitor mixtures) and analyzed by Western blotting by using the appropriate antibodies as indicated.
Oligonucleotides and Transfection.
CD146-specific siRNA was synthesized by Invitrogen using sequences as previously described (18, 53). Scrambled nontargeting control siRNA was used as a negative control. Lipofectamine 2000-mediated transfection was used according to the manufacturer’s instructions. To down-regulate CD146 expression, 50 nM siRNA targeting CD146 was transfected into hCMEC/D3 cells. The cotransfection of CD146-siRNA (50 nM) and CD146 expression plasmid (pcDNA3.1-CD146 plasmid; 2 μg) was performed to restore CD146 expression. The empty vector (pcDNA3.1-empty) was used as an internal control.
Isolation and Culture of Mouse Brain Pericytes.
Mice 6–8 wk of age were euthanized shortly before brain extraction, and brains were placed upside-down in cold DMEM. Subsequently, brains were diced with a sterilized razor blade, and the minced brain tissue was aspirated by using a 10-mL pipette into a 15-mL universal tube. The mixture was centrifuged at 290 × g for 5 min and then digested with 0.1% collagenase IV and 30 U/mL DNase I for 90 min at 37 °C. Approximately 7 mL of 22% BSA was added to the digested brain tissue, and the mix was centrifuged at 1,360 × g for 10 min. The myelin layer on top of the vial was carefully removed, and the vascular tubes as well as the cell pellets were washed two times with 2 mL EC growth medium (ECGM) and centrifuged at 290 × g for 5 min. Cell pellets were resuspended in 6 mL ECGM and plated into three wells of a six-well plate coated with collagen type I. After the cells reached confluence, the cultures were harvested with trypsin and passaged twice. Pericyte growth medium was then used for culturing of brain pericytes.
Isolation and Culture of Mouse Brain Capillary ECs.
Mice 6–8 wk of age were euthanized, and brains were extracted. The midbrain, cerebellum, and olfactory bulb were removed. To remove the meninges, brains were gently rolled along a piece of filter paper placed in a glass Petri dish and cooled by ice. The visible white matter was removed with forceps, and the cleaned cortex was added into 0.5 mL DMEM per brain. The brains were then diced with a sterilized razor blade, and the minced brain tissue was digested with 0.1%/1 mg/mL collagenase II and 30 U/mL DNase I for 90 min at 37 °C. The mixture was then centrifuged at 205 × g for 5 min and washed with DMEM. Next, 25% BSA was added and centrifuged at 1,360 × g for 10 min. The myelin on top of the vial was carefully removed, and the vascular tubes and cell pellets were washed with DMEM and centrifuged at 205 × g for 5 min. The capillaries were resuspended in 1 mL per brain DMEM and digested with 200 mg/mL Dispase and 0.1%/1 mg/mL collagenase II for 1 h at 37 °C. After spinning for 10 min to remove the enzymes, cell suspensions were plated onto six-well plates coated with rat tail collagen type I. EBM2 media was used to culture the cells. After the cells reached confluence, the primary cells were digested and sorted by FACS with the labeling of anti-CD31 for further experimental procedures. In parallel, the cerebral capillaries isolated from these mice and the primary cells raised from the capillaries were stained with TJ markers (claudin-5 and ZO-1) to identify their features of the BBB.
Tracer Experiments.
Permeability studies to tracers of different sizes were performed in 96-well culture plates with 0.4-µm transwell culture inserts by using hCMEC/D3 cells or primary BECs purified from Cd146WT or Cd146EC-KO mice. Cells were cultured in the upper chamber of the transwell culture insert for 48 h. Tracers of different sizes including dextran-70kD-FITC (1 mg/mL), dextran-4kD-FITC (1 mg/mL), cadaverine-Alexa 488 (0.2 mg/mL), and cadaverine-Alexa 555 (0.2 mg/mL) were then added to the upper chamber of the transwell culture insert at 100 µL per well, respectively, and medium without tracers was added to the bottom chamber. The plate was incubated for 1 h at 37 °C. Subsequently, solution from the bottom chamber (200 µL) was transferred into a 96-well plate, and absorption spectra of dye solutions were measured with a spectrophotometer.
Brain Water Content.
Briefly, mice were killed and brains were extracted. The wet weight of brain tissues was determined immediately. After drying for 24 h at 100 °C, the dry weight of brains was measured. Percentage water content was determined by the following formula: water content (%) = (wet weight − dry weight) × 100/wet weight.
MRI Experiments.
MRI data were acquired by using a Biospec 94/30 preclinical system (Bruker) operating at 400 MHz (9.4 T) and Paravision 6 (Bruker BioSpin). A T2-weighted anatomical scan was acquired using a spin-echo [turbo rapid acquisition with relaxation enhancement (RARE)] sequence with the following parameters: field of view (FOV), 20 × 20 mm2; matrix dimension (MD), 133 × 133; slice thickness, 0.15 mm; repetition time (TR), 10,884 ms; echo time (TE), 33 ms; flip angle (FA), 90°; bandwidth, 261 kHz; RARE factor, 8; and number of averages (NEX), 3. The reconstructed images have an isotropic voxel size of 150 μm. Another T2-weighted image with high in-plane resolution (0.083 × 0.083 mm2) but large slice thickness (0.3 mm) was acquired for the purpose of visual inspection. Other parameters included FOV, 20 × 20 mm2; MD, 244 × 244; TR, 5,000 ms; TE, 30 ms; FA, 90°; bandwidth, 145 kHz; RARE factor, 8; and NEX, 3. The imaging plane was coronal.
Ventricle Volume Measurement.
The DICOM images from the scanner were converted to NIFTI file format for further analysis. All images were aligned to a template in standard space (54) by using rigid registration. Then, on the T2 images with isotropic voxel size, the DARTEL toolbox in SPM (55) was used to segment the brain into gray matter, white matter, and cerebrospinal fluid (CSF), as well as to create a study-specific mouse brain template. Ventricles, consisting of left/right lateral ventricles, the third and fourth ventricles, were manually segmented on the template of CSF. By using this manual segmentation as masks, the ventricle volumes were calculated from the modulated CSF image of each individual.
Real-Time PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen). RNA (2 μg) was reverse-transcribed into cDNA by random primers. Real-time PCR analysis was performed on a Corbett 6200 device using SYBR Green PCR mix (Toyobo). Threshold cycle (Ct) values of GAPDH were subtracted from Ct values of the genes of interest (ΔCt). All primers used were synthesized by Invitrogen by using the sequences listed in Table S3.
Table S3.
Real-time PCR primer sequences used in this study
| Target gene | Sequence (5′-3′) |
| h-CD146 | AACACAGTGGGCGCTATGAA |
| AACTCGAGGTCCTGGCTACT | |
| h-ZO-1 | ACCGGAGTCTGCCATTACAC |
| AACGTTGAATGCTTGCTGCT | |
| h-CLAUDIN-5 | TCTCTCCTGTCTGAAGGCCA |
| GGCTTCCCAGACCTCTCAATC | |
| h-GAPDH | CTCTGCTCCTCCTGTTCGAC |
| TGAAGGGGTCATTGATGGCA | |
| m-Cd146 | CGGGTGTGCCAGGAGAG |
| GGCGGTGCTCATATTCACCA | |
| m-Claudin-5 | CTTTGTTACCTTGACCGGCG |
| TACTTCTGTGACACCGGCAC | |
| m-Zo-1 | CGCGGAGAGAGACAAGATGT |
| CTTCAGCTGGCCCTCCTTTT | |
| m-Occludin | CCGGCCGCCAAGGTTC |
| CATGCATCTCTCCGCCATAC | |
| m-Gapdh | GGTTGTCTCCTGCGACTTCA |
| TAGGGCCTCTCTTGCTCAGT |
Cell Proliferation and Migration Assays.
PDGF-B–induced pericyte proliferation was performed by using the CCK-8 Cell Counting Kit according to the manufacturer’s instructions. Briefly, pericytes (3,000 cells) were seeded into a 96-well plate. Anti-CD146 AA98 or mouse IgG (50 μg/mL) was added to the plate. At 48 h after stimulation with PDGF-B (20 ng/mL), the relative cell number was determined by measuring the optical density of CCK-8 at 450 nm.
Cell-migration experiments were assessed by using a 96-well Boyden chamber (Corning Costar) containing a filter with a pore size of 8 μm. Pericytes (6,000 cells) were grown in DMEM (with 1% FBS) in the upper chamber. Mouse IgG or anti-CD146 AA98 (50 μg/mL) was added during migration, and PDGF-B (20 ng/mL) was added to the upper chamber for stimulation. Cell migration was initiated by filling the lower chamber with DMEM (with 10% FBS) and incubating for 12 h. The cells migrating to the lower membrane were stained with crystal violet and counted by using a microscope.
Cell Lysis and Protein Extraction.
Cells were washed with cold PBS solution twice and harvested by scraping in ice-cold lysis buffer [8 M urea, 100 mM Tris⋅HCl, pH 8.5, and protease inhibitor mixture (Roche)]. Cells were sonicated on ice, and cell lysates were centrifuged at 16,000 × g for 15 min at 4 °C to remove cell debris. Supernatants were collected and stored at −80 °C until use. Cell lysates were denatured according to the filter assistant sample preparation (FASP) procedure with minor modifications. Briefly, cell lysates were transferred to Amicon 0.5 ultra filtration units, and the buffer was exchanged with UA buffer (8 M urea, 0.1 M Tris, pH 8.5) three times. Protein samples were reduced with 10 mM DTT at 37 °C for 1 h and then alkylated with 50 mM iodoacetamide at room temperature for 1 h (in the dark). The buffer of the denatured protein samples was then exchanged with UA buffer three times and exchanged with 50 mM tetraethylammonium bromide (TEAB) twice. Proteins were recovered by reverse centrifugation at 1,000 × g for 2 min and resolved in 50 mM TEAB. Protein concentrations were determined in triplicate by using the bicinchoninic acid method (Thermo Scientific Pierce) according to the manufacturer’s instructions.
Protein Digestion and iTRAQ Labeling.
A total of 100 µg of lysate proteins from siRNA-treated bEND.3 cells and controls were digested with trypsin at 37 °C overnight. The tryptic peptides were then labeled with TMT reagent according to the manufacturer’s instructions. The siRNA-treated bEND.3 cells and controls were labeled as follows: controls with 126 tag and 129 tag (technical replicate), and siRNA treated with 127 tag and 130 tag (technical replication). Following labeling, the four tagged peptide samples were pooled and stored at −80 °C until use for MS analysis.
Peptide Fractionation Using High-pH Reversed-Phase Liquid Chromatography.
Following desalting using Sep-Pak Vac 1-cm3 (100 mg) C18 SPE cartridges (Waters), labeled peptides were fractionated by using YMC-Triart C18 basic reversed-phase liquid chromatography columns (250 × 4.6 mm, 5 μm particles; cat. no. TA12S05-2546WT; YMC). Briefly, peptides were separated in a binary buffer system of 2% ACN, pH 10 (pH adjusted with ammonium hydroxide, solution A), and 98% ACN, pH 10 (pH adjusted with ammonium hydroxide, solution B) in an Ultimate-3000 LC system (Thermo Scientific). The gradient of buffer B was set as follows: 0–5% for 1 min, 5–8% for 5 min, 8–18% for 35 min, 18–32% for 22 min, 32–95% for 2 min, and 95% for 7 min. Eluates were collected every 75 s, and a total number of 57 fractions were obtained. The first five fractions were discarded because most of the excess labeling reagent was eluted in these fractions. The remaining 52 fractions were combined into 13 fractions by merging fractions 1, 14, 27, and 40; fractions 2, 15, 28, and 41; and so on. Then, all fractions were dried in a vacuum concentrator and stored at −20 °C until further analysis.
Two-Dimensional LC-MS/MS Analysis.
All MS data were collected on a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled with an Easy-nLC system (Thermo Fisher Scientific). Each fraction of tryptic peptides was resuspended in 0.1% formic acid and separated on EASY-Spray column (C18; 2-μm particle size, 100-Å pore size, 75-μm i.d. × 50-cm length; Thermo Fisher Scientific). Samples were resolved with a linear gradient of solvent B (100% ACN, 0.1% formic acid); 5–50% over 76 min and 50–90% over 12 min at a flow rate of 300 nL/min. The separated peptide ions eluted from the analytic column were applied by mass spectrometer at an electrospray voltage of 2.1 kV. MS/MS spectra were acquired in a data-dependent mode for fragmentation of the 10 most abundant peaks from the full MS scan with 30% normalized collision energy. The dynamic exclusion duration was set at 20 s, and the isolation mass width was 2.5 Da. MS spectra were acquired within a mass range of 400–1,800 m/z and 70,000 resolution at m/z 200. MS/MS resolution was acquired at a resolution of 17,500.
Protein Identification and Quantification.
The LC-MS/MS data for the identification and quantification of the global proteome were analyzed by using Proteome Discoverer (version 1.4; Thermo Scientific) against the Uniprot KB mouse database (released September 2013) with 50,287 entries, to which 175 commonly observed contaminants and all of the reverse sequences were added. Search parameters included precursor ions mass tolerance of 10 ppm (monoisotopic mass), fragment ions mass tolerance of 0.02 Da (monoisotopic mass), fixed modifications of cysteine carbamidomethylation and TMTsixplex labeling at the N terminus and lysine residues, variable modification of addition of 15.999 Da on methionine (oxidation). The search results were passed through additional filters before exporting the data. For protein identification, the filters were set as follows: peptides with cutoff false discovery rate of 0.01 and significance threshold P < 0.05 (with 95% confidence). For protein quantification, filters were set as follows: normalize with protein median; only unique peptides were used to quantify proteins.
Acknowledgments
We thank Dr. Torsten Juelich, Dr. N. Joan Abbott, Dr. Yun Zhao, Dr. Qi Xie, Dr. Laixin Xia, Dr. Chen Ni, and Dr. Zhihai Qin for careful reading and editing of the manuscript; Dr. Yanxia Jia, Junfeng Hao, Jifeng Wang, Xuehui Chen, Yan Teng, Yun Feng, Junying Jia, Zhen Xiong, Zhenzhen Wu, Chaoen Hu, Sai Yang, Chengpeng Fu, and Xinyi Wu for technical support; and the Chinese Academy of Sciences-Institute of Automation Center for Advanced Imaging for MRI experiments and data analysis. This work was supported by National Basic Research Program of China 973 Programs 2015CB553705 and 2014CBA02003; National Natural Science Foundation of China Grants 31370824, 81371025, 91529306, 81371330, 91640112, 31670185, 81527805, 81671851, and 81502547; Strategic Priority Research Program of the Chinese Academy of Sciences (Grants XDA12020207 and XDA12030202); Guangdong Natural Science Fund for Distinguished Young Scholars, China (2017); and Novo Nordisk-Chinese Academy of Sciences Research Fund Grant NNCAS-2015-11.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710848114/-/DCSupplemental.
References
- 1.Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Establishment and Dysfunction of the blood-brain barrier. Cell. 2015;163:1064–1078. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greene C, Campbell M. Tight junction modulation of the blood brain barrier: CNS delivery of small molecules. Tissue Barriers. 2016;4:e1138017. doi: 10.1080/21688370.2015.1138017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelilah-Seyfried S. Claudin-5a in developing zebrafish brain barriers: Another brick in the wall. BioEssays. 2010;32:768–776. doi: 10.1002/bies.201000045. [DOI] [PubMed] [Google Scholar]
- 4.Haseloff RF, Dithmer S, Winkler L, Wolburg H, Blasig IE. Transmembrane proteins of the tight junctions at the blood-brain barrier: Structural and functional aspects. Semin Cell Dev Biol. 2015;38:16–25. doi: 10.1016/j.semcdb.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Paul D, Cowan AE, Ge S, Pachter JS. Novel 3D analysis of Claudin-5 reveals significant endothelial heterogeneity among CNS microvessels. Microvasc Res. 2013;86:1–10. doi: 10.1016/j.mvr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. 2013;19:1584–1596. doi: 10.1038/nm.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegenthaler JA, Sohet F, Daneman R. ‘Sealing off the CNS’: Cellular and molecular regulation of blood-brain barriergenesis. Curr Opin Neurobiol. 2013;23:1057–1064. doi: 10.1016/j.conb.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott NJ, Friedman A. Overview and introduction: The blood-brain barrier in health and disease. Epilepsia. 2012;53:1–6. doi: 10.1111/j.1528-1167.2012.03696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange C, Storkebaum E, de Almodóvar CR, Dewerchin M, Carmeliet P. Vascular endothelial growth factor: A neurovascular target in neurological diseases. Nat Rev Neurol. 2016;12:439–454. doi: 10.1038/nrneurol.2016.88. [DOI] [PubMed] [Google Scholar]
- 10.Daneman R, et al. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci USA. 2009;106:641–646. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen M, et al. GPR124, an orphan G protein-coupled receptor, is required for CNS-specific vascularization and establishment of the blood-brain barrier. Proc Natl Acad Sci USA. 2011;108:5759–5764. doi: 10.1073/pnas.1017192108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posokhova E, et al. GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Reports. 2015;10:123–130. doi: 10.1016/j.celrep.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armulik A, Genové G, Betsholtz C. Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Sweeney MD, Ayyadurai S, Zlokovic BV. Pericytes of the neurovascular unit: Key functions and signaling pathways. Nat Neurosci. 2016;19:771–783. doi: 10.1038/nn.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelhardt B, Liebner S. Novel insights into the development and maintenance of the blood-brain barrier. Cell Tissue Res. 2014;355:687–699. doi: 10.1007/s00441-014-1811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7:a020412. doi: 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang T, et al. CD146 is a coreceptor for VEGFR-2 in tumor angiogenesis. Blood. 2012;120:2330–2339. doi: 10.1182/blood-2012-01-406108. [DOI] [PubMed] [Google Scholar]
- 19.Duan H, et al. Targeting endothelial CD146 attenuates neuroinflammation by limiting lymphocyte extravasation to the CNS. Sci Rep. 2013;3:1687. doi: 10.1038/srep01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malpass K. Disease mechanisms in MS: Cell adhesion molecule MCAM on pathogenic T cells-a green light for CNS entry in multiple sclerosis. Nat Rev Neurol. 2012;8:592. doi: 10.1038/nrneurol.2012.204. [DOI] [PubMed] [Google Scholar]
- 21.Crisan M, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Bardin N, et al. Identification of CD146 as a component of the endothelial junction involved in the control of cell-cell cohesion. Blood. 2001;98:3677–3684. doi: 10.1182/blood.v98.13.3677. [DOI] [PubMed] [Google Scholar]
- 23.Bardin N, et al. CD146 and its soluble form regulate monocyte transendothelial migration. Arterioscler Thromb Vasc Biol. 2009;29:746–753. doi: 10.1161/ATVBAHA.108.183251. [DOI] [PubMed] [Google Scholar]
- 24.Kebir A, et al. CD146 short isoform increases the proangiogenic potential of endothelial progenitor cells in vitro and in vivo. Circ Res. 2010;107:66–75. doi: 10.1161/CIRCRESAHA.109.213827. [DOI] [PubMed] [Google Scholar]
- 25.Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Cristante E, et al. Identification of an essential endogenous regulator of blood-brain barrier integrity, and its pathological and therapeutic implications. Proc Natl Acad Sci USA. 2013;110:832–841. doi: 10.1073/pnas.1209362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilhelm I, Nyúl-Tóth Á, Suciu M, Hermenean A, Krizbai IA. Heterogeneity of the blood-brain barrier. Tissue Barriers. 2016;4:e1143544. doi: 10.1080/21688370.2016.1143544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge S, Song L, Pachter JS. Where is the blood-brain barrier ... really? J Neurosci Res. 2005;79:421–427. doi: 10.1002/jnr.20313. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Zvi A, et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weksler B, Romero IA, Couraud PO. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. 2013;10:16. doi: 10.1186/2045-8118-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Q, et al. Impaired tumor angiogenesis and VEGF-induced pathway in endothelial CD146 knockout mice. Protein Cell. 2014;5:445–456. doi: 10.1007/s13238-014-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler EA, Bell RD, Zlokovic BV. Central nervous system pericytes in health and disease. Nat Neurosci. 2011;14:1398–1405. doi: 10.1038/nn.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013;330:150–162. doi: 10.1016/j.canlet.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 34.Solovey AN, et al. Identification and functional assessment of endothelial P1H12. J Lab Clin Med. 2001;138:322–331. doi: 10.1067/mlc.2001.118519. [DOI] [PubMed] [Google Scholar]
- 35.Boneberg EM, Illges H, Legler DF, Fürstenberger G. Soluble CD146 is generated by ectodomain shedding of membrane CD146 in a calcium-induced, matrix metalloprotease-dependent process. Microvasc Res. 2009;78:325–331. doi: 10.1016/j.mvr.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Ponta H, Sherman L, Herrlich PA. CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 37.Zemskov EA, et al. Regulation of platelet-derived growth factor receptor function by integrin-associated cell surface transglutaminase. J Biol Chem. 2009;284:16693–16703. doi: 10.1074/jbc.M109.010769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loukinova E, et al. Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP). Evidence for integrated co-receptor function betwenn LRP and the PDGF. J Biol Chem. 2002;277:15499–15506. doi: 10.1074/jbc.M200427200. [DOI] [PubMed] [Google Scholar]
- 39.Porsch H, Mehić M, Olofsson B, Heldin P, Heldin CH. Platelet-derived growth factor β-receptor, transforming growth factor β type I receptor, and CD44 protein modulate each other’s signaling and stability. J Biol Chem. 2014;289:19747–19757. doi: 10.1074/jbc.M114.547273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathiisen TM, Lehre KP, Danbolt NC, Ottersen OP. The perivascular astroglial sheath provides a complete covering of the brain microvessels: An electron microscopic 3D reconstruction. Glia. 2010;58:1094–1103. doi: 10.1002/glia.20990. [DOI] [PubMed] [Google Scholar]
- 41.Brucklacher-Waldert V, Stuerner K, Kolster M, Wolthausen J, Tolosa E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain. 2009;132:3329–3341. doi: 10.1093/brain/awp289. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Generation and characterization of a panel of monoclonal antibodies against distinct epitopes of human CD146. Hybridoma (Larchmt) 2008;27:345–352. doi: 10.1089/hyb.2008.0034. [DOI] [PubMed] [Google Scholar]
- 43.Hartmann DA, et al. Pericyte structure and distribution in the cerebral cortex revealed by high-resolution imaging of transgenic mice. Neurophotonics. 2015;2:041402. doi: 10.1117/1.NPh.2.4.041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Attwell D, Mishra A, Hall CN, O’Farrell FM, Dalkara T. What is a pericyte? J Cereb Blood Flow Metab. 2016;36:451–455. doi: 10.1177/0271678X15610340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renier N, et al. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Sharpe J, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 47.Luo Y, et al. Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene. 2012;31:306–321. doi: 10.1038/onc.2011.244. [DOI] [PubMed] [Google Scholar]
- 48.Song N, et al. Overexpression of platelet-derived growth factor-BB increases tumor pericyte content via stromal-derived factor-1alpha/CXCR4 axis. Cancer Res. 2009;69:6057–6064. doi: 10.1158/0008-5472.CAN-08-2007. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Rodriguez AB, Acaz-Fonseca E, Viveros MP, Garcia-Segura LM. Changes in cannabinoid receptors, aquaporin 4 and vimentin expression after traumatic brain injury in adolescent male mice. Association with edema and neurological deficit. PLoS One. 2015;10:e0128782. doi: 10.1371/journal.pone.0128782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing S, et al. Targeting endothelial CD146 attenuates colitis and prevents colitis-associated carcinogenesis. Am J Pathol. 2014;184:1604–1616. doi: 10.1016/j.ajpath.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 51.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdel-Wahab O, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng C, et al. Endothelial CD146 is required for in vitro tumor-induced angiogenesis: The role of a disulfide bond in signaling and dimerization. Int J Biochem Cell Biol. 2009;41:2163–2172. doi: 10.1016/j.biocel.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 54.Johnson GA, et al. Waxholm space: An image-based reference for coordinating mouse brain research. Neuroimage. 2010;53:365–372. doi: 10.1016/j.neuroimage.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]