Significance
Leukotriene B4 (LTB4) is a one of the most potent chemotactic agents known to date and participates in leukocyte recruitment during the innate immune response. Leukotriene A4 hydrolase/aminopeptidase (LTA4H) catalyzes the committed step in LTB4 biosynthesis. Here we report high-resolution crystal structures of LTA4H in complex with its highly labile substrate LTA4, which reveal the structural basis for the enzyme’s unique epoxide hydrolase mechanism. Moreover, we show that LTA4H undergoes domain movements, which gates the hydrophobic cavity for entrance of LTA4 followed by induced fit. Our results provide new insights to the mechanism of LTA4H and structure-based drug design.
Keywords: LTA4 hydrolase, LTB4, innate immunity, drug design, X-ray crystallography
Abstract
Human leukotriene (LT) A4 hydrolase/aminopeptidase (LTA4H) is a bifunctional enzyme that converts the highly unstable epoxide intermediate LTA4 into LTB4, a potent leukocyte activating agent, while the aminopeptidase activity cleaves and inactivates the chemotactic tripeptide Pro-Gly-Pro. Here, we describe high-resolution crystal structures of LTA4H complexed with LTA4, providing the structural underpinnings of the enzyme’s unique epoxide hydrolase (EH) activity, involving Zn2+, Y383, E271, D375, and two catalytic waters. The structures reveal that a single catalytic water is involved in both catalytic activities of LTA4H, alternating between epoxide ring opening and peptide bond hydrolysis, assisted by E271 and E296, respectively. Moreover, we have found two conformations of LTA4H, uncovering significant domain movements. The resulting structural alterations indicate that LTA4 entrance into the active site is a dynamic process that includes rearrangement of three moving domains to provide fast and efficient alignment and processing of the substrate. Thus, the movement of one dynamic domain widens the active site entrance, while another domain acts like a lid, opening and closing access to the hydrophobic tunnel, which accommodates the aliphatic tale of LTA4 during EH reaction. The enzyme–LTA4 complex structures and dynamic domain movements provide critical insights for development of drugs targeting LTA4H.
Leukotriene A4 hydrolase/aminopeptidase (LTA4H; EC 3.3.2.6) is a 69-kDa cytosolic bifunctional zinc metalloenzyme that converts the transient epoxide LTA4 (5S-5,6-oxido-7,9-trans-11,14-cis-eicosatetraenoic acid) into the dihydroxy acid LTB4 (5S,12R-dihydroxy-6,14-cis-8,10-trans-eicosatetraenoic acid) via a unique epoxide hydrolase (EH) reaction (1). LTB4, a potent chemotactic and immune-activating agent, is implicated in acute and chronic inflammatory diseases (2, 3), cardiovascular disorders (4, 5), and carcinogenesis (6). In addition to its proinflammatory EH activity, LTA4H may also counteract inflammation by its aminopeptidase activity, which inactivates by cleavage another neutrophil attractant, the tripeptide Pro-Gly-Pro (PGP) (7). In addition, the fatty acid epoxide LTA4 is also the key precursor for biosynthesis of lipoxins, a family of anti-inflammatory, proresolving lipid mediators (8). Hence, LTA4H is a checkpoint in lipid mediator biosynthesis that regulates both the initiation and resolution phases of inflammation.
LTA4H consists of N-terminal, catalytic, and C-terminal domains whose interface delimits a deep cleft harboring an l-shaped pocket, with a wide polar arm that binds PGP and a narrow hydrophobic arm that is believed to accommodate LTA4 (9). The elbow of the pocket includes H295, H299, and E318, which coordinate a Zn2+ ion essential for both LTA4H activities.
LTA4H is the only EH reported to form a nonvicinal distant 5,12 diol (10), requiring an atypical catalytic mechanism, the details of which have remained elusive in the absence of a crystal structure with bound substrate (11). Acquirement of an enzyme-substrate complex is complicated by the fact that LTA4 is a highly labile allylic epoxide with a half-life of about 10 s at neutral pH, physical properties that until now have precluded determination of its binding at the active site of LTA4H. In the present report, we describe successful trapping and structure determination of LTA4H-LTA4 complexes providing insights to the EH mechanism. Moreover, we show that LTA4H undergoes domain movements, with structural alterations indicating gated substrate entrance into the active site followed by induced fit to achieve optimal alignment and catalysis.
Results and Discussion
In this study, we describe six different structures of LTA4H from five distinct crystal forms. These visualize two conformational states of the enzyme (Tables S1 and S2). The mutants we investigated include variants E271A (12), D375N (13), and R563A (11), which lack EH activity. An orthorhombic structure of cocrystallized D375N, as well as orthorhombic and trigonal structures of soaked E271A, were obtained in a closed conformational state with captured LTA4 in the active site. In contrast, trigonal structures of both cocrystallized and apo D375N, as well as an orthorhombic structure of R563A, show an open conformation of LTA4H without bound ligand.
Table S1.
Structures of LTA4H mutants complexed with LTA4
| # | Mutants | PDB ID code | Space group | Protein–ligand complex formation | Ligand in the active site | Conformation of dynamic domains |
| 1 | D375N | 5NI6 | Orthorhombic | Cocryst | LTA4 | Closed |
| 2 | E271A | 5NI2 | Orthorhombic | Soaking | LTA4 | Closed |
| 3 | E271A | 5NI4 | Trigonal | Soaking | LTA4 | Closed |
Table S2.
Structures of LTA4H mutants in open conformation
| # | Mutants | PDB ID code | Space group | Protein–ligand complex formation | Ligand in the active site | Conformation of dynamic domains | State of hydrophobic tunnel |
| 1 | D375N | 5NIA | Trigonal | — | — | Open | Closed |
| 2 | D375N | 5NID | Trigonal | Cocryst | — | Open | Open |
| 3 | R563A | 5NIE | Orthorhombic | — | — | Open | 2 Open/1 Closed* |
The structure of R563A contains three molecules in the asymmetric unit.
LTA4 Binding and Turnover in the Active Site of LTA4H.
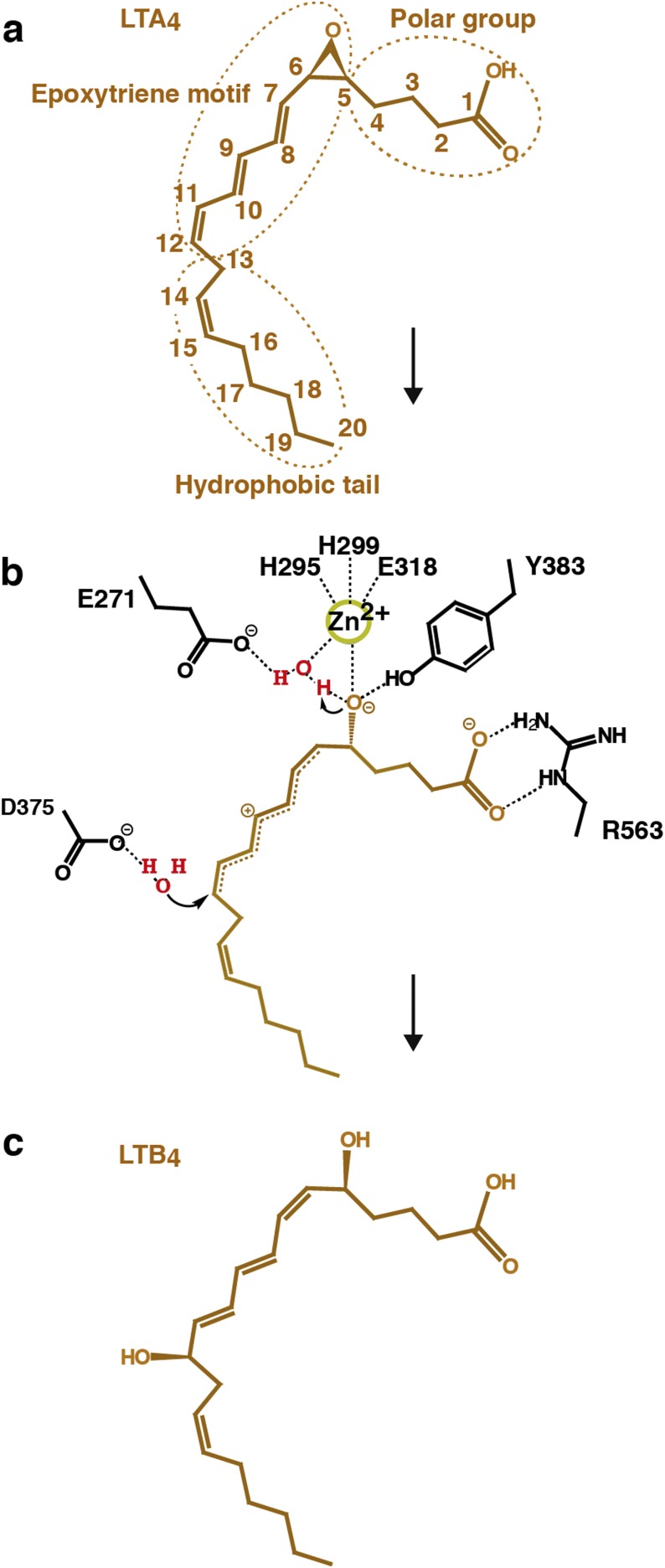
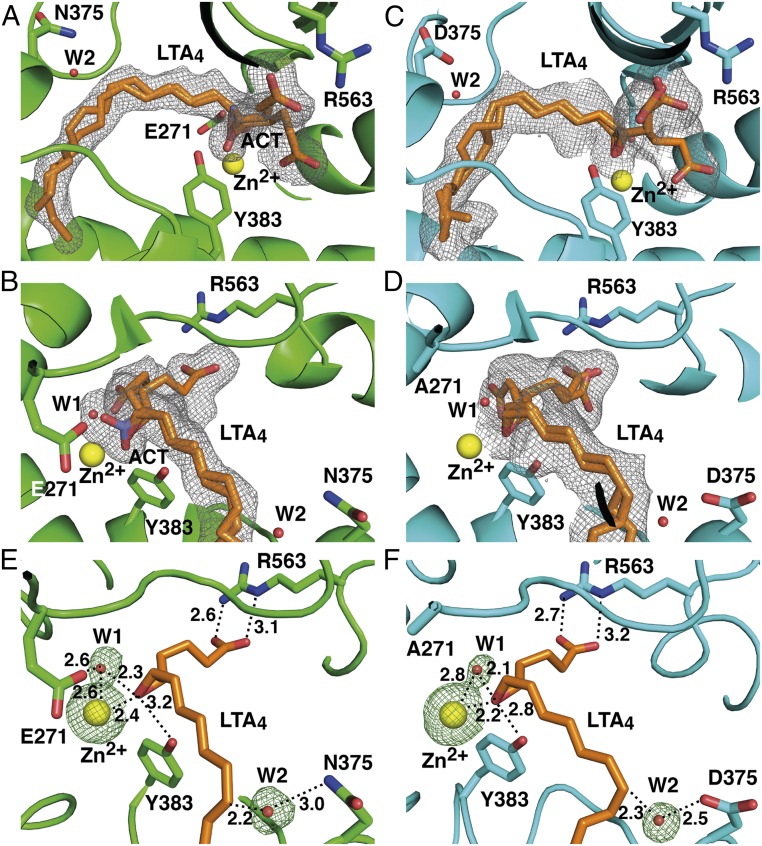
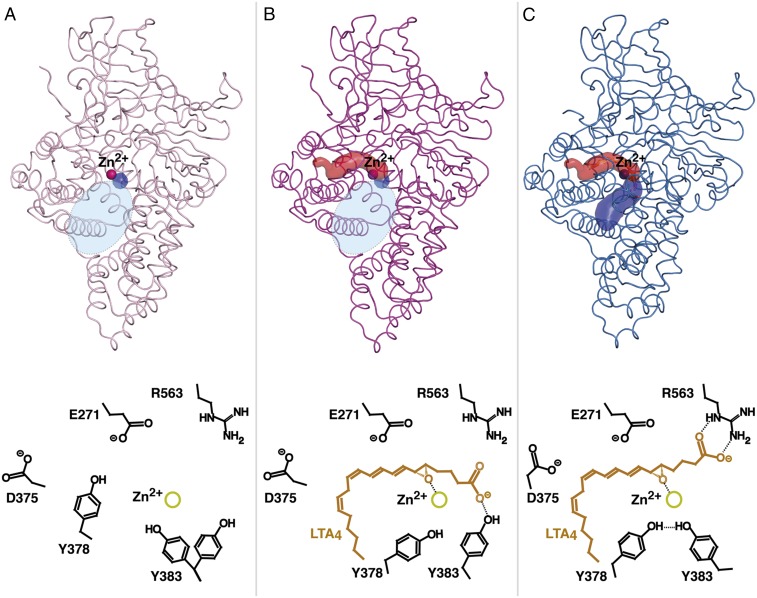
Structures of D375N and E271A showed difference density for LTA4 in the active site cavity (Table S1). The ligand, which consists of a hydrophobic tail, epoxytriene, and polar carboxylate group (Fig. S1A), was modeled in two conformations in the structures of D375N and trigonal E271A, and in three conformations in the structure of orthorhombic E271A. The conformations differ mostly in the position of the C1 carboxylate that is either bound by the guanidine group of R563 or unbound facing toward the active site entrance (Fig. 1 A–D). The positions of the hydrophobic tail and epoxytriene motif (C5–C12) of LTA4 are similar in all LTA4-bound LTA4H structures. The aliphatic tail of the substrate occupies the hydrophobic cavity with its ω-end abutting three structural waters at the bottom of the cavity. The epoxide oxygen is coordinated by Zn2+ and the hydroxyl group of Y383 (Fig. 1 E and F). Y383 locates opposite to one of the catalytic waters (W1), which is coordinated by Zn2+ and E271, and shields the unstable epoxide ring from nucleophilic attack by nearby water molecules. Residues surrounding the conjugated triene system of LTA4 (Y378, Y267, M270, and Q136) also protect the ligand from nonenzymatic hydrolysis. The second catalytic water (W2) is found close to C12 of LTA4 and coordinated by asparagine in D375N-LTA4 (Fig. 1E), and by aspartic acid in E271A-LTA4 (Fig. 1F).
Fig. S1.
Proposed mechanism for enzymatic conversion of LTA4 into LTB4. (A) Chemical structure of LTA4. (B) Formation of a carbonium intermediate after SN1 acid-induced opening of the epoxide moiety. During the reaction, one catalytic water assists in the opening of the epoxide moiety, and the second catalytic water is responsible for the stereospecific insertion of an (R)-hydroxyl group at C12 of the carbonium ion. This process, together with rearrangement of the geometry of the conjugated triene system leads to formation of the bioactive LTB4. (C) Chemical structure of LTB4.
Fig. 1.
Binding of LTA4 in the active site of LTA4H. (A and B) Different views of the active site of orthorhombic D375N (residues are in green) complexed with LTA4 (orange) and acetate (ACT, violet). Yellow and red spheres represent Zn2+ and catalytic waters, respectively. mFo-DFc polder omit map contoured at 3.0 σ is shown in gray mesh around LTA4 and ACT. (C and D) Different views of the active site of orthorhombic E271A-LTA4 complex (residues are in cyan). (E) LTA4 bound with its C1 carboxylate anchored to R563 in the active sites of D375N and (F) E271A. mFo-DFc omit map for Zn2+ and catalytic waters is shown in green mash at 3.0 σ. Distances are labeled in Å.
Summarizing the structural information, we conclude that the EH reaction strictly depends on the correct alignment of LTA4 in the active site cavity. As the ω-end enters the hydrophobic cavity, the C1 carboxylate group anchors to R563, while Zn2+ and Y383 coordinate the epoxide oxygen. At the same time, W1, polarized by Zn2+ and E271, donates a proton to promote SN1 acid-induced opening of the epoxide ring. A carbonium ion is formed, whose positive charge is delocalized over the triene system with its C6-C7 bond in a procis configuration stabilized by the l-shape of the tight pocket (Fig. S1B). At the final step of the reaction, W2, perfectly poised to control stereospecificity, is activated by D375 and attacks C12 to generate a hydroxyl group in R configuration. In this way, LTA4H converts LTA4 into a dihydroxy acid containing a ∆6-cis-∆8,10-trans geometry of the conjugated triene system and a 12R hydroxyl group, two key structural features of LTB4 that confer bioactivity (Fig. S1C). It is noteworthy that our conclusions reported here rationalize previous functional experiments with site-directed mutagenesis (11–13).
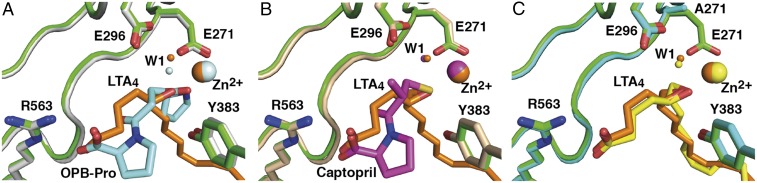
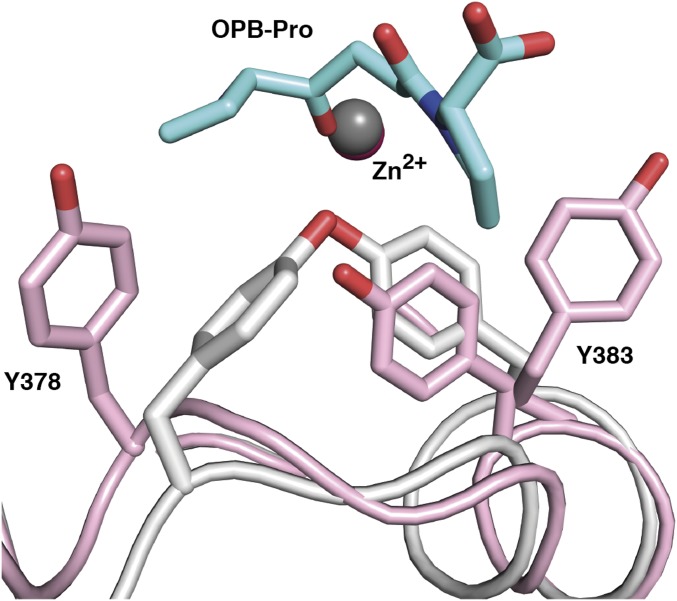
Comparison of the structures of LTA4H-LTA4 and a previously reported structure of LTA4H complexed with a PGP analog (OPB-Pro) (14) shows that a single catalytic water, W1, is involved in both activities of LTA4H (Fig. 2A). This water is coordinated to both E271 and E296, creating hydrogen bonds to their carboxylate groups. Depending on activity, W1 is activated by E296 for peptide bond hydrolysis or by E271 for epoxide ring opening. This water molecule is also present at the same position in a structure of LTA4H in complex with captopril (Fig. 2B). In complexed E271A-LTA4, in the absence of E271, W1 remains hydrogen bonded only to the side chain of E296 (Fig. 2C).
Fig. 2.
A catalytic water has the same position in structures of LTA4H complexed with different ligands. Superposition of D375N-LTA4 (residues are in green; LTA4, catalytic water and Zn2+ are in orange) onto (A) LTA4H-OPB-Pro (PDB ID code: 4MS6) (residues are in gray; OPB-Pro, catalytic water and Zn2+ are in light cyan), onto (B) structure of LTA4H-captopril (PDB ID code: 4DPR) (residues are in beige; captopril, catalytic water and Zn2+ are in magenta), and onto (C) E271A-LTA4 (residues are in cyan; LTA4, catalytic water and Zn2+ are in yellow).
Dynamic Gating of LTA4 in the Active Site of LTA4H.
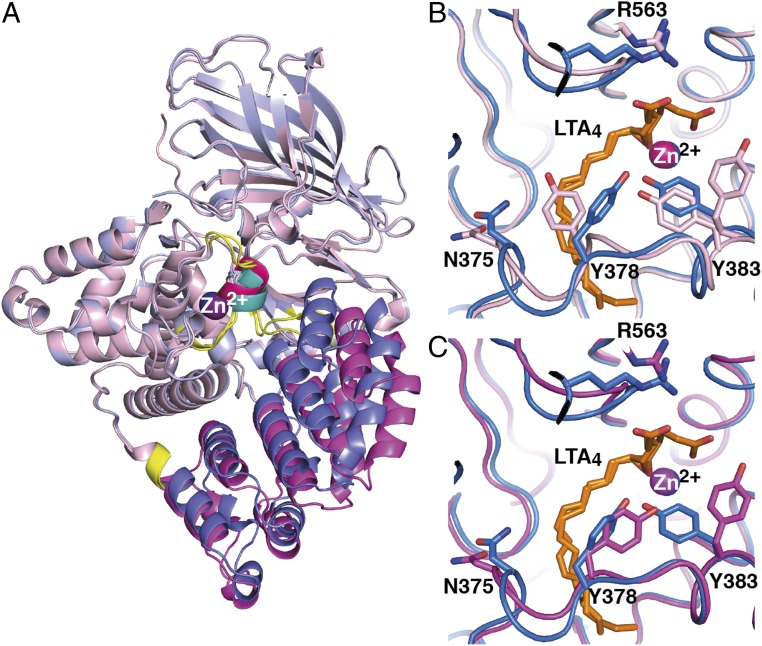
Structures of the two mutants D375N and R563A reveal for the first time an “open” domain conformation of LTA4H distinct from all previously reported structures of the enzyme (Table S2). Comparison of “closed” and open conformations of our present structures (Tables S1 and S2) delineated three dynamic domains: the fixed domain (FD), the first moving (relative to FD) domain (MD1), and the second moving domain (MD2), with bending regions between them (Fig. 3A and Movie S1). The FD and MD1 can rotate relative to each other by ∼16–22°, governing the width of the entrance to the active site cavity. During dynamic domain motion, the residues V260, T465, and N466 act as a hinge, and MD2 moves by 3–4 Å and acts as a lid separating the exterior hydrophilic part of the active site from the interior hydrophobic tunnel. In the open conformation, Y378 from the lid seals off the entrance to the hydrophobic cavity, which accommodates the tail of LTA4 during EH reaction (Figs. 3B and 4A and Movie S2). In this capacity, Y378 becomes a molecular gate for entry of LTA4 into the hydrophobic tunnel and, simultaneously, prevents access of water molecules that could otherwise interfere with subsequent binding of the lipid substrate, as previously reported for the soluble bovine β-lactoglobulin (15). Of note, we could not find any structural obstacles for peptide substrate binding and turnover in the active site during lid displacement (Fig. S2).
Fig. 3.
Dynamic domain motion in the structure of LTA4H. (A) Closed conformation of LTA4H with its FD in light blue and MD1 and MD2 in blue and teal, respectively. The same regions are colored pink, magenta, and hot pink for the open conformation. The hinge regions are in yellow. Zn2+ is in blue in the closed conformation and in magenta in the open conformation. (B) Y378 seals off the hydrophobic tunnel. Superposition of the active site and region of MD2 of closed D375N with bound LTA4 (residues and Zn2+ are in blue, LTA4 is in orange) onto open D375N (in pink) with blocked tunnel, and onto (C) open D375N (in magenta) with open tunnel.
Fig. 4.
Tunnel formations (Upper; the structures and corresponding Zn2+ are colored as in Fig. 3 B and C) and proposed mechanism of substrate binding (Lower) during dynamic domain motion between open and closed D375N. (A) In the open conformation (polar part of binding pocket is shown as tiny blue spot; wide space between domains at the entrance of the active site is indicated as an oval in shaded light blue) Y378 blocks the entrance of the hydrophobic cavity. (B) During substrate binding, lid region moves and opens the hydrophobic tunnel (red surface representation) for LTA4 entrance. When LTA4 slides into the hydrophobic pocket, its carboxylate group is guided by Y383. (C) During domain motion, the ω-end reaches the bottom of the cavity, the epoxide binds to Zn2+, and the C1 carboxylate switches from Y383 to R563. The closed conformation affords productive substrate alignment and allows epoxide activation and subsequent hydrolysis according to an SN1 mechanism. The polar arm of the binding pocket that is formed as a result of convergence of domains is shown as blue surface.
Fig. S2.
Superposition of the structures of open D375N (pink cartoon representation) onto closed LTA4H (gray) in complex with OPB-Pro (cyan) does not reveal structural blockage for PGP binding. Zn2+ is represented by gray sphere and locates essentially at the same position in the open and closed conformations of the enzyme.
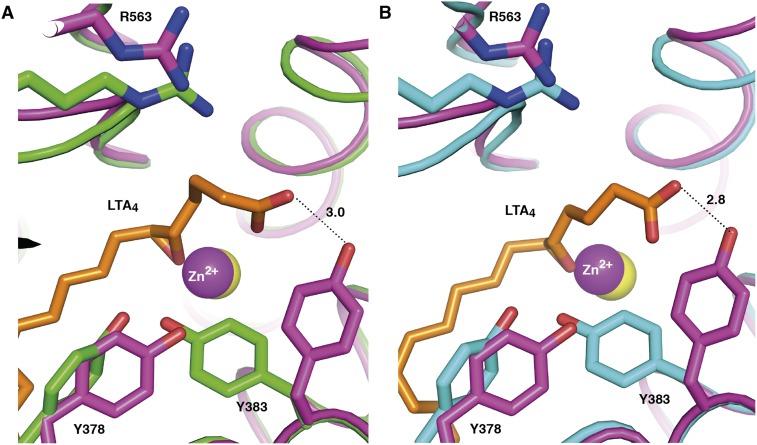
The structure of open D375N cocrystallized with LTA4 shows no density for the ligand, but the internal lid of MD2 has changed position, uncovering the hydrophobic cavity for substrate entrance (Figs. 3C and 4B). Moreover, superposition of open conformation of D375N and closed conformation of D375N and E271A suggests that in the open conformation of LTA4H, Y383 may anchor the polar group of the substrate during LTA4 entrance (Fig. S3 A and B). This hypothesis explains the presence of unbound carboxylate of LTA4 observed in complex structures of D375N-LTA4 and E271A-LTA4 (Fig. 1 A–D). After domain closure, the carboxylate group of LTA4 switches from the hydroxyl group of Y383 to the guanidinium group of R563. In a concerted manner, Y383 changes conformation and forms hydrogen bonds to the hydroxyl group of Y378 and the epoxide oxygen, which are molecular events of a substrate-mediated induced fit to achieve optimal alignment of LTA4 in the active site cavity (Fig. 4C).
Fig. S3.
The alternative conformation of LTA4 with an unbound C1 carboxylate, observed in closed LTA4H, may make a hydrogen bond to the hydroxyl group of Y383 in the context of the open enzyme. Superposition of open D375N (residues and Zn2+ are in magenta) onto (A) orthorhombic D375N (residues are in green, Zn2+ is in yellow) in complex with LTA4 (orange) and (B) onto orthorhombic E271A-LTA4 (residues are in cyan, Zn2+ is in yellow). Distances are labeled in Å.
Changing of Coordination Mode of E318 in the Zn2+-Binding Site.
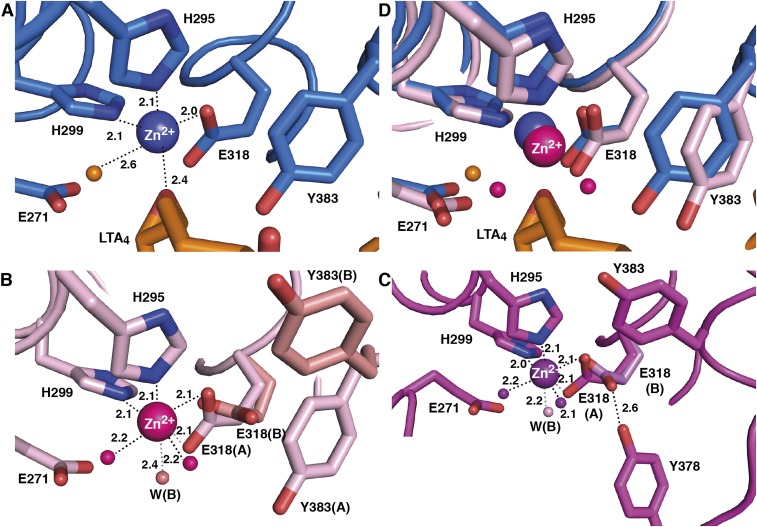
The open and closed conformations of LTA4H display different coordination modes of the carboxylate group of the Zn2+ ligand E318. In closed structures, Zn2+ is coordinated by five bonds to surrounding ligands (Fig. S4A), whereas in open structures, we observe six Zn2+–ligand interactions arranged in octahedral geometry, involving bidentate binding to the carboxylate of E318 (Fig. S4 B and C). Thus, during movement between open and closed conformations, we observe a carboxylate shift (16) from bidentate to monodentate coordination (Fig. S4D). This shift can occur against a relatively low-energy barrier and may reflect reduction of Zn2+-positive charge during binding of the negatively charged epoxide oxygen of LTA4 (17).
Fig. S4.
Geometry of Zn2+-binding site in open and closed conformations of LTA4H. (A) Monodentate carboxylate group coordination in the closed D375N-LTA4 (residues and Zn2+ are in blue, LTA4 and catalytic water are in orange). Distances are labeled in Å. (B) Bidentate carboxylate group coordination in open D375N (residues, Zn2+ and coordinating waters are in pink) with blocked tunnel and (C) in open D375N (residues, Zn2+ and coordinating waters are in magenta) with open tunnel. Alternative conformation of residues and water molecules in open D375N (B) and D375N (C) are colored salmon and light magenta, respectively. (D) Superposition of Zn2+-binding site in open and closed conformations of D375N.
The Side Chain of E318 Helps the Y378 Gate Stay Open.
Interestingly, analysis of the open structures also reveals two conformational states of the side chain of E318, one of which appears to prevent the Y378 gate from closing. Thus, in one state, the carboxylate group of E318 makes two bonds to Zn2+, as described earlier. In the second state, the side chain of E318 is slightly rotated and has changed position, retaining only one bond to the metal ion, which instead binds an additional water molecule to complete the octahedral geometry (Fig. S4 B and C). In the open structure with blocked hydrophobic tunnel, Y378 is not within interacting distance to the E318 carboxylate, which leaves room for alternative position of the side chain of Y383 (Fig. S4B). In contrast, when the hydrophobic tunnel is open, the hydroxyl group of Y378 interacts with the E318 carboxylate in its second, rotated conformation, a hydrogen bond that can help stabilize the Y378 gate in open position and promote entrance of LTA4 into the hydrophobic tunnel. Also, when the Y378 gate is open, Y383 has only space to allow one position distant from Y378 (Fig. S4C).
Implications for Rational Design of LTA4H Inhibitors.
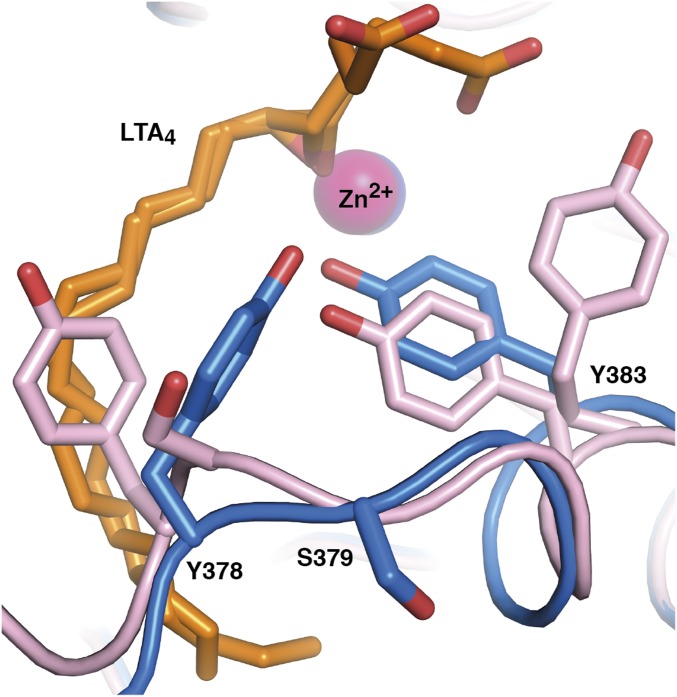
As a result of the role of LTB4 in inflammation, LTA4H is an important drug target. The enzyme’s substrate LTA4 is also a precursor for the lipoxins, a family of proresolving lipid mediators, and thus inhibition of LTA4H may enhance lipoxin synthesis by shunting of LTA4. Indeed, inhibitors of LTA4H have been developed in the past but have not reached the clinic, perhaps because they have all blocked the enzyme’s aminopeptidase activity and details of LTA4 binding and enzyme dynamics have not been known. Notably, the results of the present study define the chemical landscape for drug design. Thus, small molecules should take advantage of the hydrophobic tunnel, reach beyond the Y378 lid, and anchor polar residues such as D375 or the carbonyl of W311, D312, F362, K364, or L365 without compromising the aminopeptidase active site. While exploiting these residues, it is important to exclude S379 from the list of possible contacts for drug molecule stabilization. S379 is a polar residue of the hydrophobic pocket but is juxtaposed to Y378 on the mobile lid and completely changes its position during lid displacement (Fig. 5). A successful LTA4H inhibitor may have three modes of action: it will dampen the early inflammatory response by inhibition of LTB4 synthesis and promote resolution of inflammation by preservation of PGP inactivation and shunting of LTA4 into lipoxin generation.
Fig. 5.
Residue S379 changes position during lid movement. Superposition of open D375N (residues and Zn2+ are in pink) with blocked hydrophobic tunnel onto closed LTA4H (residues and Zn2+ are in blue) in complex with LTA4 (orange).
Concluding Remarks.
LTA4H catalyzes hydrolysis of the unstable epoxide intermediate LTA4 into LTB4, which is a potent chemotactic factor for human neutrophils and plays a role in the mounting of an inflammatory response. In the present report, we describe the structural characterization of LTA4H–LTA4 complexes providing the structural basis for LTB4 synthesis. We also show that LTA4H undergoes domain movements, which gate the hydrophobic cavity for entrance of LTA4. Our results give insights to general enzymology, biochemistry of innate immunity, and structure-based design of anti-inflammatory drugs.
Materials and Methods
DNA Constructs.
LTA4H mutant E271A was generated as described (12); mutation D375N was introduced with a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). Oligonucleotides were from Plasmid TAG Copenhagen A/S, and constructs were verified by DNA sequencing (SeqLab). A codon-optimized gene for N-terminally 6His-tagged LTA4H mutant R563A was synthesized by GenScript.
Protein Expression and Purification.
All mutants were expressed in Escherichia coli JM101. Cells grew in LB medium with ampicillin (0.1 mg⋅mL−1) at 37 °C to an optical density (λ = 600) of 0.6. Protein expression was induced by addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside. After overnight cultivation at 20 °C, the cells were harvested by 10 min centrifugation at 3,000 × g. The cells were disrupted by sonication, and the lysate was centrifuged at 60,000 × g for 18 min. To perform immobilized ion affinity chromatography, the crude extract was applied onto a disposable chromatography column (BioRad) with affinity resin charged with Ni2+ and equilibrated with a standard buffer consisting of 25 mM Tris⋅HCl at pH 7.8, 200 mM NaCl, and 10% (vol/vol) glycerol. Elution of unbound and weakly bound fractions was performed in two steps with standard buffers containing 20 and 40 mM imidazole. C-terminally 6His-tagged proteins were eluted with standard buffer containing 250 mM imidazole. After immobilized ion affinity chromatography, the proteins were dialyzed against 25 mM Tris⋅HCl buffer at pH 7.8 and were purified by ion-exchange chromatography followed by size-exclusion chromatography. Ion-exchange chromatography was performed on MonoQ HR5/50 GL column (GE Healthcare) that was equilibrated with 25 mM Tris⋅HCl at pH 7.8, and LTA4H mutants were eluted with a linear gradient of KCl (0–0.4 mM) in 25 mM Tris⋅HCl at pH 7.8. Size-exclusion chromatography was performed using HiLoad 16/600 Superdex 200 pg column (GE Healthcare) equilibrated with gel-filtration buffer (25 mM Tris⋅HCl at pH 7.8 and 50 mM NaCl). All eluted fractions were analyzed with SDS/PAGE on a PhastSystem (GE Healthcare), using 10–15% gradient gel. Fractions containing proteins were concentrated in Amicon Ultra units (Millipore).
Saponification of LTA4 Methyl Ester.
To obtain LTA4, LTA4 methyl ester (Med Chem 101) in tetrahydrofuran was saponified with 1 M LiOH [6% (vol/vol)] for 48 h at 4 °C.
Crystallization and Data Collection.
Mutants E271A and D375N were concentrated to 9 and 2.5 mg/mL, respectively, and crystallized by liquid/liquid diffusion in capillaries (Vitrex Medical A/S) at 4 °C, essentially as described for wild-type LTA4H (9).
Rhomboid-shaped orthorhombic crystals of E271A appeared in 3 d in 22% (wt/vol) PEG 8000, 100 mM imidazole at pH 6.7, 5 mM YbCl3. After 15 s soaking with 1 mM LTA4, specimens were flash-cooled in liquid nitrogen and used for X-ray diffraction experiments under cryogenic conditions (100 K) at beamline ID23-1 (18) of the European Synchrotron Radiation Facility (Grenoble, France). To preserve LTA4 from nonenzymatic hydrolysis, just before soaking, the ligand was exposed to a stream of nitrogen gas to evaporate tetrahydrofuran, and subsequently dissolved in 100 mM borate buffer pH 10 mixed with 20% (vol/vol) glycerol.
To cocrystallize mutant D375N with LTA4, protein was preincubated with 0.4 mM ligand for 30 min on ice. Plate-shaped orthorhombic crystals of the D375N-LTA4 complex could only be obtained in the presence of sodium acetate, which was added at 100 mM concentration to 22% (wt/vol) PEG 8000, 100 mM imidazole at pH 7.0, 5 mM YbCl3. Crystals grew over the course of 3 mo and were cryoprotected with 25% (vol/vol) glycerol in crystallization solution before flash-cooling in liquid nitrogen. For X-ray experiments, crystals were placed into a nitrogen gas stream at 100 K, and data were collected at a wavelength of 13.5 KeV (0.91841 Å) at beamline BL14.1 (19) operated by the Helmholtz-Zentrum Berlin at the BESSY II electron storage ring.
The mutants E271A, D375N and R563A were used for crystallization experiments by vapor-diffusion method, as well. All experiments were performed at room temperature with a ratio of 1 μL protein to an equal amount reservoir solution.
Optimized crystallization conditions of Morpheus screen (20) turned out to be successful for crystallization of mutants E271A (8 mg/mL) and D375N (7 mg/mL) by hanging drop technique. Rhomboid-shaped trigonal crystals of E271A grew in 5 d in the drops containing 8% (wt/vol) PEG 20 000, 15% (wt/vol) PEG MME 550, 100 mM Mes/imidazole at pH 6.9, and 20 mM of each alcohol [1,6-hexanediol, 1-butanol, (R,S)-1,2-propanediol, 2-propanol, 1,4-butanediol, 1,3-propanediol], and were used for soaking experiments with 1 mM LTA4 as described earlier. Trigonal crystals of mutant D375N and cocrystallized D375N were found in 5 d in drops consisting of 8% (wt/vol) PEG 20 000, 15% (wt/vol) PEG MME 550, 100 mM Hepes/Mops at pH 7.7–8.0, 15 mM CaCl2, and 15 mM MgCl2. Before flash-cooling in liquid nitrogen, crystals of D375N were cryo-protected with crystallization solution containing 15% (vol/vol) glycerol. Crystals of both mutants E271A and D375N were transferred into a nitrogen gas stream at 100 K and X-ray exposed at a wavelength of 12.7 KeV (0.97625 Å) at beamline ID-29 (21) (the European Synchrotron Radiation Facility).
Rod-shaped orthorhombic crystals of mutant R563A (4.1 mg/mL) appeared in 1 wk in the sitting drop plate in optimized conditions [20% (wt/vol) PEG MME 5000, 100 mM Mes monohydrate at pH 6.0 and 150 mM ammonium sulfate] of Crystal Screen HT (Hampton Research). Crystals were cryoprotected with 10% (vol/vol) glycerol in crystallization solution and flash-cooled in liquid nitrogen. X-ray data were collected under cryogenic conditions (100 K) at a wavelength of 14.2 KeV (0.8729 Å) at beamline ID23-2 (22) at the European Synchrotron Radiation Facility.
Data Processing and Structure Determination.
All datasets were integrated and scaled using XDS (23).
To obtain unbiased electron density for the ligand-binding site of LTA4H, the orthorhombic structure of the E271A mutant was determined de novo by single-wavelength anomalous diffraction at 12.7 KeV (0.97625 Å). AutoSHARP (24) located 2 Yb3+ ions and 1 Zn2+ ion, which were used to calculate experimental phases (phasing power, 1.014; overall figure of merit, 0.26) that were improved with SOLOMON (25), using an optimized solvent content of 49.4%. An initial model was built by ARP/wARP (26), which placed and docked in sequence 583 residues (R = 26.0%; Rfree = 29.8%). After rebuilding in Coot (27), the structure was refined with the phenix.refine module of the PHENIX package (28), using experimental phase restraints (maximum likelihood Hendrickson–Lattman target). Analysis of the resulting maps showed clear positive difference density in the active site cavity of the enzyme for the LTA4 ligand, which was placed using model and restraint files generated by PRODRG (29). Protein geometry was validated using MolProbity (30).
The structures of D375N and R563A, as well as the trigonal structure of E271A from a crystal grown by vapor-diffusion, were solved by molecular replacement with PHASER (31), using as search model the partially refined orthorhombic structure of E271A (before addition of the LTA4 ligand model). Rebuilding and refinement were performed as described earlier, using an ML target.
To prevent the bulk solvent mask from penetrating the active site cavity of the enzyme, omit mFo-DFc difference map for LTA4 in the active site of mutants E271A and D375N was generated using program phenix.polder (32). Omit map for catalytic waters and Zn2+ was calculated with the FFT program (33) from CCP4 program suite (34).
Domain motion in LTA4H was analyzed with DynDom (35), which identified three dynamic domains, corresponding to residues 1–260, 269–372, and 380–465 (FD); residues 261–268 and 466–610 (MD1); and residues 373–377 (MD2). The bending regions include residues 260–270 and 465–466 for MD1 and residues 370–373 and 378–381 for MD2.
Least squares fits of structures were performed using the McLachlan algorithm (36), as implemented in the program ProFit (www.bioinf.org.uk/software/profit/). Animated visualization of the trajectory from open to closed conformation of LTA4H was performed with Morph (37) PyMOL feature. The tunnels were calculated and visualized with CAVER (38) PyMOL plugin. Figures were made with PyMOL (Schrödinger, LLC) and ChemDraw Prime 15.1 (PerkinElmer).
Data collection and refinement statistics are presented in Table S3 (orthorhombic and trigonal structures of E271A, orthorhombic structure of D375N) and Table S4 (trigonal structures of D375N and orthorhombic structure of R563A).
Table S3.
Data collection and refinement statistics
| Crystal (PDB ID code) | E271A-LTA4 (5NI2) | E271A-LTA4 (5NI4) | D375N-LTA4 (5NI6) |
| Data collection | |||
| No. of crystals | 1 | 1 | 1 |
| Space group | P21221 | P32 | P212121 |
| Cell dimensions | |||
| a, b, c, Å | 67.71, 84.61, 133.43 | 139.56, 139.56, 87.95 | 78.13, 87.45, 99.46 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Molecules, A.U.* | 1 | 3 | 1 |
| Resolution, Å | 39.37–1.50 (1.56–1.50)† | 49.81–1.90 (1.96–1.90) | 43.23–1.54 (1.60–1.54) |
| Rmerge‡ | 0.099 (2.077) | 0.189 (1.609) | 0.105 (1.729) |
| Rpim§ | 0.041 (0.858) | 0.081 (0.695) | 0.045 (0.753) |
| I/σI¶ | 9.25 (0.67) | 6.61 (0.93) | 10.35 (1.06) |
| Wilson B factor, Å2 | 28.32 | 26.66 | 20.64 |
| Total reflections | 812,861 (80,689) | 970,034 (95,990) | 657,381 (59,881) |
| Unique reflections | 122,621 (11,695) | 152,082 (15,217) | 100,716 (9,575) |
| Completeness, % | 99.5 (96.4) | 99.9 (99.6) | 99.5 (95.8) |
| Redundancy | 6.6 (6.7) | 6.4 (6.3) | 6.5 (6.3) |
| CC(1/2)# | 0.996 (0.288) | 0.994 (0.351) | 0.998 (0.432) |
| Refinement | |||
| Resolution, Å | 39.37–1.50 | 49.81–1.90 | 43.23–1.54 |
| Reflections | 122,196 (11,695) | 152,057 (15,207) | 100,690 (9,566) |
| Free reflections | 6,121 (603) | 8,107 (802) | 5,063 (507) |
| Rwork‖/Rfree** | 0.1722/0.1963 | 0.1615/0.2000 | 0.1581/0.1908 |
| Ramachandran plot | |||
| Favored, % | 98.18 | 97.8 | 97.69 |
| Allowed, % | 1.82 | 2.20 | 2.31 |
| Outliers, % | 0.00 | 0.00 | 0.00 |
| No. of atoms | |||
| Protein | 4,914 | 14,907 | 5,013 |
| Ligand&ion | 72 | 176 | 57 |
| Water | 671 | 2,082 | 840 |
| B-factors | |||
| Protein | 40.53 | 31.09 | 25.66 |
| Ligand&ion | 37.24 | 28.65 | 33.25 |
| Water | 43.63 | 38.51 | 37.79 |
| R.m.s deviations | |||
| Bond lengths, Å | 0.006 | 0.005 | 0.006 |
| Bond angles, ° | 0.75 | 0.68 | 0.83 |
A.U., asymmetric unit.
Values in parentheses are for highest-resolution shell.
Rmerge, ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity for an observation of a reflection and 〈I(hkl)〉 is the average intensity of all symmetry-related observations of a reflection.
Rpim, Σhkl √(1/n − 1) Σi|Ii(hkl) - 〈I(hkl)〉|/ΣhklΣi Ii(hkl).
I/σI, signal to noise ratio.
CC(1/2), percentage of correlation between intensities from random half-datasets.
‖Rwork, Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|.
Rfree, same as Rwork calculated from free reflections excluded from refinement.
Table S4.
Data collection and refinement statistics
| Crystal (PDB ID code) | D375N (5NIA) | D375N (5NID) | R563A (5NIE) |
| Data collection | |||
| No. of crystals | 1 | 1 | 1 |
| Space group | R3:H | R3:H | P212121 |
| Cell dimensions | |||
| a, b, c Å | 153, 153, 74.88 | 153.93, 153.93, 76.79 | 51.97, 132.15, 320.12 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Molecules, A.U.* | 1 | 1 | 3 |
| Resolution, Å | 44.17–1.76 (1.83–1.76)† | 44.44–1.57 (1.62–1.57) | 49.47–2.60 (2.70–2.60) |
| Rmerge‡ | 0.104 (1.785) | 0.059 (1.882) | 0.193 (1.109) |
| Rpim§ | 0.043 (0.752) | 0.023 (0.785) | 0.081 (0.576) |
| I/σI¶ | 9.28 (1.02) | 13.67 (0.96) | 5.90 (0.94) |
| Wilson B factor, Å2 | 29.80 | 29.65 | 41.96 |
| Total reflections | 438,699 (40,632) | 655,660 (64,529) | 358,982 (19,332) |
| Unique reflections | 64,241 (6,342) | 95,099 (9,558) | 64,086 (5,413) |
| Completeness, % | 99.8 (98.3) | 99.9 (99.7) | 92.8 (80.0) |
| Redundancy | 6.8 (6.4) | 6.9 (6.8) | 5.6 (3.6) |
| CC(1/2)# | 0.998 (0.394) | 0.999 (0.374) | 0.984 (0.471) |
| Refinement | |||
| Resolution, Å | 44.17–1.76 | 44.44–1.57 | 49.47–2.60 |
| Reflections | 64,238 (6,341) | 95,071 (9,539) | 64,055 (5,403) |
| Free reflections | 3,271 (266) | 4,840 (484) | 3,104 (264) |
| Rwork‖/Rfree** | 0.1801/0.2074 | 0.1821/0.2097 | 0.2372/0.2935 |
| Ramachandran plot | |||
| Favored, % | 96.38 | 97.70 | 96.19 |
| Allowed, % | 3.45 | 2.30 | 3.70 |
| Outliers, % | 0.16 | 0.00 | 0.11 |
| No. atoms | |||
| Protein | 4,947 | 4,950 | 14,534 |
| Ligand&ion | 1 | 1 | 3 |
| Water | 360 | 412 | 155 |
| B-factors | |||
| Protein | 48.41 | 50.94 | 57.12 |
| Ligand&ion | 31.63 | 26.08 | 49.46 |
| Water | 41.48 | 47.04 | 29.91 |
| R.m.s. deviations | |||
| Bond lengths, Å | 0.015 | 0.010 | 0.003 |
| Bond angles, ° | 1.23 | 0.96 | 0.62 |
A.U., asymmetric unit.
Values in parentheses are for highest-resolution shell.
Rmerge, ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity for an observation of a reflection and 〈I(hkl)〉 is the average intensity of all symmetry-related observations of a reflection.
Rpim, Σhkl √(1/n − 1) Σi|Ii(hkl) - 〈I(hkl)〉|/ΣhklΣi Ii(hkl).
I/σI, signal to noise ratio.
CC(1/2), percentage of correlation between intensities from random half-datasets.
‖Rwork, Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|.
Rfree, same as Rwork calculated from free reflections reflections excluded from refinement.
Supplementary Material
Acknowledgments
We thank Michaela Mårback for help with site-directed mutagenesis and are grateful to Luca Jovine and Agnes Rinaldo-Matthis (Karolinska Institutet) for valuable discussions and advice. We acknowledge the European Synchrotron Radiation Facility and Helmholtz Zentrum Berlin, as well as BiostructX-783, for access to synchrotron radiation sources. We also thank the Protein Science Facility (Karolinska Institutet) for access to infrastructure for protein crystallization and excellent assistance. The work was supported by grants from the Swedish Research Council (10350), Linneus Grant CERIC, and Novo Nordisk Foundation (NNF15CC0018346), and a Distinguished Professor Award from Karolinska Institutet.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5NI2, 5NI4, 5NI6, 5NIA, 5NID, and 5NIE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710850114/-/DCSupplemental.
References
- 1.Haeggström JZ, Funk CD. Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 2.Samuelsson B. Leukotrienes: Mediators of immediate hypersensitivity reactions and inflammation. Science. 1983;220:568–575. doi: 10.1126/science.6301011. [DOI] [PubMed] [Google Scholar]
- 3.Peters-Golden M, Henderson WR., Jr Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 4.Allen S, Dashwood M, Morrison K, Yacoub M. Differential leukotriene constrictor responses in human atherosclerotic coronary arteries. Circulation. 1998;97:2406–2413. doi: 10.1161/01.cir.97.24.2406. [DOI] [PubMed] [Google Scholar]
- 5.Bäck M, Hansson GK. Anti-inflammatory therapies for atherosclerosis. Nat Rev Cardiol. 2015;12:199–211. doi: 10.1038/nrcardio.2015.5. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snelgrove RJ, et al. A critical role for LTA4H in limiting chronic pulmonary neutrophilic inflammation. Science. 2010;330:90–94. doi: 10.1126/science.1190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thunnissen MM, Nordlund P, Haeggström JZ. Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation. Nat Struct Biol. 2001;8:131–135. doi: 10.1038/84117. [DOI] [PubMed] [Google Scholar]
- 10.Arand M, Cronin A, Adamska M, Oesch F. Epoxide hydrolases: Structure, function, mechanism, and assay. Methods Enzymol. 2005;400:569–588. doi: 10.1016/S0076-6879(05)00032-7. [DOI] [PubMed] [Google Scholar]
- 11.Rudberg PC, Tholander F, Andberg M, Thunnissen MMGM, Haeggström JZ. Leukotriene A4 hydrolase: Identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates. J Biol Chem. 2004;279:27376–27382. doi: 10.1074/jbc.M401031200. [DOI] [PubMed] [Google Scholar]
- 12.Rudberg PC, Tholander F, Thunnissen MM, Haeggström JZ. Leukotriene A4 hydrolase/aminopeptidase. Glutamate 271 is a catalytic residue with specific roles in two distinct enzyme mechanisms. J Biol Chem. 2002;277:1398–1404. doi: 10.1074/jbc.M106577200. [DOI] [PubMed] [Google Scholar]
- 13.Rudberg PC, Tholander F, Thunnissen MMGM, Samuelsson B, Haeggstrom JZ. Leukotriene A4 hydrolase: Selective abrogation of leukotriene B4 formation by mutation of aspartic acid 375. Proc Natl Acad Sci USA. 2002;99:4215–4220. doi: 10.1073/pnas.072090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stsiapanava A, et al. Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor. Proc Natl Acad Sci USA. 2014;111:4227–4232. doi: 10.1073/pnas.1402136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qvist J, Davidovic M, Hamelberg D, Halle B. A dry ligand-binding cavity in a solvated protein. Proc Natl Acad Sci USA. 2008;105:6296–6301. doi: 10.1073/pnas.0709844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rardin RL, Tolman WB, Lippard SJ. Monodentate carboxylate complexes and the carboxylate shift–Implications for polymetalloprotein structure and function. New J Chem. 1991;15:417–430. [Google Scholar]
- 17.Ryde U. Carboxylate binding modes in zinc proteins: A theoretical study. Biophys J. 1999;77:2777–2787. doi: 10.1016/S0006-3495(99)77110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurizzo D, et al. The ID23-1 structural biology beamline at the ESRF. J Synchrotron Radiat. 2006;13:227–238. doi: 10.1107/S0909049506004341. [DOI] [PubMed] [Google Scholar]
- 19.Mueller U, et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur Phys J Plus. 2015;130:141. [Google Scholar]
- 20.Gorrec F. The MORPHEUS protein crystallization screen. J Appl Cryst. 2009;42:1035–1042. doi: 10.1107/S0021889809042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Sanctis D, et al. ID29: A high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J Synchrotron Radiat. 2012;19:455–461. doi: 10.1107/S0909049512009715. [DOI] [PubMed] [Google Scholar]
- 22.Flot D, et al. The ID23-2 structural biology microfocus beamline at the ESRF. J Synchrotron Radiat. 2010;17:107–118. doi: 10.1107/S0909049509041168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 25.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 26.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 30.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebschner D, et al. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read RJ, Schierbeek AJ. A phased translation function. J Appl Cryst. 1988;21:490–495. [Google Scholar]
- 34.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 36.McLachlan AD. Rapid comparison of protein structures. Acta Crystallogr A. 1982;38:871–873. [Google Scholar]
- 37.Schrödinger LLC. Schrödinger; New York: 2016. The Pymol Molecular Graphics System, Version 1.8. [Google Scholar]
- 38.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.