Abstract
GLI1 is a key downstream transcription effector of the Hedgehog (Hh) signaling pathway that is involved in promoting cell growth, differentiation and tissue patterning in embryonic development. GLI1 over-activation and its nuclear localization has also been linked to the increased aggressiveness of a number of cancers. It has previously been demonstrated that DYRK1A (dual-specificity tyrosine-regulated kinase 1A) can phosphorylate GLI1 and promote GLI1 nuclear localization and its transcriptional activity. Utilizing recombinant human GLI1 and DYRK1A proteins and phospho-peptide mass spectrometry, we demonstrated that GLI1 is phosphorylated by DYRK1A at Ser408, a phospho-site that falls within the putative nuclear localization sequence (NLS) of GLI1 suggesting a possible mechanistic role in modulating its translocation. Further, we showed that the Ser408 site on GLI1 was not phosphorylated in the presence of the selective DYRK1A inhibitor harmine. The data described herein provide the first identification of a DYRK1A-mediated site of phosphorylation on GLI1 within its NLS and may serve as a valuable mechanism for further understanding Hh signaling modulation.
Keywords: Hedgehog, GLI1, DYRK1A, phosphorylation, nuclear localization, mass spectrometry
1. Introduction
The hedgehog (Hh) pathway has a critical role in early embryogenesis where it promotes differentiation and the patterning of tissues [1]. The downstream transcription factor GLI1 (GLIoma-associated oncogene homolog 1) acts as the terminal effector of Hh signaling [2-4], with its status controlling proliferation, differentiation and survival. Activated GLI1 translocates into the nucleus and stimulates the transcription of Hh pathway target genes, including GLI1, PTCH1 and many pro-survival pathways [5]. The Hh pathway has also been implicated in a wide range of cancers [6], with nuclear expression of GLI1 correlating with poor prognosis in pancreatic cancer [7], brain gliomas [8] and basal-like breast cancer [9].
Nuclear localization of GLI1 is important for its ability to serve as a transcription factor [10], and substantially increases GLI1 mediated transcriptional activity [11]. A number of phosphorylation events have been shown to regulate the activity of GLI1 either positively or negatively [12, 13]. The kinase DYRK1A (dual-specificity tyrosine phosphorylation-regulated kinase 1a) has been shown to activate GLI1 via a phosphorylation event, leading to the translocation of GLI1 from the cytoplasm to the nucleus [14-17]. Co-expression of DYRK1A and GFP-GLI1 promotes nuclear localization of GLI1-GFP in NIH3T3 cells [15, 17], and in an Ewing sarcoma tumor line [14]. This activity is dependent on the kinase activity of DYRK1A as a kinase-null mutant of DYRK1A does not induce nuclear accumulation of GLI1 [15, 16]. Studies using recombinant GLI1 truncation mutants and [32P]ATP labeling localized DYRK1A-dependent phosphorylation of GLI1 on Ser/Thr residues in both the N-terminal and C-terminal portions of GLI1 [15]. Although it has been demonstrated that DYRK1A can phosphorylate GLI1 and promote GLI1 nuclear localization, the exact DYRK1A-mediated sites of phosphorylation within the C-terminal region of GLI1 were unknown.
Here, we have used an in vitro kinase assay and phospho-peptide mass spectrometry analysis to identify site(s) of direct phosphorylation of GLI1 by DYRK1A and have determined that DYRK1A phosphorylates GLI1 at Ser408 within its nuclear localization sequence.
2. Materials and methods
2.1 Materials
Human recombinant GLI1 protein (#TP301110; C-terminal MYC/DDK-tagged) and expression vector ((#RC201110; pCMV6 vector containing C-terminal myc-DDK tagged-GLI1) were from Origene (Rockville, MD). Human recombinant DYRK1A protein (GST-tagged; PV3997) and harmine were from ThermoFisher Scientific (Waltham, MA). Anti-Flag agarose affinity gel (cat #: A2220) was from Sigma Aldrich (St. Louis, MO). Rabbit anti-human GLI1 was from Origene (TA310536) and anti-rabbit IRDye 800 from LI-COR (Lincoln, NE).
2.2 Preparation of GLI1 protein and western blotting
HEK-293 cells (ATCC, CRL-1573) at 80-90% confluence in T150 flasks were transfected with pCMV6 GLI1-myc-DDK plasmid (15 μg) using Turbofectin (Origene) and cells grown in Eagle's Minimum Essential Medium with 10% FBS for 24 h at 37°C. Cells were then lyzed and the cell extract centrifuged at 14,000 rpm for 15 min. Anti-FLAG affinity gel in TBS was then added to the clarified cell extract and gently mixed for 2 h at 4°C. The resin was then washed with TBS/0.05% Tween 20, bound protein eluted at pH 3.0 and. elution fractions neutralized.
For Western blotting, proteins were electrophoresed on NU-PAGE Bis-Tris 4-12% gels (ThermoFisher) and after transfer to Immobilon, membranes were blocked for 1 h with 5% nonfat milk. Membranes were then incubated with rabbit anti-GLI1 at 1:5,000 (overnight at 4°C) After washing, membranes were incubated with secondary anti-rabbit IRDye 800 antibody (1:15,000) for 1 h and after washing, blots visualized on a LI-COR Odyssey.
2.3 In vitro kinase assay and gel slice isolation
Purified recombinant human GLI1 protein (2 μg) was incubated with recombinant human DYRK1A protein (1 μg) in kinase buffer (25 mM Tris-HCl pH 7.5 plus phosphatase inhibitors) with ATP (1 mM) for 30 min at 30°C. Controls included no DYRK1A, no ATP and plus harmine (1 μM). Samples were electrophoresed on 4-12% Bis-Tris SDS-PAGE gels for 1 h at 120 V, bands detected with Coomassie blue staining and the appropriate gel bands excised.
2.4 Mass spectrometry analysis
Mass spectrometry was performed at the UNC-CH Proteomics Center (Chapel Hill, NC, USA). Detailed methodology is described in the accompanying Data in Brief article [18]. Briefly, gel slices were divided into two pieces and each piece treated with reducing agent, alkylated and digested with trypsin. TiO2 beads were used to enrich for phosphopeptides. The tryptic peptides were separated by reversed-phase hplc on a C18AQ column (Bruker-Michrom Biosciences) using a Nano-Acquity HPLC system (Waters Corp) interfaced to an electrospray ionization/LTQ Velos-Orbitrap ion trap mass spectrometer (Thermo Fisher Scientific). Initial MS scans were recorded over m/z range 400–2000 with the most abundant ions selected for collision-activated dissociation. All files were searched using MASCOT [19] (Matrix Science) against the protein of interest (GLI1, Origene ID RC201110; Supplementary Fig. 1). The search parameters included peptide mass tolerance of 10 ppm, fragment ion tolerance of 0.6 mass unit. Each sample was run twice (n = 2).
3. Results and discussion
A number of studies have demonstrated that translocation of the Hh pathway transcription factor GLI1 to the nucleus and its transcriptional activity are positively regulated by phosphorylation by the kinase DYRK1A [14, 15, 17]. It was previously reported using truncated versions of recombinant GLI1 and [32P]ATP labeling that DYRK1A phosphorylated GLI1 within residues 1-225 of the N-terminal and within residues 394-1106 of the C-terminal portions of the protein [15]. However, the exact site(s) of phosphorylation were not identified. Herein, using phospho-peptide mass spectrometry analysis, we have determined a site of direct phosphorylation on GLI1 by DYRK1A at Serine 408, which is within the C-terminal fragment identified by Mao [15], and more interestingly falls within the nuclear localization sequence of GLI1 [20].
To identify the site(s) of DYRK1A-mediated phosphorylation on GLI1 we used an in vitro kinase assay based on a previously published procedure measuring DYRK1A-mediated phosphorylation of Tau [21]. The recombinant human GLI1 protein used for our studies was generated by transfection of a human myc-DDK-tagged GLI1 plasmid into HEK-293 cells and affinity purification of the GLI1 protein by anti-FLAG resin. Expression and purification was confirmed by Western blotting (Fig. 1A). SDS-PAGE analysis of Myc-DDK-tagged GLI1 showed the protein to have an estimated molecular weight ∼150,000 Da (Figs. 1B).
Fig. 1. Expression and purification of recombinant human GLI1 protein.
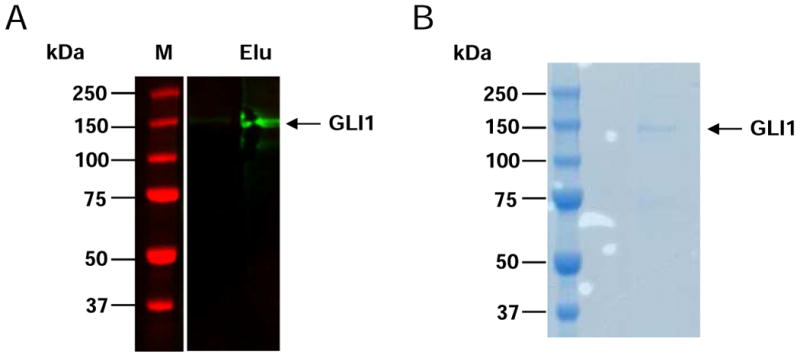
FLAG-tagged GLI1 was expressed in HEK-293 cells and isolated using anti-FLAG-M2 affinity resin. (A) Western blotting with anti-GLI1 antibody. M = molecular weight markers; Elu = elution. (B) SDS-PAGE of purified FLAG-tagged GLI1.
We incubated GLI1 protein in the absence and presence of recombinant DYRK1A protein, and GLI1/DYRK1A with and without ATP (study 1). Samples were then run on SDS-PAGE and gel bands excised (Fig. 2A). Gel slices were treated with trypsin to generate tryptic peptides, phospho-peptides enriched by TiO2 beads and analyzed by mass spectrometry. Details on sequence coverage and all phospho-peptides identified across all samples are detailed in the accompanying Data in Brief article [18]. Based on the GLI1 sequence from NCBI (Supplementary Fig. 1), approximately 50-70% coverage was observed and between 76 to 161 peptides identified for the samples (see Table 1 in [18]). A number of high confidence phospho-peptides were identified with the majority present in all samples, suggesting those were related to basal GLI1 phosphorylation. Phosphorylation was observed on serine (Ser) and threonine (Thr) residues with a predominance for Ser, agreeing with previous observations that DYRK1A has a preference for Ser over Thr [22]. The phospho-peptides identified by at least 4 PSM (peptide spectrum matches) are detailed in Table 2 in [18].
Fig. 2. In vitro DYRK1A kinase assays, gel slice isolation and phospho-mapping analysis of GLI1.
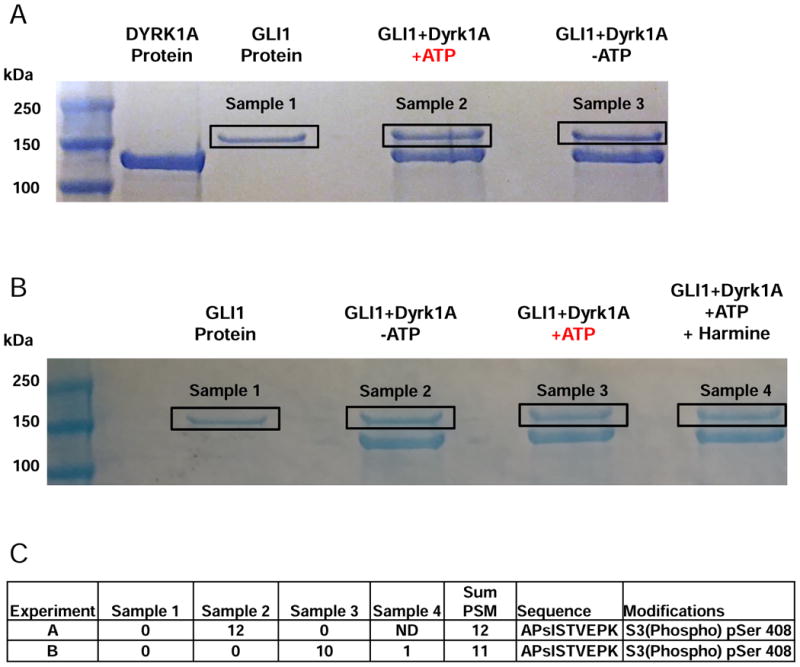
Two (panels A and B) independent experiments were conducted. (A) Purified GLI1 was incubated in kinase buffer with (or without) DYRK1A and in the presence or absence of ATP. (B) Purified GLI1 was incubated in kinase buffer with (or without) DYRK1A, ATP, or with harmine. The protein bands corresponding to GLI1 (indicated by the black rectangles) were excised from the gels and treated with trypsin. Phospho-peptides were enriched and subjected to mass spectrometry for peptide identification and detection of phospho-sites. (C) Number of PSMs for APsISTVEPK phospho-peptide detected for each sample from A and B. ND = not done.
The major difference between the samples was phosphorylation of the peptide APsISTVEPK at serine 408 (pSer408). This phosphorylated peptide was detected 6 times in each of the analyses of sample 2 (Fig. 2A and Table 3 in [18]) but not in any other run. In contrast, the unphosphorylated peptide (APSISTVEPK) was detected in all samples at ∼10 to 13 times per run (see Table 3 in [18].). The MS/MS fragmentation spectra for the unphosphorylated and phosphorylated APSISTVEPK peptide showing the increased mass difference due to a phosphorylation event are shown in Fig. 3 (A and B respectively) and detailed in (see Figures 1 and 2 in [18]). The pSer408 site falls within the GLI1 fragment (Fig. 4; residues 394-1106) previously identified using DYRK1A and [32P]ATP labeling [15], and more specifically within the nuclear localization signal (NLS) sequence (residues 380-420 [20]) (Fig. 4). A second phospho-peptide GPsPSFGVQPGPHDSAR (pSer146) was also detected only in the GLI1 + DYRK1A +ATP sample (see Table 2 in [18]). This site is not within any identified domain but may reflect the N-terminal phosphorylation event previously identified by [32P]ATP labeling within residues 1-225 [15].
Fig. 3. MS/MS fragmentation spectra for the unphosphorylated and phosphorylated APSISTVEPK GLI1 peptide.
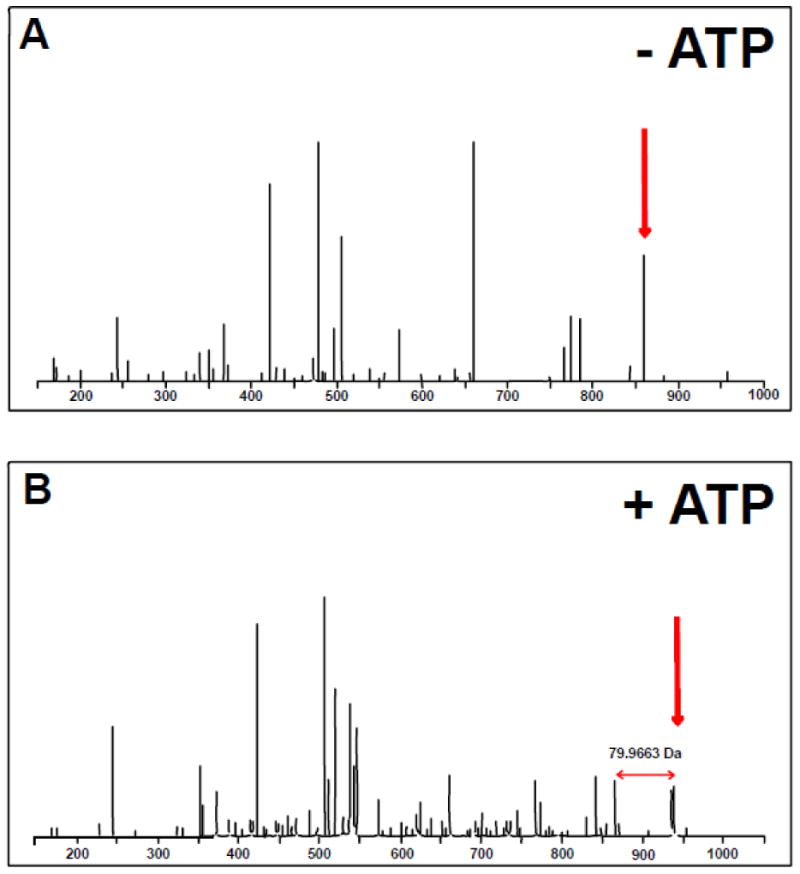
Mass spectra obtained without (A) and with (B) ATP addition during the in vitro kinase assay. The appearance of an additional peak with ATP is highlighted (red arrows). The mass shift corresponds to a difference of 79.9663 Da or 1 phosphorylation event.
Fig. 4. Schematic of GLI1 domains and identified DYRK1A phosphorylation sites.
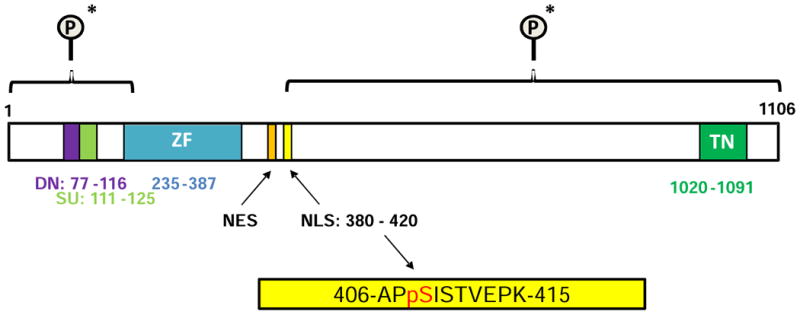
The DYRK1A-mediated GLI1 phospho-peptide (residues 406-415) spanning the Ser408 site is highlighted in yellow and falls within the NLS. The two fragments of GLI1 (N-terminal (1-225) and C-terminal (394-1106)) shown previously by Mao et al. [15] to be phosphorylated by DYRK1A are shown bracketed (*). Dn – degron degradation signal; SU – SUFU-binding domain; ZF – Zinc finger DNA binding domain; NES – Nuclear export signal; NLS – Nuclear localization signal; TN - Transactivation domain. Residue numbering and domain designations based on Carpenter and Lo 2013 [20].
Although the DYRK1A protein was essentially pure as judged by Coomassie staining on SDS-PAGE (Fig. 1B), we could not totally rule out the presence of another kinase contributing to the phosphorylation event. Hence, to further confirm our findings and to ensure that the observed phosphorylation event was mediated by DYRK1A and not a contaminating kinase we also included an additional sample in which GLI1 + DYRK1A + ATP was also incubated in the absence and presence of the highly selective DYRK1A inhibitor harmine [23-25]. As before, samples were incubated for 30 min at 30 °C, run on SDS-PAGE, gel bands excised (Fig. 2B) and tryptic phospho-peptides generated for mass spectrometry (study 2). Again, a number of high confidence phosphorylation sites were identified with those phospho-peptides identified by at least 4 PSM detailed (see Table 4 in [18]). As in study 1, the major difference between the samples was phosphorylation of the tryptic peptide APpSISTVEPK (pSer408) with 10 of 11 peptides (MH+ [Da] = 1108.52836790125) observed only in sample 3 (Fig. 2B, GLI1+DYRK1A+ATP and see Table 4 in [18]), not in the other samples and only once in the sample in which the DYRK1A inhibitor harmine was included demonstrating that this phosphorylation event is mediated by DYRK1A. The second peptide (GPsPSFGVQPGPHDSAR) previously detected in study 1 was not detected above the 4 PSM threshold in this follow up experiment.
Defining a substrate recognition motif for DYRK1A has been challenging with a diversity of sequences identified (see [26]). An in vitro kinase substrate screen found that DYRK1A phosphorylates peptides that have a small hydrophobic residue at the P+1 position and a preference for arginine at P-2 to P-4 [27]. Another study found a preference by DYRK1A for arginine at P-3 and to be “proline-directed”, with a consensus phosphorylation motif for DYRK1A defined as RPX(S/T)P [22]. For the residues surrounding the Ser408 site we identified in GLI1, there are some elements of the consensus sequence with an arginine at position P-3 and an isoleucine at position P+1 (see Supplementary Fig. 1), although isoleucine is classified as being a large volume hydrophobic residue [28]. In general, DYRK1A has been observed to have a broad specificity with a number of DYRK1A substrates having been identified that contain none of these specificity determinants [27].
Deciphering how phosphorylation promotes nuclear import is challenging [29]. Several critical cell cycle regulators and transcription factors are phosphorylated at sites within or adjacent to classical NLS sequences [30]. NLS classically consist of either a single cluster of basic residues (monopartite) [31] or two clusters separated by a 10-12 residue stretch of amino acids (bipartite) [32]. Phosphorylation within or adjacent to the NLS sequence increases the binding affinity for importin α [29]. There is much data on the importance to Hh signaling of NLS-mediated control of GLI proteins in non-vertebrate and vertebrates (reviewed [33]). The transcription factor mediating Hh signaling in Drosophila, Cubitus interruptus (Ci), has a bipartite NLS that was shown to be critical for its nuclear accumulation [34]. Importin α3 was shown to be a nuclear transport receptor for Ci [35], with binding of the GLI1-importin α3 complex by importin β resulting in nuclear transport [33]. Sequence analysis predicted two candidate NLS in GLI1 with mutation of a predicted monopartite at residues 79-84 not affecting GLI1 localization [36]. The other predicted NLS has been localized to the fifth zinc finger and its C-terminal flanking region (spanning 364-410) [37]. Mutation of both these predicted NLSs promoted cytoplasmic accumulation of GLI1 [36]. The GLI1-NLS has been further defined as comprising residues 380-420 [20, 38] (Fig. 4). We have identified a DYRK1A phosphorylation site on GLI1 at Ser408 that falls within this NLS sequence.
Phosphorylation of general nuclear translocation signals at three amino acid Ser-Pro-Ser (SPS) clusters can also promote nuclear accumulation in the absence of canonical NLS [39]. DYRK1A has also been shown to phosphorylate GLI1 in the N-terminal domain at two SPS clusters (S102/104 and S130/32) and that this phosphorylation promotes nuclear transport of GLI1 [40]. Further, when both SPS clusters were mutated, GLI transcriptional activity was not enhanced by DYRK1A [40]. Direct phosphorylation of Ser102 was shown by mass spectrometry [40]. We also identified a tryptic phospho-peptide spanning this SPS (TsPSSLVAFINSR; see Table 2 in [18]). However, in our study, this phosphorylated peptide was detected in both the +ATP and −ATP samples suggesting basal phosphorylation.
Multiple phosphorylation events control GLI1 activity [13, 41, 42]. GLI1 can be retained in the cytoplasm by suppressor of FUSED (SUFU) binding, the major negative regulator of signaling by Hh, or by phosphorylation by PKA at residue Thr 374 of GLI1 [11]. SMO-independent non-canonical activation of GLI1 by mTOR/S6K1 signaling has been reported with S6K1 phosphorylation of GLI1 at Ser84 inhibiting SUFU binding [43]. SUFU binds GLI directly within residues 116-125 of GLI1 [44]. The SUFU binding site falls between the two SPS clusters identified by Schnieder et al. [40] but interestingly, although DYRK1A was shown to promote GLI1/SUFU dissociation, it was independent of the SPS clusters [40]. Direct phosphorylation of GLI1 by the AMP-activated protein kinase (AMPK) in medulloblastoma [45] has been shown to lead to GLI1 destabilization [46]. Three sites for AMPK phosphorylation of GLI1 were identified in the medulloblastoma study; Ser 102, Ser 408 and Thr 1074. Interestingly, two of these sites were those identified by us herein (Ser408) or by others (Ser102; [40]) as DYRK1A-mediated sites on GLI1. In an independent study, phosphorylation of GLI1 by AMPK at Ser408 was shown to be inhibitory leading to GLI1 ubiquitination and proteasomal degradation [45]. Downregulation of GLI1 expression by AMPK has also been observed in heptacellular cancer [47].
In summary, we have followed up on those earlier studies demonstrating that DYRK1A potentiates the nuclear translocation and transcriptional activity of GLI1 [14-17] by using mass spectrometry to identify a site of DYRK1A-mediated phosphorylation on GLI1 at serine 408 which is localized within the nuclear localization sequence of GLI1. This direct DYRK1A-GLI1 interaction within the NLS may prove to be a critical mechanism for understanding GLI1 regulation and requires further study to delineate this activity from those observed at the SPS site on GLI1 and that observed by AMPK at the same site. Future experiments will assess the effect on GLI1 translocation of mutating the Ser408 site. With DYRK1A phosphorylation predicted to activate GLI1 [15], this phospho-site (Ser408) has the potential to be a targetable site for cancers mediated by high levels of GLI1 transcriptional activity.
Supplementary Material
Supplementary Fig. 1. Amino acid sequence of recombinant human GLI1 indicating location of Ser408. Sequence of the C-terminal MYC/DDK-tagged human GLI1 based on NCBI Reference Sequence: NP_005260.1. The tryptic peptide spanning Ser408 (highlighted in red) is shown boxed. The arginine residue at P-3 is shown circled. The myc-DDK tag is shown underlined.
Highlights.
We describe a novel direct DYRK1A phosphorylation site on GLI1 at Ser408.
Harmine a selective DYRK1A inhibitor, blocks DYRK1A-mediated GLI1 phosphorylation.
The Ser408 phospho-site is within the GLI1 nuclear localization sequence (380-420).
This phosphorylation site provides a potential anti-Hh therapeutic target.
Acknowledgments
This study was supported in part by grants from the NIH (R15CA208651, U54CA156735, R41CA174097, P20CA202924) and the Department of Defense (W81XWH-13-1-0141). This research is based in part upon work conducted using the UNC Michael Hooker Proteomics Center, which is supported in part by the NIH-NCI Grant No. CA016086 to the Lineberger Comprehensive Cancer Center. We thank David Smalley and Laura Herring from the UNC Michael Hooker Proteomics Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Kasper M, Regl G, Frischauf AM, Aberger F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer. 2006;42:437–445. doi: 10.1016/j.ejca.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Lo HW. The Human Glioma-Associated Oncogene Homolog 1 (GLI1) Family of Transcription Factors in Gene Regulation and Diseases. Current Genomics. 2010;11:238. doi: 10.2174/138920210791233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang J, Hui Cc. Hedgehog signaling in development and cancer. Developmental Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 7.Yang SH, Hsu CH, Lee JC, Tien YW, Kuo SH, Cheng AL. Nuclear expression of glioma-associated oncogene homolog 1 and nuclear factor-κB is associated with a poor prognosis of pancreatic cancer. ONCOLOGY. 2013;85:86–94. doi: 10.1159/000353452. [DOI] [PubMed] [Google Scholar]
- 8.Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A. Cyclopamine-Mediated Hedgehog Pathway Inhibition Depletes Stem-Like Cancer Cells in Glioblastoma. Stem cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Yang W, Yang Q, Zhou S. Nuclear localization of GLI1 and elevated expression of FOXC2 in breast cancer is associated with the basal-like phenotype. Histology and Histopathology. 2012;27:475. doi: 10.14670/HH-27.475. [DOI] [PubMed] [Google Scholar]
- 10.Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt B, Toftgård R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear–cytoplasmic shuttling of Gli-1. Nature Cell Biology. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 11.Sheng T, Chi S, Zhang X, Xie J. Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. Journal of Biological Chemistry. 2006;281:9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- 12.Aberger F, Ruiz i Altaba A. Seminars in Cell & Developmental Biology. Elsevier; 2014. Context-dependent signal integration by the GLI code: The oncogenic load, pathways, modifiers and implications for cancer therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Research. 2013;23:186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC, Peaceman D, Özdemirli M, Rodriguez O, Macdonald TJ. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. The Journal of Clinical Investigation. 2011;121:148. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, Wu G, Wu D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. Journal of Biological Chemistry. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 16.Shimokawa T, Rahman MFU, Tostar U, Sonkoly E, Ståhle M, Pivarcsi A, Palaniswamy R, Zaphiropoulos PG. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biology. 2013;10:321. doi: 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgård R, Zaphiropoulos PG. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. Journal of Biological Chemistry. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehe BK, Lamson DR, Tarpley M, Onyenwoke RO, Graves LM, Williams KP. Data on phospho-peptide mass spectrometry analysis for DYRK1A-mediated phosphorylation of GLI1. Data in Brief. 2017 doi: 10.1016/j.dib.2017.09.057. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins D, Pappin D, Creasy D, Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter RL, Lo HW. Hedgehog pathway and GLI1 isoforms in human cancer. Discovery Medicine. 2012;13:105. [PMC free article] [PubMed] [Google Scholar]
- 21.Frost D, Meechoovet B, Wang T, Gately S, Giorgetti M, Shcherbakova I, Dunckley T. β-carboline compounds, including harmine, inhibit DYRK1A and tau phosphorylation at multiple Alzheimer's disease-related sites. PLoS One. 2011;6:e19264. doi: 10.1371/journal.pone.0019264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. Journal of Biological Chemistry. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 23.Adayev T, Wegiel J, Hwang YW. Harmine is an ATP-competitive inhibitor for dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) Archives of Biochemistry and Biophysics. 2011;507:212–218. doi: 10.1016/j.abb.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Göckler N, Jofre G, Papadopoulos C, Soppa U, Tejedor FJ, Becker W. Harmine specifically inhibits protein kinase DYRK1A and interferes with neurite formation. FEBS Journal. 2009;276:6324–6337. doi: 10.1111/j.1742-4658.2009.07346.x. [DOI] [PubMed] [Google Scholar]
- 25.Bain J, Plater L, Elliott M, Shpiro N, Hastie C, Mclauchlan H, Klevernic I, Arthur J, Alessi D, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papadopoulos C, Arato K, Lilienthal E, Zerweck J, Schutkowski M, Chatain N, Müller-Newen G, Becker W, de la Luna S. Splice variants of the dual specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. Journal of Biological Chemistry. 2011;286:5494–5505. doi: 10.1074/jbc.M110.157909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soundararajan M, Roos AK, Savitsky P, Filippakopoulos P, Kettenbach AN, Olsen JV, Gerber SA, Eswaran J, Knapp S, Elkins JM. Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure. 2013;21:986–996. doi: 10.1016/j.str.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pommié C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. Journal of Molecular Recognition. 2004;17:17–32. doi: 10.1002/jmr.647. [DOI] [PubMed] [Google Scholar]
- 29.Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Communication and Signaling. 2010;8:1. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 32.Robbins J, Dilwortht SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 33.Hatayama M, Aruga J. 4 Gli Protein Nuclear Localization Signal. Vitamins and hormones. 2012;88:73. doi: 10.1016/B978-0-12-394622-5.00004-3. [DOI] [PubMed] [Google Scholar]
- 34.Wang QT, Holmgren RA. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development. 1999;126:5097–5106. doi: 10.1242/dev.126.22.5097. [DOI] [PubMed] [Google Scholar]
- 35.Seong KH, Akimaru H, Dai P, Nomura T, Okada M, Ishii S. Inhibition of the nuclear import of cubitus interruptus by roadkill in the presence of strong hedgehog signal. PloS one. 2010;5:e15365. doi: 10.1371/journal.pone.0015365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui Cc. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 37.Hatayama M, Tomizawa T, Sakai-Kato K, Bouvagnet P, Kose S, Imamoto N, Yokoyama S, Utsunomiya-Tate N, Mikoshiba K, Kigawa T. Functional and structural basis of the nuclear localization signal in the ZIC3 zinc finger domain. Human molecular genetics. 2008;17:3459–3473. doi: 10.1093/hmg/ddn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter RL, Lo HW. Identification, functional characterization, and pathobiological significance of GLI1 isoforms in human cancers. Vitamins and hormones. 2012;88:115. doi: 10.1016/B978-0-12-394622-5.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Molecular cell. 2008;31:850–861. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Schneider P, Bayo-Fina JM, Singh R, Dhanyamraju PK, Holz P, Baier A, Fendrich V, Ramaswamy A, Baumeister S, Martinez ED. Identification of a novel actin-dependent signal transducing module allows for the targeted degradation of GLI1. Nature communications. 2015;6 doi: 10.1038/ncomms9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell reports. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C [igr]/[lgr] regulates the growth of basal cell carcinomas. Nature. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. Journal of Biological Chemistry. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 45.Di Magno L, Basile A, Coni S, Manni S, Sdruscia G, D'Amico D, Antonucci L, Infante P, De Smaele E, Cucchi D, Ferretti E, Di Marcotullio L, Screpanti I, Canettieri G. The energy sensor AMPK regulates Hedgehog signaling in human cells through a unique Gli1 metabolic checkpoint. Oncotarget. 2016;7:9538–9549. doi: 10.18632/oncotarget.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li YH, Luo J, Mosley YY, Hedrick VE, Paul LN, Chang J, Zhang G, Wang YK, Banko MR, Brunet A, Kuang S, Wu JL, Chang CJ, Scott MP, Yang JY. AMP-Activated Protein Kinase Directly Phosphorylates and Destabilizes Hedgehog Pathway Transcription Factor GLI1 in Medulloblastoma. Cell reports. 2015;12:599–609. doi: 10.1016/j.celrep.2015.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q, Liu X, Zheng X, Yao Y, Wang M, Liu Q. The transcriptional activity of Gli1 is negatively regulated by AMPK through Hedgehog partial agonism in hepatocellular carcinoma. International journal of molecular medicine. 2014;34:733–741. doi: 10.3892/ijmm.2014.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Amino acid sequence of recombinant human GLI1 indicating location of Ser408. Sequence of the C-terminal MYC/DDK-tagged human GLI1 based on NCBI Reference Sequence: NP_005260.1. The tryptic peptide spanning Ser408 (highlighted in red) is shown boxed. The arginine residue at P-3 is shown circled. The myc-DDK tag is shown underlined.


