Growing evidence in vertebrates predicts that cellular heme levels in animals are not only maintained by a cell’s internal capacity for heme synthesis in a cell-autonomous manner, but also by an inter-organ heme trafficking network through cell-nonautonomous regulation. Using C. elegans, a genetically and optically amenable animal model for visualizing heme-dependent signaling, we show that HRG-7, a protein with homology to aspartic proteases, mediates inter-organ signaling between the intestine and extra-intestinal tissues. Intestinal HRG-7 functions as a secreted signaling factor during heme starvation in extra-intestinal tissues and is regulated through a DBL-1/BMP-dependent signal from neurons. Given the evidence that vertebrate homologs exist for each of the components of the HRG-7-mediated signaling pathway, it is conceivable that the cell-nonautonomous signaling framework that we uncovered in C. elegans may have functional relevance for inter-organ regulation of iron and heme metabolism in humans.
Heme is an iron-containing porphyrin that is required as a prosthetic group in a variety of proteins crucial for cellular functions including globins for gas-binding, cytochromes for electron transfer, and guanylate cyclase for cellular signaling1, 2. It is generally accepted that cellular requirements for heme in animals are fulfilled by the cell’s internal capacity to regulate and synthesize its own heme, that is, cell-autonomous regulation3. However, several lines of evidence in humans, mice and zebrafish support the existence of a cell-nonautonomous regulation by a systemic heme communication system even though they are capable of intracellular heme synthesis4–8.
C. elegans is ideal to determine if such systemic signaling pathways exist as it overcomes several obstacles encountered in mammalian models9. Worms are heme auxotrophs permitting external control of intracellular heme levels, can be manipulated and observed live at subcellular resolution, and are optically transparent for in vivo monitoring of heme signals between tissues during development9, 10. Herein, we show that HRG-7 mediates non-cell autonomous heme signaling by functioning as a secreted factor and communicating intestinal heme status with extra-intestinal tissues. Reciprocally, a DBL-1/BMP-dependent signal from the neurons transcriptionally regulates both hrg-7 and intestinal heme transport. Our work provides a cell biological model for how organs communicate their heme status to regulate metabolism at the organismal level11.
RESULTS
A genome-wide RNAi screen identifies HRG-7 as a regulator of systemic heme homeostasis
Our previous studies revealed that C. elegans imports heme into the intestine by the concerted functions of HRG-4 on the plasma membrane and HRG-1 on endo-lysosomal membranes, which mobilizes heme from vesicles, while the ABCC-type transporter MRP-5 is the major intestinal heme exporter. Loss of hrg-4 results in reduced heme transport into the intestine, whereas loss of mrp-5 causes heme accumulation within the intestine while extra-intestinal tissues become heme-deprived8, 12. Depletion of either gene results in robust upregulation of GFP in the transgenic C. elegans heme sensor strain IQ6011 (Phrg-1::GFP), which shows strong GFP expression under low heme conditions (Figure 1a). While this result is expected for depletion of hrg-4, it is paradoxical for mrp-5 depletion as intestinal heme accumulation should normally suppress this GFP reporter12–14. One potential explanation for GFP reporter upregulation is that the heme sensor strain responds not only to intestinal heme deficiency, as in the case of hrg-4 depletion, but also to heme starvation signals emanating from the extra-intestinal tissues due to mrp-5 deficiency8, a concept originally proposed to exist for the regulation of inter-tissue copper and iron homeostasis in mammals15–17.
Figure 1. Heme homeostasis is regulated by the aspartic protease homolog hrg-7.
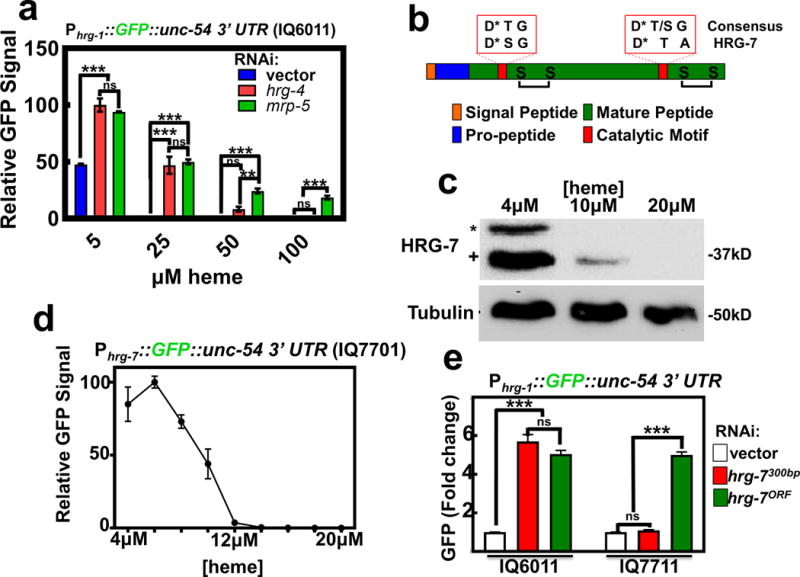
a) GFP fluorescence quantified from strain IQ6011 (Phrg-1::GFP) fed dsRNA against control vector, hrg-4, or mrp-5 at 5, 25, 50, and 100 μM heme. GFP was quantified using COPAS BioSort. GFP is represented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA). See Supplementary Table 3 for statistics source data. b) Cartoon depicting general features of aspartic proteases. * indicates catalytic Asp residues. c) Immunoblot analysis of HRG-7 expression in N2 grown in increasing heme concentrations. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. * indicates pro-HRG-7. + indicates mature HRG-7. Unprocessed blots are shown in Figure S6. Data representative of three independent experiments. d) GFP expression in strain IQ7701 (Phrg-7::GFP) grown in varying heme concentrations. GFP was quantified with the COPAS BioSort. GFP is represented as relative intensity on a scale of 1-100. Graph represents the mean and SEM of three biological independent experiments. N=100 worms per treatment per experiment. e) GFP fluorescence quantified from IQ6011 (Phrg-1::GFP) and IQ7711 (Pvha-6::HRG-7PR:ICS::mCherry; Phrg-1::GFP) fed dsRNA against control vector, hrg-7300bp, and hrg-7ORF at 10 μM heme. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (one-way ANOVA). See Supplementary Table 3 for statistics source data.
In order to identify factors involved in inter-tissue heme homeostasis, we exploited the Phrg-1::GFP heme sensor worms to perform a feeding RNAi screen by systematically depleting 18,533 genes that encompasses ~93% of the worm genome. To eliminate genes that regulated GFP non-specifically, we rescreened 1,641 positive hits in the Pvha-6::GFP worms, which expresses intestinal GFP independent of heme. GFP was quantified using COPAS BioSort flow cytometry; genes were considered candidates if knockdown resulted in regulation of Phrg-1::GFP by ≥ 2 fold (log scale) compared to Pvha-6::GFP; we found 177 genes that preferentially regulated Phrg-1::GFP (Supplementary Table 4). A comparison of these candidate genes with the 288 heme-responsive genes that we had previously identified from microarray experiments14 revealed three overlapping genes between the two datasets: mrp-5, R193.2, and C15C8.3. Both mrp-5 and R193.2 encode multiple transmembrane domain-containing proteins while C15C8.3 encoded a soluble protein containing a signal peptide, an indication that the protein could potentially be secreted and thus capable of cell-cell communication. We termed C15C8.3 as heme responsive gene 7 (hrg-7).
HRG-7 is a putative member of the A1 family of aspartic proteases. Like other members of this family, HRG-7 contains a predicted signal peptide, a pro-region that is cleaved during protein maturation, conserved cysteines for disulfide bonds, and an active site containing aspartic acids essential for proteolytic cleavage (Figure 1b). Antibodies generated against the carboxy-terminal 17 amino acid residues of HRG-7 detected two forms of HRG-7 (corresponding to the pro and mature forms of the predicted proteins) on immunoblots of lysates from worms grown in low heme but not in 20 μM heme (Figure 1c). Consistent with the effect of heme on HRG-7, genomic sequence alignment of hrg-7 from three Caenorhabditis species (C. elegans, C. briggsae, and C. remanei) revealed a conserved cis-regulatory element termed HEme Responsive Element (HERE) in the 5’ upstream region (Supplementary Figure 1a)18. The hrg-7 HERE, as previously demonstrated for the hrg-1 promoter, is flanked by GATA elements that are responsible for intestinal gene expression in C. elegans19. Indeed, transgenic worms bearing the Phrg-7::GFP transcriptional reporter (strain IQ7701) showed strong GFP expression in the intestine only under low heme conditions that was suppressed either by heme in a dose-dependent manner (Figure 1d, Supplementary Figure 1b) or by mutagenesis of the HERE (Supplementary Figure 1c).
To determine whether HRG-7 regulates intestinal heme homeostasis, we expressed an RNAi-resistant form of hrg-7 (hrg-7PR) from the intestinal-specific vha-6 promoter in the Phrg-1::GFP heme sensor worms. The hrg-7PR transgene contains a recoded portion within its open-reading frame (ORF) such that it is resistant to dsRNA directed against the first 300 bp (hrg-7300bp), but it is susceptible to dsRNA directed against the remainder of the ORF (hrg-7ORF) (Supplementary Figure 1d & 1e). Thus, hrg-7PR expression permits interrogation of transgene function in the presence or absence of endogenous hrg-7. Intestinally-expressed hrg-7PR restored Phrg-1::GFP expression to WT levels when hrg-7 was depleted by hrg-7300bp, but not with hrg-7ORF (Figure 1e). These results show that HRG-7 is required to maintain intestinal heme homeostasis under low heme conditions.
HRG-7 is secreted from the intestine for cell-nonautonomous regulation of intestinal heme homeostasis
Because hrg-7 depletion in the Phrg-1::GFP heme sensor worms resulted in a heme deficiency readout even in the presence of excess heme, we examined whether intestinal heme uptake was aberrant when hrg-7 is depleted. RNAi depletion of heme importers, hrg-1 and hrg-4, resulted in the expected alteration in accumulation of the fluorescent heme analog zinc mesoporphyrin (ZnMP), while hrg-7 knockdown showed no effect on the ZnMP signal (Supplementary Figure 2a)13. We next examined HRG-7 protein localization in transgenic worms expressing a Phrg-7::HRG-7::mCherry translational reporter. Unexpectedly, little or no signal was detected for HRG-7::mCherry within the intestine of worms grown in low heme. Instead, HRG-7::mCherry localized to distinct punctate structures near the anterior and posterior regions of the worm (Figure 2a). HRG-7::mCherry also accumulated within coelomocytes, macrophage-like cells that take up proteins and compounds from the body cavity, an attribute that is typically exploited as a marker for secretion into the pseudocoelom (Figure 2a)20. This unexpected localization was not due to ectopic expression of the hrg-7 transgene as HRG-7 accumulated in similar extra-intestinal structures when directed from the intestinal-specific vha-6 promoter (Supplementary Figure 2b & 2c)21, 22. Co-expression with the coelomocyte marker Punc-122::GFP verified HRG-7::mCherry secretion (Supplementary Figure 2c). Although fluorescent-tagged secreted proteins could accumulate non-specifically in coelomocytes, the distinct punctate localization of HRG-7::mCherry was highly specific as there was no overlap with either full-length or signal peptide HRG-3::mCherry, another secreted protein that delivers intestinal heme to extra-intestinal tissues (Supplementary Figure 2d)23.
Figure 2. HRG-7 is secreted and functions in a cell-nonautonomous manner.
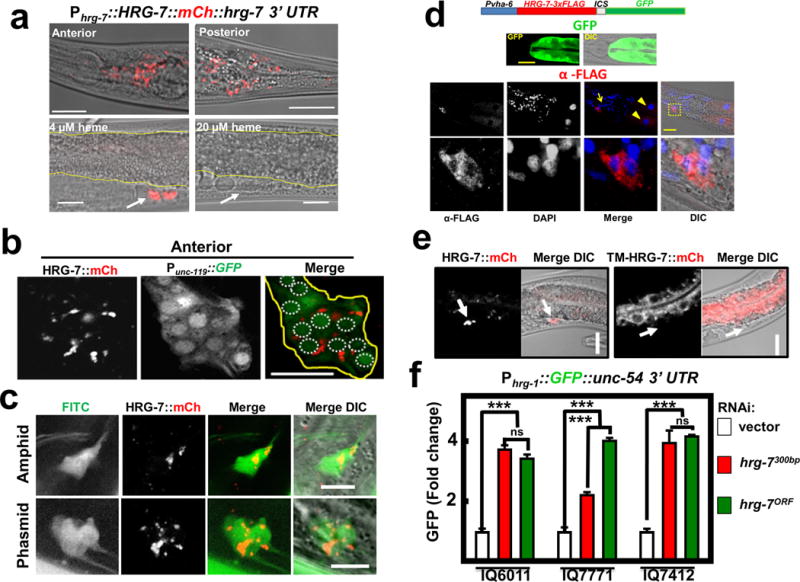
a) mCherry expression in IQ7777 (Phrg-7::HRG-7::mCherry). Intestine is indicated by yellow lines, coelomocytes are indicated by a white arrow. Scale bar = 20 μm. Images are representative of three independent experiments. b) Coexpression of the pan-neuronal GFP marker Punc-119::GFP and Pvha-6::HRG-7::mCherry. Image is representative of the anterior of adults. Yellow outline indicates nerve ring. White dotted circles indicate nerve ring nuclei. Scale bar = 10 μM. Images are representative of three independent experiments. c) IQ7670 (Pvha-6::HRG-7::mCherry) were pulsed with 10 μg /ml FITC for 3 hours, then analyzed by confocal microscopy. Scale bar = 5 μM. Images are representative of three independent experiments. d) Fluorescence and immunofluorescence images of worms expressing Pvha-6::HRG-7-3XFLAG::ICS::mCherry. GFP expression was analyzed in live worms. HRG-7-3XFLAG expression was analyzed in fixed worms incubated with M2 α-FLAG antibody followed by incubation with Tetramethyl Rhodamine 5 (and 6)-Isothiocyanate (TRITC) conjugated goat-α-mouse antibody and DAPI. Arrow indicates neuron nuclei. Arrowhead indicates intestine nuclei. Bottom panels are zoomed-in images of boxed region indicated in the middle panels. Images are representative of two independent experiments. Scale bar = 20μm. e) mCherry fluorescence in worms expressing Pvha-6::HRG-7PR::mCherry or Pvha-6::TM-HRG-7PR::mCherry. Arrows indicate middle coelomocytes near the vulva. Scale bar = 20μm. Images are representative of three independent experiments. f) GFP fluorescence quantified from IQ6011 (Phrg-1::GFP), IQ7771 (Pvha-6::HRG-7PR::mCherry; Phrg-1::GFP), and IQ7412 (Pvha-6::TM-HRG-7PR::mCherry; Phrg-1::GFP) fed dsRNA against vector, hrg-7300bp, and hrg-7ORF at 10 μM heme. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (one-way ANOVA). See Supplementary Table 3 for statistics source data.
To identify the tissue location of secreted HRG-7, we generated double transgenic worms co-expressing Pvha-6::HRG-7::mCherry plus a marker for either body wall muscle, hypodermis, pharyngeal muscle, or neurons (Figure 2b & Supplementary Figure 2e). HRG-7::mCherry co-localized strongly with only the pan-neuronal GFP maker (Punc-119::GFP) (Figure 2b). Reconstruction of 0.5 μm-thin confocal sections revealed that secreted HRG-7::mCherry was located within puncta in a bundle of head neurons near the nerve ring. In C. elegans, chemosensory neurons appear to have a critical role in nutrient homeostasis and project cilia to the external medium permitting detection by uptake of 5-fluorescein isothiocyanate (FITC) dye24–26. HRG-7::mCherry co-localized with these FITC-positive neurons in both the anterior (amphids) and posterior (phasmids) of the worm, indicating that HRG-7 preferentially localizes to sensory neurons (Figure 2c). To corroborate HRG-7 localization, we generated Pvha-6::HRG-7-3xFLAG::ICS::GFP. In this strain, gfp and hrg-7-3xFLAG are initially transcribed as a single polycistronic pre-mRNA but the intercistronic (ICS) sequence is SL2 trans-spliced generating two mRNAs that are translated independently, making this transgenic construct ideal for separating mRNA expression from protein localization27. Immunohistochemistry with a commercial monoclonal FLAG antibody detected HRG-7::3xFLAG in both the intestine and neurons, but the GFP fluorescence was limited to the intestine as expected (Figure 2d). These results further confirm that HRG-7 is synthesized in the intestine but is secreted and localizes to neuronal structures.
To determine whether intestinally produced HRG-7 functions intracellularly (autonomous) or extracellularly (non-autonomous), we tethered HRG-7::mCherry to intestinal cells by mutating the signal peptidase cleavage site (TM-HRG-7), as retention of the signal peptide should act as a transmembrane anchor. Predictably, TM-HRG-7 was not secreted, but rather retained within the worm intestine (Figure 2e). We then evaluated if TM-HRG-7 was capable of suppressing the heme deficiency signal in heme-sensor worms by using RNAi-resistant transgenes. The Phrg-1::GFP sensor was significantly repressed only when HRG-7::mCherry was secreted but not when it was tethered to intestinal cells implying that HRG-7 functions extracellularly to regulate intestinal heme responsiveness (Figure 2f).
We next determined the source of the heme deficiency signal in hrg-7 mutant worms by depleting hrg-7 in combination with either the heme importer hrg-4 or the heme exporter mrp-5 in the Phrg-1::GFP heme-sensor strain. Co-RNAi knockdown of either hrg-4 and hrg-7 or hrg-4 and mrp-5 resulted in an enhanced heme deficiency signal phenotype that was not observed when hrg-7 and mrp-5 were co-depleted (Supplementary Figure 2f). Because mrp-5 depletion causes extra-intestinal heme deficiency secondary to intestinal heme overload, we infer that the lack of a synergistic effect in hrg-7 and mrp-5 double RNAi worms is due to both genes regulating intestinal heme responsiveness by altering extra-intestinal heme homeostasis.
Functionally mature HRG-7 is produced solely by the intestine
To evaluate HRG-7 secretion, we expressed HRG-7::mCherry from the inducible hsp-16.2 promoter. The hsp-16.2 promoter is activated in several tissues, including the intestine, following a transient exposure of worms to 37°C28. Approximately 180 min post heat-shock, HRG-7::mCherry was clearly visible in the pseudocoelom and coelomocytes but not within the intestine (Figure 3A & 3B, Supplementary Figure 3a) affirming that HRG-7 is secreted from the intestine shortly after protein translation.
Figure 3. HRG-7 secretion and maturation is regulated by specific trafficking factors.
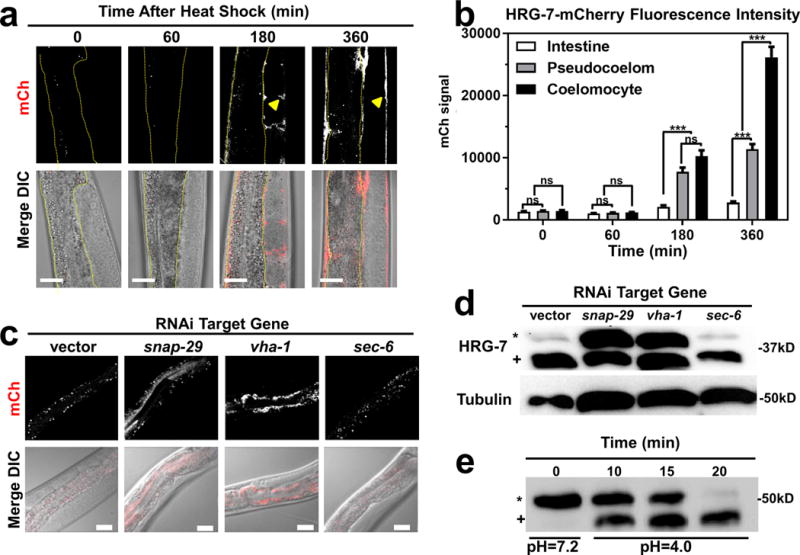
a) mCherry expression in strain IQ7170 (Phsp-16.2::HRG-7::mCherry) placed in 37°C for 30 minutes, then 20°C for 60, 180, and 360 minutes. Dotted yellow lines outline the intestine. Arrowheads indicate HRG-7::mCherry in the pseudocoelom. Scale bar = 20 μm. Images are representative of three independent experiments. b) Quantification of mCherry in coelomocytes, pseudocoelom, and intestine of IQ7170 following heat shock. For quantification, 10 images, consisting of 10-15 Z stacks per image, were used for each timepoint. ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA). mCherry signal is in arbitrary unites. Graph and statistics are from a single experiment. The experiment was repeated one time with similar results. c) mCherry expression in strain IQ7670 fed dsRNA against vector, snap-29, vha-1, and sec-6. The small puncta visible in all panels are autofluorescent gut granules in the intestine. Scale bar = 20 μm. Images are representative of three independent experiments. d) Immunoblot analysis of strain N2 fed dsRNA against vector, snap-29, vha-1, and sec-6. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. * indicates pro-HRG-7. + indicates mature HRG-7. Unprocessed blots are shown in Figure S6. Data are representative of three independent experiments. e) Immunoblot analysis of lysate prepared from strain IQ7370 (Pvha-6::HRG-7::3xFLAG) at pH 7.2 and pH 4. Membranes were probed with monoclonal anti-FLAG antibody and then incubated with HRP-conjugated anti-mouse secondary antibody. * indicates pro-HRG-7-3xFLAG. + indicates mature HRG-7-3xFLAG. Unprocessed blots are shown in Figure S6. Data are representative of three independent experiments.
To gain a better understanding of factors that regulate HRG-7 production and export, we RNAi-depleted 45 genes that encode known regulators of endocytosis and secretion in the Pvha-6::HRG-7::mCherry reporter strain29. We found that depletion of 11 trafficking factors caused HRG-7::mCherry to mis-localize (Supplementary Table 1). For example, depletion of genes encoding the SNARE (snap-29) or vacuolar ATPase (vha-1) components resulted in accumulation of HRG-7::mCherry either in the intestine (Figure 3c) or as an immature form in which the pro-region was not cleaved (Figure 3d). Consistent with aberrant HRG-7 secretion and processing, RNAi of snap-29 also resulted in increased GFP expression in the Phrg-1::GFP heme sensor strain (Supplementary Figure 5e). These results demonstrate that HRG-7 maturation and secretion require endosomal fusion and acidification. Because genetic disruption of the vacuolar ATPase complex can affect both secretory and acidification processes30, we analyzed the dynamics of HRG-7 maturation in vitro. Lysates from transgenic worms expressing Pvha-6::HRG-7-3xFLAG were exposed to either neutral or acidic pH at 25° C for 30 min. While HRG-7-3xFLAG remained in the pro-form at pH 7.2, at pH 4 the majority of pro-HRG-7-3xFLAG was processed by cleavage within 20 min (Figure 3e), supporting a role for an acidic environment for HRG-7 maturation.
To examine whether the intestinal source of HRG-7 could be substituted by expressing hrg-7 either directly in the recipient tissue (neuron) or from another tissue (body wall muscle), we expressed hrg-7PR::ICS::mCherry from the unc-119 or myo-3 promoters, respectively. The SL2 trans-spliced mCherry expression revealed that the transgene was indeed expressed in the appropriate tissue, but mature HRG-7 protein could only be detected when expressed from the intestine (Supplementary Figure 3b & 3c). Consequently, only intestinal hrg-7 was capable of restoring GFP expression in the Phrg-1::GFP heme sensor worms (Supplementary Figure 3d). Together, these results imply that the intestine is the sole tissue source of functional HRG-7.
The heme-signaling function of HRG-7 does not require conserved active site aspartates
The hrg-7(tm6801) mutant contains an in-frame deletion of 40-amino acids encompassing a conserved tyrosine flap shown to be critical for the function of aspartic proteases (Figure 4a; Supplementary Figure 4a & 4b). The hrg-7(tm6801) mutants have undetectable levels of functionally-mature HRG-7 protein as identified by immunoblotting (Supplementary Figure 4c). Longer exposure of the immunoblots revealed mutant HRG-7 migrating around the same molecular weight as the wild type protein. This would be expected if the mutant HRG-7 retained the 30 amino acid pro-peptide, offsetting much of the size reduction expected for a 40 amino acid deletion. We speculate that the higher molecular weight band in the hrg-7(tm6801) mutant could be misfolded aggregates of mutant HRG-7. hrg-7(tm6801) worms showed a severe heme-dependent growth defect and a striking heme deficiency signal in the Phrg-1::GFP heme-sensor worms, phenotypes that were fully suppressed by co-expression of a transgene encoding the wildtype HRG-7 protein (Figure 4b and Supplementary Figure 4d). Importantly, qRT-PCR confirmed that endogenous hrg-1 mRNA was also significantly upregulated in the hrg-7(tm6801) mutants (Figure 4c), reproducing the GFP upregulation observed in the Phrg-1::GFP heme sensor worms (Figure 1e and Supplementary Figure 4d).
Figure 4. Conserved aspartic acid residues are dispensable for HRG-7 function in heme homeostasis.
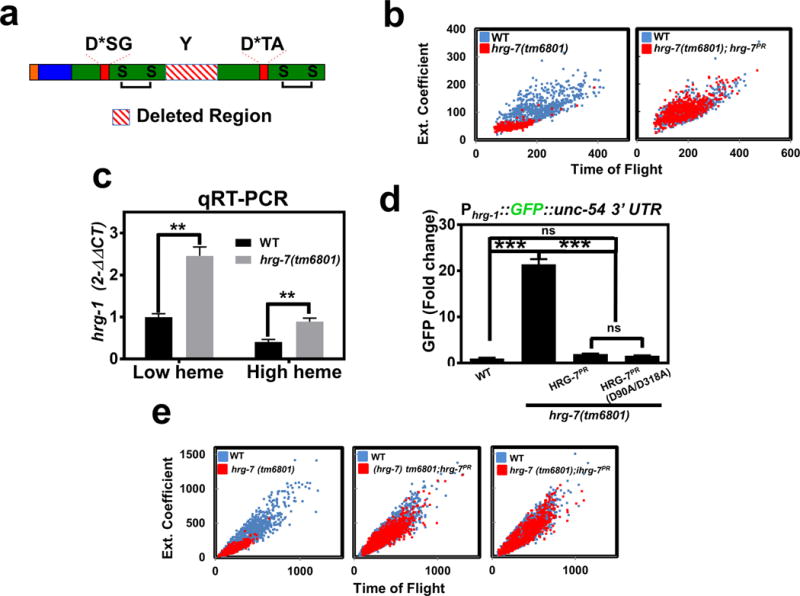
a) Cartoon of HRG-7 depicting the truncation resulting from the tm6801 allele, which encompasses the conserved tyrosine of the flap region. b) Size quantification (400-800 worms per strain) of wildtype worms, hrg-7(tm6801) mutant worms, or hrg-7(tm6801) mutants expressing transgenic Pvha-6::hrg-7PR grown on RP523 bacteria supplemented with 1 μM heme for two generations. F1 worms were harvested as wild type were becoming L4 larvae. Graph is representative of a single experiment. Two experiments were repeated independently with similar results. c) qRT-PCR of hrg-1 in hrg-7(tm6801) mutant worms or wildtype broodmates fed OP50 (low heme) or OP50 with 50 μM heme (high heme). Graph represents the mean and SEM of three biological independent experiments. hrg-1 expression was normalized to gpd-2. Data is presented as fold change compared to hrg-1 expression in wildtype broodmates grown with low heme. ***P<0.001 (unpaired two-tailed t-test). See Supplementary Table 3 for statistics source data. d) GFP fluorescence quantified from IQ6011 expressing WT hrg-7, hrg-7(tm6801) and transgenic Pvha-6::hrg-7PR, or hrg-7(tm6801) and transgenic Pvha-6::hrg-7PR(D90A/D318A) and grown on OP50. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to wild type (WT) worms. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (one-way ANOVA). See Supplementary Table 3 for statistics source data. e) Size quantification of strains (500-1000 worms per strain) from panel F grown on RP523 bacteria supplemented with 4 μM heme for two generations. Graph is representative of a single experiment. Two experiments were repeated independently with similar results.
Typical aspartic acid proteases utilize a catalytic dyad of two aspartic acids for proteolysis, a feature shared by other A1 aspartic protease family members that include cathepsin D, cathepsin E, pepsin, and renin. In silico homology modeling predicts HRG-7 to be a bi-lobed structure with each lobe contributing an active site aspartic acid residue and a conserved disulfide bond (Supplementary Figure 4b). To determine whether the signaling function of HRG-7 was dependent on these aspartic acids, we mutated D90 and D318 to alanines. Surprisingly, a D90A/D318A double mutant fully restored GFP expression to wildtype levels in the Phrg-1::GFP heme sensor worms crossed into hrg-7(tm6801) mutants (Figure 4d). Furthermore, the D90A/D318A mutant was comparable to wildtype in suppressing the growth defects of hrg-7(tm6801) mutants (Figure 4e). The phenotypic rescue by the D90A/D318A allele was not due to overexpression of the transgene as the steady state level of the mutant protein was even lower than endogenous HRG-7 (Supplementary Figure 4e). Together, these results demonstrate that HRG-7 is essential for heme-dependent growth and that the signaling function of HRG-7 is not dependent on conserved aspartates in the putative active site.
DBL-1/BMP regulates hrg-1 and hrg-7 through neuron-to-intestine signaling
Our results show that intestinal HRG-7 is secreted and localizes distally to sensory neurons and that the loss of HRG-7, or obstructing its secretion, causes aberrant intestinal heme homeostasis. In C. elegans, the intestines are physically separated from most extra-intestinal tissues by the pseudocoelom, a fluid-filled body cavity that bathes the internal organs31, 32. This raised the possibility that an extra-intestinal regulatory signal might reciprocally regulate intestinal heme homeostasis through a secreted, diffusible factor(s). To identify this regulatory factor, we screened 117 genes that represented 80% of the predicted secreted morphogens and neuropeptides in C. elegans using RNAi depletion in the RNAi hypersensitive heme sensor strain Phrg-1::GFP; lin-15b(n744) (IQ6015). The lin-15b(n744) mutation allows for more efficient knockdown in tissues that are normally resistant to RNAi, such as some neurons and the pharynx33. Quantification of intestinal GFP fluorescence for each of the 117 gene knockdowns (n=120 worms) using the COPAS Biosorter identified dbl-1, encoding a bone morphogenic protein-5 (BMP5) homolog, as the top hit (Figure 5a, Supplementary Table 2). Consistent with our RNAi screen results and published microarray results from dbl-1 mutant worms, hrg-1 mRNA was significantly upregulated in dbl-1(nk3) mutant worms compared to wildtype broodmate controls (Figure 5b)34.
Figure 5. DBL-1/BMP regulates hrg-1 and hrg-7 through neuron-to-intestine signaling.
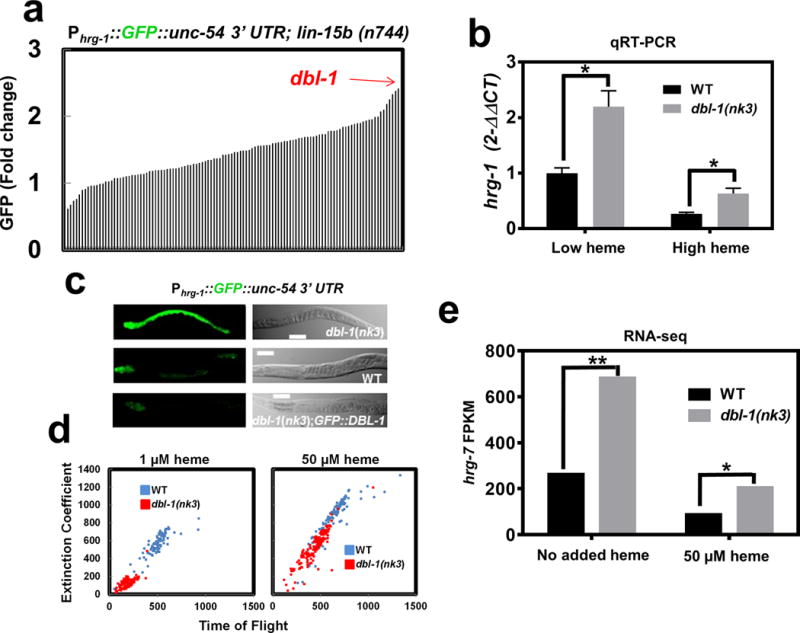
a) GFP quantification (mean of 120 worms) in strain IQ6015 [Phrg-1::GFP; lin-15b(n744)] fed dsRNA against 117 genes encoding secreted signaling factors. Y-axis is GFP fold change compared to worms fed control vector. GFP was quantified using COPAS BioSort. b) qRT-PCR of hrg-1 in dbl-1(nk3) mutant worms or WT broodmates fed OP50 (low heme) or OP50 with 50 μM heme (high heme). Graph represents the mean and SEM from five biological independent experiments. hrg-1 expression was normalized to gpd-2. Data is presented as fold change compared to hrg-1 expression in wildtype broodmates grown with low heme. ***P<0.001, **P<0.01, *P<0.05 (unpaired two-tailed t-test). See Supplementary Table 3 for statistics source data. c) Fluorescence images of IQ6011, IQ6311, and IQ6312 grown on OP50. Scale bar = 20 μm. Images are representative of three independent experiments. d) Size analyses (100-150) worms per treatment) quantified from IQ6011 and IQ6311 fed RP523 E. coli grown in l μM or 50 μM heme for 96 or 72 hours, respectively. Size was analyzed using COPAS BioSort. Graph is representative of a single experiment. Two experiments were repeated independently with similar results. e) FPKM values for hrg-7 from RNA-seq of dbl-1(nk3) mutants compared to WT broodmates. Graph represents the mean of FPKM values obtained from two biological independent experiments. ***P<0.001, **P<0.01, *P<0.05 (False Discovery Rate). See Supplementary Table 3 for statistics source data.
DBL-1 is a secreted signaling factor for which circulating levels of this morphogen are a better indicator of its function than tissue-specific expression levels35. Transcriptional reporter fusions expressing an integrated Pdbl-1::GFP transgene showed dbl-1 expression is restricted to neurons (Supplementary Figure 5a)36. However, a dbl-1::GFP translational fusion has also been reported to be expressed in neurons as well as body wall muscles and the pharyngeal region37, although we and others observe only neuronal expression (Supplementary Figure 5a). Indeed, analyses of several integrated lines expressing a recently synthesized GFP-tagged DBL-1 showed only neuronal expression38.
We analyzed heme sensing in dbl-1(nk3) mutants using the Phrg-1::GFP reporter gene. Loss of dbl-1 resulted in an up-regulation of intestinal Phrg-1::GFP over a broad range of heme concentrations when compared to wildtype broodmates (Figure 5c and Supplementary Figure 5b). By contrast, overexpression of extrachromosomal copies of a transgene encoding GFP::DBL-1 suppressed Phrg-1::GFP expression in the dbl-1(nk3) mutants. Notably, these worms showed an enhanced suppression of Phrg-1::GFP expression in low heme conditions compared to wildtype broodmates, validating a published report that DBL-1 function is dose-dependent37. Furthermore, the gfp::dbl-1 transgene expression was restricted to neurons and neuron support cells (Supplementary Figure 5c), indicating that nervous system production of DBL-1 in the dbl-1(nk3) mutants is sufficient to restore hrg-1-mediated intestinal heme homeostasis (Supplementary Figure 5b and Figure 5c).
To evaluate heme-dependent growth phenotypes in dbl-1(nk3) mutants, worms were fed RP523, an E.coli mutant unable to synthesize heme23. Worms lacking dbl-1 activity were significantly and persistently growth retarded when grown on RP523 supplemented with 1 μM heme, a phenotype that was fully suppressed with 50 μM heme supplementation (Figure 5d). RNA-seq analysis of total RNA extracted from dbl-1(nk3) mutants and the corresponding wildtype broodmate controls fed E. coli supplemented with or without heme showed significantly elevated levels of hrg-7 mRNA in dbl-1(nk3) mutants (Figure 5e); hrg-1 mRNA was also elevated by this analysis in dbl-1(nk3) mutant, confirming RT-qPCR results reported above.
Intestinal expression of hrg-1 and hrg-7 are regulated via the transcription factor SMA-9
To gain insight into how DBL-1 regulates intestinal hrg-1 expression, we compared regulators that significantly altered Phrg-1::GFP expression by ≥ 2 fold with known genetic interactors of dbl-1. Only one gene was found to be common in both datasets: that gene encodes the transcription factor SMA-9. Previously, sma-9 has been shown to genetically interact with dbl-1 to regulate body size39, but unlike neuronal dbl-1, sma-9 is expressed in most somatic tissue including the intestine (Supplementary Figure 5d)39. RNAi depletion of sma-9 phenocopied dbl-1 RNAi in the up-regulation of GFP in the Phrg-1::GFP heme sensor worms (Figure 6a and Figure 6b), and in the Phrg-7::GFP transcriptional reporter (Figure 6c). Importantly, GFP up-regulation by sma-9 depletion was abrogated in dbl-1(nk3) mutants (Figure 6a), and co-RNAi of dbl-1 and sma-9 did not show an additive effect on GFP levels (Figure 6b). Together, these results imply a role for DBL-1 in the repression of hrg-1 expression via sma-9.
Figure 6. DBL-1/BMP regulates hrg-1 and hrg-7 through SMA-9.
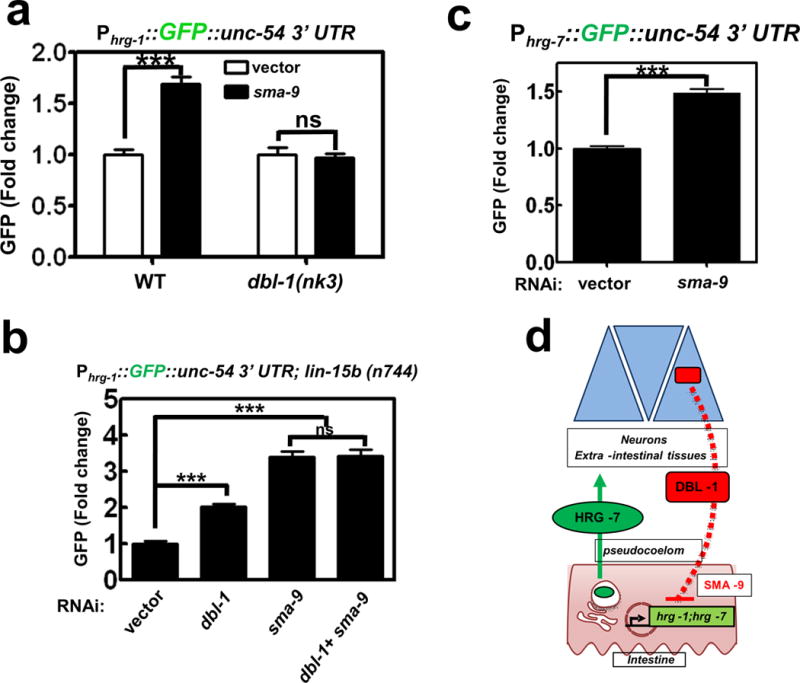
a) GFP fluorescence quantified from IQ6011 and IQ6311 fed vector control or dsRNA against sma-9 at 10 μM heme. GFP was quantified using COPAS BioSort. GFP is presented of fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (unpaired two-tailed t-test). See Supplementary Table 3 for statistics source data. b) GFP fluorescence quantified from IQ6015 fed dsRNA against vector control, dbl-1, or sma-9 alone or in combination. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=100 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (one-way ANOVA). See Supplementary Table 3 for statistics source data. c) GFP fluorescence quantified from IQ7701 worms fed dsRNA against vector control or sma-9 at 25 μM heme. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N=120 worms per treatment per experiment. ***P<0.001, **P<0.01, *P<0.05 (unpaired two-tailed t-test [AU please state type of t-test]). See Supplementary Table 3 for statistics source data. d) Model of the inter-organ communication for organismal regulation of heme homeostasis. When heme conditions are sufficiently low, hrg-1 and hrg-7 expression is upregulated to ensure adequate heme distribution throughout the animal. HRG-7 leaves the intestine and is perceived by neurons either directly or indirectly. DBL-1 secreted from neurons represses hrg-1 and hrg-7 through a SMA-9-mediated pathway.
DISCUSSION
In C. elegans, a natural heme auxotroph, organs must have a mechanism to communicate and coordinate their heme status with intestinal heme uptake and transport. In the current study, we have demonstrated that a heme-dependent, inter-organ communication system exists in C. elegans and that this system is facilitated by signaling between the intestine and neurons (Figure 6d). How does HRG-7 get targeted to the neurons? RNAi depletion of the C. elegans fibroblast growth factor (FGF) homolog, let-756, in the Phrg-1::GFP heme sensor strain harboring the hrg-7 (tm6801) mutation causes a significant increase in GFP (Supplementary Figure 5e) consistent with the possibility that LET-756 could directly interact with HRG-7 as demonstrated in a yeast two-hybrid screen40. But why should neurons influence systemic heme homeostasis when intestines are capable of regulating heme transport in a cell-autonomous manner? In C. elegans, sensory neurons are responsible for sensing nutrient availability and altering metabolism in peripheral tissues through an inter-tissue signaling pathway26. Furthermore, neuronal expression of hemoproteins such as soluble guanylate cyclases and globins dictate chemosensation and behavioral responses to food, oxygen, and other animals41, 42. Thus, it is conceivable that heme levels may alter behavioral responses to nutrient foraging and gas-sensing in worms.
Our studies have also uncovered an unanticipated role for an aspartic protease homolog in heme homeostasis. Typically, aspartic proteases described in helminths are involved in digesting dietary proteins as a nutritional source43, 44, while we show that HRG-7 is secreted from the intestine to mediate inter-organ signaling. Several features of proteases make them attractive candidates for signaling. They are involved in interactions with their cognate substrates or inhibitors. Many are secreted. Some are inactive until removal of a pro-peptide allowing for a rapid cellular response to stimuli45. Our results also reveal that mutating the conserved aspartic acid residues in the putative active site has no effect on HRG-7-dependent signaling or growth, raising the possibility that HRG-7 proteolytic activity may be irrelevant. Indeed, functional, intact active site proteases can have signaling functions that are independent of their enzymatic activity46. For example, renin binds a receptor that activates downstream signaling independent of its proteolytic processing of angiotensinogen47. Cathepsin D has been shown to act as a mitogen in tumor cells even after the active site is abolished48. Since homologs for HRG-7/cathepsin E49, DBL-1/BMP5, SMA-9/SHN, and HRG-1/SLC48A1 are also present in mammals, it is conceivable that an analogous cell-nonautonomous signaling pathway may exist in humans to regulate iron and heme metabolism.
METHODS
Animal model
Worms were maintained in liquid mCeHR2 or on Nematode Growth Medium agar plates. Experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. Adult hermaphrodites were used for analyses unless noted otherwise.
Cell Lines
No cell lines were used in this study.
RNAi interference by feeding
RNAi was performed as previously described14. For evaluation of GFP expression, GFP levels in gravid adults were observed visually using a Leica Microsystems MZ16FA stereoscope. The intensity and pattern of GFP in gravid worms feeding on bacteria producing dsRNA against each library clone was compared to the intensity and pattern of GFP in same-stage worms feeding on bacteria transformed with the empty vector. Three parameters were quantified with COPAS BioSort: time of flight (length), extinction (optical density) and GFP intensity. GFP intensity was normalized to time of flight.
GFP Quantification using COPAS BioSort
Worms were rinsed off NGM plates or pelleted in axenic media and washed three times with M9 buffer. 40-50 worms suspended in 100 μl M9 buffer were placed in individual wells of a 96 well plate, which was then analyzed with the COPAS BioSort. Photo-multiplier tube settings were 400 volts for IQ7701 and IQ6015, and 200 volts for all other strains in this study. GFP data obtained from the COPAS is presented as relative GFP on a scale from 1-100, or fold change compared to controls. The list of 177 genes determined to regulate Phrg-1::GFP by COPAS analyses can be found in Supplementary Table 4.
Cloning
To generate the Phrg-7::GFP transcriptional fusion, ~0.7 kb of the 5’ flanking region of hrg-7 was ligated into the HindIII and BamHI restriction sites within the multiple cloning sequence (MCS) of Fire vector pPD95.67. All other constructs were generated using Multisite Gateway recombination (Invitrogen). Promoters, coding regions, and 3’ untranslated regions were amplified with sequence-specific Gateway attb primers. PCR products were first recombined into donor plasmids, and then three donor plasmids were recombined into expression plasmids, according to the manufacturer’s instructions (Invitrogen).
Generation of transgenic worms
Reporter constructs mixed in a 2:1 ratio with the unc-119 rescue construct (18 μg total DNA), were introduced into unc-119(ed-3) worms by microparticle bombardment using the PDS-1000 particle delivery system (Bio-Rad). Transgenic lines were isolated after two weeks.
Immunoblotting
For detection of HRG-7 or HRG-7-3xFLAG, between 2,500 and 5,000 gravid or young adult worms were lysed in phosphate-buffered saline (PBS) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 4 mM benzamidine, 2 μg/ml leupeptin, and 1 μg/ml pepstatin) and Lysing Matrix C beads (MP Biomedicals) in a FastPrep-24 Beadbeater (MP Biomedicals). Worm lysates were centrifuged 3 times at 10,000 x g for 10 minutes., then total protein concentration in the supernatants was measured using the Pierce BCA assay kit (Thermo Scientific). Unboiled samples were mixed with Laemmli sample buffer and 50 μg protein / lane was separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was incubated overnight in 4° C with anti-HRG-7 polyclonal antibody custom produced in rabbits by Thermo Scientific Pierce Protein Research or M2 anti-FLAG monoclonal antibody (Sigma, no. F3165) at a concentration of 1:1000. Goat anti-rabbit HRP-conjugated secondary was used for anti- HRG-7, or rabbit anti-mouse HRP-conjugated secondary for anti-FLAG at a 1:10,000 dilution, and blots were developed in SuperWest Pico Chemiluminescent Substrate (Thermo Scientific). Bio-Rad Image Lab software was used to quantify blots.
DiI, FITC pulse
To stain amphid and phasmid neurons, worms were removed from plates with M9, washed twice with M9, and resuspended in 100 μl M9. A stock solution of 2 mg/ml 5-fluorescein isothiocyanate (FITC) in dimethyl formamide (DMSO) was diluted 1: 200 into the mixture. The tubes were wrapped in aluminum foil to avoid light exposure and incubated for 3 hours at room temperature. The worms were washed three times with M9 to remove excess dye, then immobilized in 10 mM levamisole on a 2% agarose pad and imaged by fluorescence microscopy using a Zeiss LSM710 laser scanning confocal microscope.
Heme response assay
Hemin chloride (Frontier Scientific) was dissolved in 0.3M ammonium hydroxide to a stock concentration of 10 mM. For heme response assays, eggs were obtained from worm strains maintained in mCeHR-2 medium with 20 μM heme or on NGM plates seeded with OP50. The following day, synchronized L1 larvae were placed in mCeHR-2 medium or on NGM plates seeded with OP50 supplemented with varying heme concentrations. After 72 hours, GFP or mCherry fluorescence was quantified with the COPAS BioSort and analyzed by fluorescent microscopy.
C. elegans growth on RP523 bacteria
The heme-deficient E. coli strain RP523 was grown overnight in LB supplemented with 4 μM heme50. The following day, cultures were placed in fresh LB with 1 μM or 50 μM heme and grown for 5.5 hours to an OD600 of 0.2. Cultures were seeded on NGM agar plates overnight. The following day, synchronized L1 larvae were placed on the RP523 seeded plates and incubated at 20°C until analyses.
HRG-7 recoding
To generate hrg-7 RNAi resistant transgenes, the first 300 bp of endogenous sequence was recoded by base pair substitution at degenerate sites using the C. elegans codon adapter (http://worm-srv3.mpi-cbg.de/codons/cgi-bin/optimize.py) and the Genscript rare codon usage tool (http://www.genscript.com/cgi-bin/tools/rare_codon_analysis)51. Any remaining contiguous stretches of six identical nucleotides between the recoded and endogenous sequences were changed manually, where applicable, to ensure RNAi resistance. The sequences were then sent to Genscript (Piscataway, NJ) for synthesis.
Worm lysate acid titration
To evaluate HRG-7 processing, IQ7370 (Pvha-6::HRG-7::3xFLAG) worms were lysed in PBS + 1% n-Dodecyl β-D-maltoside (DDM). The pH of each sample was lowered with acetate solution (1 part 0.2 M sodium acetate and 9 parts 0.2 M glacial acetic acid). The pH was brought to 8.0 with 0.1 M NaOH before running samples on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel.
Heat shock and coelomocyte mCherry quantification
Synchronized L1 worms expressing Phsp-16.2::HRG-7::mCherry (IQ7170) were placed on NGM plates seeded with OP50. After 72 hours, the plates were placed in a 37°C incubator for 30 min. The plates were then transferred to a 20°C incubator for 0, 60, 180, or 360 minutes. At the end of the time course, worms were washed off the plates with M9 and placed on ice. mCherry fluorescence was analyzed using a Zeiss LSM 710 confocal microscope. For quantification of mCherry fluorescence in coelomocytes, coelomocyte boundaries were established on merged z-stack DIC images. The corresponding area of fluorescence images was quantified using the Region of Interest (ROI) Function in Zeiss Zen software. A ROI of equal volumetric area to the area of a coelomocyte was used to quantify mCherry in the pseudocoelom and intestine. Ten images, consisting of about 10-15 z-stacks per image, were used for each time point.
ZnMP uptake
To assess uptake of ZnMP, synchronized L1 worms were grown on NGM plates seeded with OP50 or RNAi bacteria until they reached L4 stage. Worms were washed off plates and rinsed three times in M9 buffer to remove bacteria. Worms were incubated in mCeHR2 medium with 10 μM ZnMP overnight, rinsed to remove excess ZnMP, and imaged as described previously10. Briefly, worms were paralyzed with 10 mMlevamisole and imaged using a Leica DMIRE2 epifluorescence/DIC microscope. Images were obtained using a Retiga 1300 cooled mono 12-bit camera and quantified using SimplePCI software (Compix Inc).
RNA-seq
Total RNA was extracted from 30,000 late L4 worms per condition using the Trizol method. cDNA libraries were constructed with the TruSeq kit (Illumina). Single-end 50 base reads were generated using a HiSeq 2500 (Illumina) and aligned to ce10 reference genome using Tophat2, version 2.1.0. Differentially expressed genes were found using Cufflinks 2, version 2.2.1 with the cutoff of 0.05 on False Discovery Rate (FDR).
Immunofluorescence
A mixed stage population were fixed and stained as previously described52. Incubation with primary M2 anti-FLAG antibody (Sigma, no. F3165) was followed by incubation with secondary Tetramethyl Rhodamine 5 (and 6)-isothiocyanate (TRITC) conjugated goat-anti-mouse antibody (Jackson ImmunoResearch).
qRT-PCR
RNA was isolated from late 10,000 L4 worms per condition using the Trizol method. cDNA was synthesized using SuperScript III First-Strand Synthesis System (Invitrogen). qRT-PCR was performed with SsoAdvanced Univeral SYBR Green Supermix (Bio-Rad). Results were normalized with the CT value of gpd-2. Relative fold changes in hrg-1 expression were determined using the 2-ΔΔCt calculations based on average CT values. Primers used were 5′qCehrg-1 AATGGCAGGATGGTCAGAAAC, 3′qCehrg-1 CGATGAATGAAAGGAACGATACG5′, qgpd-2 TGCTCACGAGGGAGACTAC, 3′qgpd-2 CGGTGGACTCAACGACATAG.
Bioinformatics
Protein and DNA alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2). Signal peptides were predicted with the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP). Interaction networks were analyzed with geneMANIA (www.genemania.org). Homology models were generated with I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER).
Statistics and reproducibility
Statistical significance was determined with GraphPad Prism, version 7.02 (GraphPad Software, Inc.). Statistical significance for RNA-seq was determined by Cufflinks 2, version 2.2.1 with the cutoff of 0.05 on False Discovery Rate (FDR). Statistical tests are justified. No statistical method was used to predetermine sample size. All data are presented as the mean ± the standard error of the mean. Data meets the assumptions of the tests. Variance is similar among compared groups. For C. elegans quantification data, the number of times each experiment was repeated is stated in figure legends. RNAi screens were performed one time, and candidate genes were validated at least three other times. For western blots, immunofluorescence, and transgenic expression data, all experiments were performed at least two times.
Supplementary Material
Acknowledgments
We thank Andrew Chisholm, Kelly Liu, Tina Gumienny, Antony Jose and David Hall for critical discussions and advice; Tetsunari Fukushige for preparing the RNAseq libraries and extensive discussions; National Bioresource Project and S. Mitani for the hrg-7 strain; Hanna Fares for strain NP97 (Punc-122::GFP). This work was supported by funding from the National Institutes of Health DK074797 and a supplement to DK074797 (I.H.); support was also provided by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (M.K.). The genome-wide RNAi screen was funded by the Roche Foundation for Anemia Research (I.H.).
Footnotes
Author Contributions. Experimental design and execution were as follows: C. elegans experiments J.S., K.P., T.S., S.B., X.Y., J.Z., K.M., M.K., and I.H.; Homology modeling and RNA-seq analyses S.Y.; J.S. and I.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information. The RNAseq data have been deposited with the Gene Expression Omnibus at NCBI under accession number GSE76455. I.H. is the President and Founder of Rakta Therapeutics Inc. (College Park, MD), a company involved in the development of heme transporter-related diagnostics. He declares no other competing financial interests.
Code availability
Bioinformatic analyses using Clustalw is available at http://www.ebi.ac.uk/Tools/msa/clustalw2. Network analyses of dbl-1 with GeneMania is available at http://genemania.org/ by using the search term “dbl-1”.
Data availability
RNA-Seq data has been deposited in the Gene Expression Omnibus (GEO) under accession code GSE76455. Source data for Figures 1a, 1e, 2f, 4c, 4d, 5b, 5e, 6a, 6b, 6c, and Supplementary Figures 2a, 2f, 3d, 4d have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on request.
References
- 1.Hamza I. Intracellular trafficking of porphyrins. ACS Chem Biol. 2006;1:627–629. doi: 10.1021/cb600442b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Severance S, Hamza I. Trafficking of heme and porphyrins in metazoa. Chem Rev. 2009;109:4596–4616. doi: 10.1021/cr9001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korolnek T, Hamza I. Macrophages and iron trafficking at the birth and death of red cells. Blood. 2015;125:2893–2897. doi: 10.1182/blood-2014-12-567776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keel SB, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 5.Cao C, O’Brien KO. Pregnancy and iron homeostasis: an update. Nutr Rev. 2013;71:35–51. doi: 10.1111/j.1753-4887.2012.00550.x. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, et al. Kinetics and specificity of feline leukemia virus subgroup C receptor (FLVCR) export function and its dependence on hemopexin. J Biol Chem. 2010;285:28874–28882. doi: 10.1074/jbc.M110.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haldar M, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan X, et al. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1609865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair J, Hamza I. Lessons from bloodless worms: heme homeostasis in C. elegans. Biometals. 2015;28:481–489. doi: 10.1007/s10534-015-9841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao AU, Carta LK, Lesuisse E, Hamza I. Lack of heme synthesis in a free-living eukaryote. Proc Natl Acad Sci U S A. 2005;102:4270–4275. doi: 10.1073/pnas.0500877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korolnek T, Zhang J, Beardsley S, Scheffer GL, Hamza I. Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 2014;19:1008–1019. doi: 10.1016/j.cmet.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajagopal A, et al. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severance S, et al. Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001044. doi: 10.1371/journal.pgen.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemeth E, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 16.Kim BE, et al. Cardiac copper deficiency activates a systemic signaling mechanism that communicates with the copper acquisition and storage organs. Cell Metab. 2010;11:353–363. doi: 10.1016/j.cmet.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kautz L, et al. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet. 2014;46:678–684. doi: 10.1038/ng.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinclair J, Hamza I. A novel heme response element mediates transcriptional regulation in Caenorhabditis elegans. J Biol Chem. 2010 doi: 10.1074/jbc.M110.167619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGhee JD, et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Mahoney TR, et al. Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:16350–16355. doi: 10.1073/pnas.0803617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka T, Toyomura T, Honjo K, Wada Y, Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem. 2001;276:33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- 22.Pujol N, Bonnerot C, Ewbank JJ, Kohara Y, Thierry-Mieg D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem. 2001;276:11913–11921. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Samuel TK, Sinclair J, Dailey HA, Hamza I. An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell. 2011;145:720–731. doi: 10.1016/j.cell.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 25.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 26.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 27.Evans D, Blumenthal T. trans splicing of polycistronic Caenorhabditis elegans pre-mRNAs: analysis of the SL2 RNA. Mol Cell Biol. 2000;20:6659–6667. doi: 10.1128/mcb.20.18.6659-6667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding L, Candido EP. Association of several small heat-shock proteins with reproductive tissues in the nematode Caenorhabditis elegans. Biochem J. 2000;351:13–17. doi: 10.1042/0264-6021:3510013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 30.Poea-Guyon S, et al. The V-ATPase membrane domain is a sensor of granular pH that controls the exocytotic machinery. J Cell Biol. 2013;203:283–298. doi: 10.1083/jcb.201303104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, et al. Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior. Curr Biol. 2013;23:746–754. doi: 10.1016/j.cub.2013.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung WL, Wang Y, Chitturi J, Zhen M. A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development. 2014;141:1767–1779. doi: 10.1242/dev.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 34.Liang J, Yu L, Yin J, Savage-Dunn C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev Biol. 2007;305:714–725. doi: 10.1016/j.ydbio.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 35.Savage-Dunn C, Yu L, Gill K, Awan M, Fernando T. Non-stringent tissue-source requirements for BMP ligand expression in regulation of body size in Caenorhabditis elegans. Genet Res (Camb) 2011;93:427–432. doi: 10.1017/S0016672311000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki Y, et al. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 37.Morita K, Chow KL, Ueno N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development. 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- 38.Schultz RD, Bennett EE, Ellis EA, Gumienny TL. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS One. 2014;9:e101929. doi: 10.1371/journal.pone.0101929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J, et al. The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development. 2003;130:6453–6464. doi: 10.1242/dev.00863. [DOI] [PubMed] [Google Scholar]
- 40.Popovici C, et al. Direct and heterologous approaches to identify the LET-756/FGF interactome. BMC Genomics. 2006;7:105. doi: 10.1186/1471-2164-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persson A, et al. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- 42.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 43.Williamson AL, et al. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J Biol Chem. 2004;279:35950–35957. doi: 10.1074/jbc.M405842200. [DOI] [PubMed] [Google Scholar]
- 44.Brinkworth RI, Prociv P, Loukas A, Brindley PJ. Hemoglobin-degrading, aspartic proteases of blood-feeding parasites: substrate specificity revealed by homology models. J Biol Chem. 2001;276:38844–38851. doi: 10.1074/jbc.M101934200. [DOI] [PubMed] [Google Scholar]
- 45.Turk B, Turk D, Turk V. Protease signalling: the cutting edge. EMBO J. 2012;31:1630–1643. doi: 10.1038/emboj.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cousin C, Bracquart D, Contrepas A, Nguyen G. Potential role of the (pro)renin receptor in cardiovascular and kidney diseases. J Nephrol. 2010;23:508–513. [PubMed] [Google Scholar]
- 48.Glondu M, et al. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene. 2001;20:6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- 49.Ueno E, Sakai H, Kato Y, Yamamoto K. Activation mechanism of erythrocyte cathepsin E. evidence for the occurrence of the membrane-associated active enzyme. J Biochem. 1989;105:878–882. doi: 10.1093/oxfordjournals.jbchem.a122772. [DOI] [PubMed] [Google Scholar]
- 50.Li JM, Umanoff H, Proenca R, Russell CS, Cosloy SD. Cloning of the Escherichia coli K-12 hemB gene. J Bacteriol. 1988;170:1021–1025. doi: 10.1128/jb.170.2.1021-1025.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redemann S, et al. Codon adaptation-based control of protein expression in C. elegans. Nat Methods. 2011;8:250–252. doi: 10.1038/nmeth.1565. [DOI] [PubMed] [Google Scholar]
- 52.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


