Abstract
Arabidopsis thaliana Regulator of G protein Signalling 1 (AtRGS1) is a protein with a predicted N-terminal 7-transmembrane (7TM) domain and a C-terminal cytosolic RGS1 box domain. The RGS1 box domain exerts GTPase activation (GAP) activity on Gα (AtGPA1), a component of heterotrimeric G protein signaling in plants. AtRGS1 may perceive an exogenous agonist to regulate the steady-state levels of the active form of AtGPA1. It is uncertain if the full length RGS1 protein exerts any atypical effects on Gα, nor has it been established exactly how AtRGS1 contributes to perception of an extracellular signal and transmits this response to a G-protein dependent signaling cascade. Such an understanding can be deduced from in vitro studies using the purified and soluble RGS1-box domain. Further studies on full-length AtRGS1 have been inhibited due to the extreme low abundance of the endogenous AtRGS1 protein in plants and lack of a suitable heterologous system to express AtRGS1. Here, we describe methods to produce full-length AtRGS1 by cell free synthesis into unilamellar liposomes and nanodiscs. The cell-free synthesized AtRGS1 exhibits GTPase activating activity on Gα and can be purified to a level suitable for biochemical analyses.
Keywords: 7-transmembrane protein, Arabidopsis Regulator of G Signaling protein 1 (AtRGS1), in vitro translation, nanodiscs, membrane scaffold protein 1D1
Introduction
Seven transmembrane (7TM) domains are found in cell surface receptors, serving as the platform in or on which an agonist binds. Animals encode a large class of 7TM proteins termed G protein-coupled receptors (GPCRs). These proteins bind to a diverse range of extracellular ligands that are physically coupled to a cytoplasmic heterotrimeric guanine nucleotide-binding complex, consisting of Gα, Gβ and Gγ subunits. Gα binds to and hydrolyses GTP to GDP at an intrinsic rate to complete the cycle. In the Gα-GTP state, the complex dissociates into Gα subunit and Gβγ dimers, each targeting cytoplasmic enzymes or structural proteins [1]. In animals, the rate-limiting step for the G cycle is GDP release from the Gα subunit; agonist binding leads to a conformational change of GPCR and sequentially increases guanine nucleotide exchange of Gα subunit. The reverse process may be accelerated by a cytoplasmic Regulator of G signaling (RGS) protein to alter the signaling dynamics and speed returning to the resting (GDP-bound) state [2].
In many organisms outside the animal clade, the rate-limiting step of the G cycle is GTP hydrolysis [3–5], therefore Gα subunit of these organisms have no need for GPCR activation [6]. One possible regulator of the G cycle in these organisms is a 7-transmembrane (7TM) receptor-like RGS protein. The prototype for this protein architecture is Arabidopsis thaliana Regulator of G Signaling protein 1 (AtRGS1). AtRGS1 contains an N-terminal 7TM domain that resembles GPCR topology and an RGS box in the cytoplasmic C-terminal domain[7] that has GTPase Activating Protein (GAP) activity. AtRGS1 is part of the Arabidopsis heterotrimeric G protein complex, encoded by one canonical Gα subunit gene (gpa1), one Gβ subunit gene and one of three Gγ subunit genes [8]. The cytoplasmic RGS domain (RGS box) of AtRGS1 accelerates Gα GTP hydrolysis activity in vitro [4,5,7]. Consistent with RGS domain GAP activity, overexpression of AtRGS1 phenotypes are similar to gpa1mutant phenotypes [7].
Little is known about the biochemical function of the full-length AtRGS1 protein relative to its interaction with Gα, nor has it been established if AtRGS1 directly binds an extracellular ligand and transmits this to a G-protein dependent signaling cascade. Thus, purified AtRGS1 protein is required for in vitro biophysical and biochemical characterization. The low abundance of endogenous AtRGS1 precludes its purification from Arabidopsis. Attempts to express recombinant AtRGS1 protein in insect cells, yeast, plant cells, and animal cells were not successful (data not shown), perhaps because of the toxic effect of expressing an active RGS protein. As an alternative expression system that does not depend on use of a living organism, cell-free protein synthesis (CFPS) has unique advantages such as tolerance of proteins with potentially deleterious regulatory functions or enzymes with unusual catalytic activities [9]. Moreover, a cell-free protein synthesis reaction allows the addition of reagents such as lipids, detergents, metals, or cofactors to improve the outcome of the translation. Among the most widely used CFPS systems, wheat germ extract (WGE), which was established and optimized by Endo and coworkers [10,11], is the highest yield eukaryotic CFPS system [9] and also shows wide-spectrum expression of eukaryotic proteins [12]. Due to these advantages, the WGE system has been used in 7TM proteins synthesis [13–15]. However, as with other integral membrane proteins, purification and obtaining a functional, folded state is challenging because of the need for a native membrane-like environment. Although AtRGS1 could be produced using CFPS, attempts to solubilize it with detergent and/or liposomes yielded insufficient amounts to support further functional investigations (this work).
As an alternative, nanodiscs provide a novel paradigm to mimic a membrane environment for membrane proteins. A nanodisc is a self-assembled discoidal lipid/protein particle with a lipid bilayer in the center surrounded by two molecules of membrane scaffold protein (MSP). MSPs are derivatives of human apolipoprotein A-1 (Apo A-1). In optimized conditions, MSPs form homogeneous discoidal structures, which provides a stable artificial lipid bilayer for membrane protein incorporation. Generally, MSPs are expressed and purified from E. coli [16], then nanodiscs are self-assembled in the presence of purified MSPs and the pre-determined appropriate lipid. In this manner, nanodiscs have been used to reconstitute the β2-adrenergic receptor [17–19], rhodopsin [20–23] and the CCR5 chemokine receptor [24]. Since the diameter of a nanodisc can be determined by the number of α-helical repeats around the lipid bilayer center [25], various MSPs can be used to obtain monomeric or homogenous dimeric GPCR [26]. Herein, we compare two protocols for the synthesis of AtRGS1 using the WGE system, one of which synthesizes AtRGS1 directly into liposomes and another directly into nanodiscs. After affinity purification and size exclusion chromatography, we show that AtRGS1 synthesized into nanodiscs has GAP activity comparable to the cytoplasmic RGS1 catalytic domain.
Materials and Methods
Preparation of plasmid DNA
The AtRGS1 open reading frame and MSP1D1 were cloned into the cell-free pEU expression vector [27] and confirmed by DNA sequencing. Prior to the in vitro transcription reactions, plasmid DNA was subjected to proteinase K treatment as described by Makino et al [28]. Briefly, purified DNA was incubated with 50 μg/mL proteinase K in 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, and 0.1% (w/v) SDS for 1 h at 37° C. Then, the plasmids were purified with a QIAprep spin miniprep kit (QIAGEN, Valencia, CA) following the manufacturer’s instruction.
Cell free translation into liposomes
Transcription was performed following the method described by Makino et al [28]. After the transcription, the reactions were centrifuged at 14,000 rpm (18,000 × g) in a benchtop centrifuge for 3 minutes at ambient temperature. The quality of the nascent RNA in supernatant was assessed using denaturing RNA agarose gel electrophoresis. For synthesis of AtRGS1 into liposomes, 5 μL RNA supernatant of AtRGS1 was added to the in vitro translation system containing 1.2 mg/mL soy liposomes. Translation reactions were performed for 14 h at room temperature (21–25°C).
Soy liposomes were prepared following the method detailed by Goren et al [29]. Briefly, one gram of soy lipid was dissolved in 3 mL of chloroform, then flushed with a stream of liquid nitrogen gas to remove the majority of the organic solvent and to allow formation of a thin film of lipid which is amenable to aqueous reconstitution into a multilamellar vesicles. To remove residual solvent, the lipid was dried under vacuum pressure for 30 minutes. The lipids were re-suspended in 67 mL of 25 mM HEPES-NaOH pH 7.5, 100 mM NaCl, with brief vortexing. Unilamellar liposomes were formed by extrusion through an Avanti mini-extruder (Avanti) 11 times with a 0.4 μm track-etched polycarbonate membrane (Nucleopore) and 11 times through a 0.1 μm membrane. For long term storage, liposomes prepared in this manner were stored at −80°C under nitrogen until further use.
Detergent solubilization screen
The effectiveness of detergent to solubilize hexahistidine tagged RGS1 was assessed by immobilised metal affinity purification (IMAC) purification. Approximately 1/2 volume of unfractionated translation reactions were diluted with 50 μL of binding buffer (50 mM sodium phosphate, 300 mM NaCl, 50 mM imidazole, 2 mM DTT, pH 8.0, detergent to the concentration given in the Results section) and incubated for 1 hour at room temperature. Binding buffer contained detergent as indicated in the Results section. Samples were centrifuged at 18,000 ×g to remove insoluble material and the soluble fraction subjected to further purification. Purifications were performed on a 96-well filter plate containing 10 μL Ni Sepharose (GE) resin (50% slurry) per well. Solubilized sample (50μL) was diluted with 50μL binding buffer and incubated with resin for 10 min. Unbound components were removed by centrifugation through the filter at 2,500 × g for 1 min. Resin was washed three times with 150 μL of wash buffer (50 mM sodium phosphate, 300 mM NaCl, 50 mM imidazole, 2 mM DTT, pH 8.0, detergent to the concentration given in the Results section). Protein was eluted with 50 μL elution buffer (500 mM imidazole in place of the 50 mM in the binding/washing buffer). 12 μL was taken for SDS-PAGE analysis (+ 6 μL of 3x SDS sample buffer) and 6 μL was loaded per well. For analysis of the pellet fraction, 1/5 volume of the translation reaction was centrifuged at 14,000 rpm (~18000 × g) for 3 min at ambient temperature. The pellet resulting from resolubilization was resuspended in 90 μL of 1× SDS sample buffer. 6 μL was loaded per well on an SDS-PAGE gel.
Cell-free translation with nanodiscs
Cell-free transcription and translation was performed following the method described in Makino et al[28] with some modifications. Briefly, proteins were synthesized in a dialysis mode reaction using WEPRO2240 (CellFree Sciences Co., Ltd.) at ambient temperature. For co-expression, 1.7 μL RNA of MSP1D1 and 3.3 μL of RNA for AtRGS1 were added into the in vitro translation system containing 0.6 mM cardiolipin (Avanti Polar Lipids, Inc. #710335C). Final volumes of the translation reaction ranged between 24–25 μL. Translation reactions were performed for 16 h. For sequential expression, 5 μL RNA of MSP1D1 was added into the in vitro translation system containing 0.6 mM cardiolipin to initiate the reaction. AtRGS1 RNA was added sequentially after 16 and 40 hours. Final volumes of the translation reaction ranged between 34–35 μL. Translation reactions were performed for 64 h.
Affinity purification
Purification was performed at ambient temperature. For nanodisc-assembled protein, the crude translation extract was centrifuged at 18,000 × g for 3 min to fractionate supernatant and pellet. For the purification, the supernatant, 75 μL of buffer (20 mM HEPES-NaOH pH 8.0, 150 mM NaCl) and 30 μL (50% slurry) of suspended StrepTactin Sepharose resin (IBA 2-1201-002) were added to the wells of a 96-well filter plate (MultiScreen HTS-HV, 0.45-μm pore, Millipore, MSHVS4510). After agitation for 30 min on a vibrating platform (~400 rpm), unbound components were removed by centrifugation at 2,500 × g for 1 min. The resin was washed 3 times by 225 μL buffer described, and the bound components were eluted twice by incubating with 35 μL elution buffer (20 mM HEPES-NaOH pH 8.0, 150 mM NaCl, 2.5 mM d-desthiobiotin (Sigma D1441-500)), and then collected by centrifugation at 2,500 × gfor 1 min.
For AtRGS1 synthesized into liposomes, 20 μL (50% slurry) of suspended TALON Metal Affinity Resin (Clontech #635501) was equilibrated in IMAC binding buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 50 mM imidazole) and used for affinity binding. After protein binding, the resin was washed 3 times by 225 μL IMAC washing buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 25 mM imidazole), and the bound components were eluted twice by 35 μL IMAC elution buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 500 mM imidazole). The supernatant (containing eluted AtRGS1) was clarified by centrifugation at 2,500 × gfor 1 min and then collected.
Nanodisc purification
AtRGS1 assembled-nanodiscs were separated from the affinity eluate by size exclusion chromatography on a Superose 12 10/300 GL column (GE healthcare) equilibrated with 20 mM Tris-HCl pH 7.6, 150 mM NaCl. Blue Dextran 2000, BSA and IgG were used to calibrate the column elution times. The affinity eluate was concentrated to 100 μL with a 10 kDa Amicon Ultra-0.5 mL Centrifugal Filter (EMD Millipore Corporation) and injected onto the column with 0.2 mL fractions collected at 4 °C.
Immunoblot analysis
Total cell free reactions and purified proteins were analyzed by immunoblot analysis. Samples were separated on SDS-PAGE gel, then transferred to a PVDF membrane (BioRad). AtRGS1 protein was detected using anti-AtRGS1 anti-sera directed against the RGS1 box domain plus the C-terminus and the linker region between 7TM domain and RGS1 box domain[30] Goat Anti-Rabbit IgG (whole molecule)– peroxidase was used as the secondary antibody (Sigma, A4914). AtRGS1 containing a hexahistidine tag was detected using a mouse anti-His tag monoclonal antibody (Roche, clone BMG-His-1, Ref 11 922 416 001) and Rabbit Anti-Mouse IgG + IgM (H+L) (315-035-044, Jackson ImmunoResearch). StrepII-tagged MSP1D1 was detected using an anti-Strep-tag II monoclonal antibody (IBA, Cat: 2-1507-001). Immunoblots were developed using Amersham ECL prime western blotting detection reagent (GE Healthcare).
Purification of AtGPA1
AtGPA1 was expressed and purified as described previously[4]. Briefly, the cDNA of AtGPA1 was cloned into the pPROEX vector. Recombinant His-tagged AtGPA1 protein expression was induced in ArcticExpress RP cells (Agilent Technologies) using 0.5 mM IPTG at 12 °C. Cells were suspended in N1 buffer (25 mM Tris-HCl pH 7.6, 100 mM NaCl, 5% glycerol, 10 mM imidazole, 10 mM MgCl2, 125 μM GDP, 12.5 mM 2-mercaptoethanol, 1 mM PMSF, 10 μM leupeptin, 0.25 mg/mL Lysozyme, 0.1% Thesit (Sigma, 88315)) and incubated at 4 °C for 30 min, then lysed by Sonic Dismembrator, Model 550, Fisher Scientific (power level 5, 0.5′/0.5′ off for 1 minute, 3 cycles in salt ice bath). The concentration of NaCl in the cell lysate was adjusted to 300 mM and incubated at 4 °C for 30 min. The soluble fraction was centrifuged at 30,000 × g 4 °C for 45 min, then incubated with TALON Metal Affinity Resin (50 μL 50% slurry per 1 g cell pellet) for 1.5 hours at 4 °C. The resin was washed with wash buffer (25 mM Tris-HCl pH 7.6, 300 mM NaCl, 5% glycerol, 10 mM imidazole, 10 mM MgCl2, 50 μM GDP, 5 mM 2-mercaptoethanol, 1 mM PMSF, 10 μM leupeptin), eluted with elution buffer (50 mM Tris-HCl pH 7.6, 300 mM NaCl, 5% glycerol, 300 mM imidazole, 10 mM MgCl2). Purified His-AtGPA1 was dialyzed against dialysis buffer (20 mM Tris-HCl pH 7.6, 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 2 μM GDP) at 4 °C for 16 hours and quantified by the Bio-Rad protein assay. The ORF of the C-terminal domain of AtRGS1 (residues 284–459) was cloned into the pDEST17 vector[4]. Recombinant His-tagged AtRGS1 was expressed and purified by the same method without MgCl2and GDP in the buffers.
Single-turnover [γ-32P] GTP hydrolysis assay
To assess GTPase activating protein (GAP) activity of AtRGS1, single-turnover GTP hydrolysis reaction were performed as previously described [4]. Briefly, purified AtGPA1 (250 nM) was preloaded with radioactive [γ-32P] GTP (Perkin Elmer, NEG004Z250UC) in TMDL buffer (50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 0.05% Thesit) for 5 minutes on ice. The hydrolysis reaction was then initiated by adding an equal volume of TMDL buffer + GTPγS (50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.05% Thesit and 400 μM GTPγS) with 1000 times diluted crude extract, or the indicated fraction of AtRGS1 in preloaded AtGPA1. At each time point, duplicate samples were denatured by mixing with 1 mL quench buffer (5% (w/v) activated charcoal, 50 mM phosphoric acid pH 2.0) to remove non-hydrolyzed [γ-32P] GTP and proteins. The charcoal-treated samples were centrifuged and the amount of phosphate in solution was measured by scintillation counting. The TMDL buffer was slightly modified to measure AtRGS1 activity within the wheat-germ crude extracts in liposomes as follows: 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM 2- mercaptoethanol, 0.05% Thesit, 1 mM PMSF, 1 μg/mL leupeptin and 5 mM ATP.
Results and Discussion
The preparation of AtRGS1 introduces two challenges. First, production of the integral membrane 7TM domain requires an expression system that is able to produce useful quantities of membrane proteins. Second, the expressed and purified protein must retain its stability and GTPase accelerating activity. The former is easily assessed by quantitation of solubilized protein and its purity. The latter is assessed by measuring the catalytic activity of the solubilized full-length AtRGS1 protein and comparing this to the known activity of the soluble RGS catalytic domain.
Cell-free expression of the full length RGS1 protein in liposomes
To obtain the RNA template for in vitro translation, coding sequences of the desired genes were cloned into the pEU vector containing an SP6 promoter, a transcriptional enhancer, and a terminator. Before use in the transcription reaction, plasmids were treated with proteinase K to remove residual RNase contamination from DNA purification. RNA was synthesized in vitro by SP6 RNA polymerase, and the quality of transcribed RNA was examined by denaturing agarose gel electrophoresis and ethidium bromide staining (Figure 1A). The ladder of RNA seen in Figure 1A was due to longer transcripts created by rounds of transcription past the terminator.
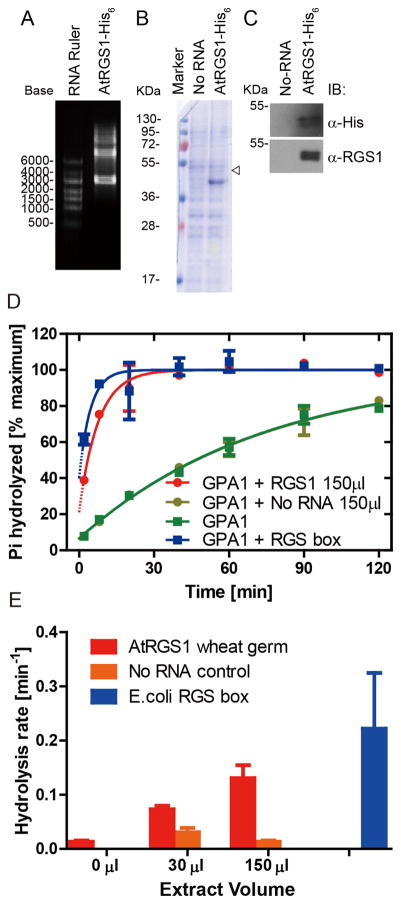
Figure 1. Synthesis of AtRGS1 from wheat germ cell-free extract in the presence of liposomes.
(A) In vitro transcription RNA products for AtRGS1- His6 were separated by 1.5% agarose gel. (B and C) Crude extract of in vitro translation for No-RNA control (No-RNA) and AtRGS1-His6 were separated by 12% SDS-PAGE and detected by Coomassie Brilliant Blue Staining (B), and immunoblot analysis with antiserum against the His6 tag (C, upper) or the C-terminal domain containing the RGS box domain (C, lower) antibodies, respectively. The open arrowhead indicates the molecular weight of AtRGS1. (D) 75 μL of AtRGS1 in liposomes were added to 700 μL of reaction mixture on ice. 100 μL of reaction was stopped at 2, 8, 20, 40, 60 and 120 min by ice cold quench buffer. Duplicate time points, except for a single time point at 120 min, were fit with an exponential One-phase association function using GraphPad Prism version 5.0. “No RNA” represents the wheat germ extract containing liposomes. RGS1 box represents 500 nM of truncated AtRGS1 (Lys284 - carboxyl terminus) used as a reference. (E) Hydrolysis rates were estimated from the presented data. Errors are 95% confidence intervals.
His-tagged AtRGS1 protein expression was studied by both Coomassie Brilliant Blue (CBB) stain assay and immunoblot analysis with anti-AtRGS1 antibody or anti-His antibody (Figure 1B and C, respectively). Although no obvious signal was found at the predicted position in the CBB-stained gel, the more sensitive western blot analysis showed a significant band migrating at about 50 KDa in the AtRGS1 lane but not in the “No-RNA” negative control lane. No other fragments were detected with these antibodies, except a diffuse band between 95~130 kDa, approximately twice the molecular weight of AtRGS1. Together, these results indicate that the full-length AtRGS1 was produced in this cell-free expression system. To assess the GTPase accelerating activity of AtRGS1 on its substrate AtGPA1, we undertook a single-turnover nucleotide hydrolysis assay (Figure 1D and E). The translation reaction containing full-length AtRGS1 and RGS1 box accelerated GTP hydrolysis. As a negative control, the “No RNA” extract had no influence on the reaction rate.
Solubility of AtRGS1 in detergent
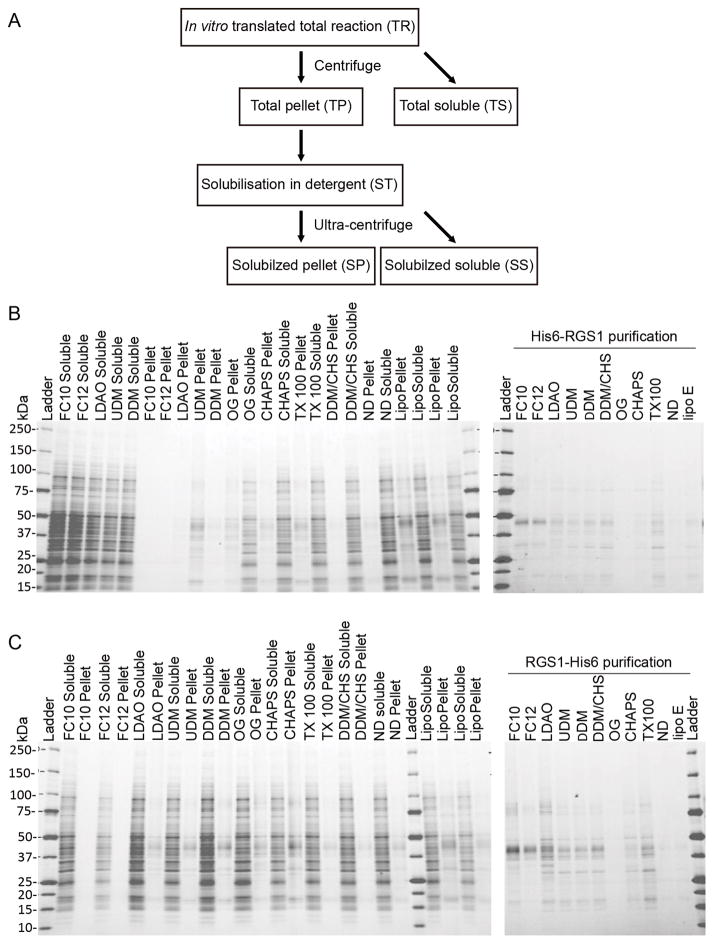
Since further functional or structural studies require purified integral membrane proteins to be solubilized, we screened a set of detergents for solubilization of AtRGS1 following the scheme outlined in Figure 2A. Aliquots of the translation reaction were added to solutions containing one of the following detergents: Fos-cholines (FC10, FC12), LDAO, maltosides (DDM and UDM), CHAPS, OG, and Triton-X100. Fos-choline detergents (FC10 and FC12) were most effective at solubilizing AtRGS1. Dodecylmaltoside (DDM) or a 5:1 (w/w) mix of DDM with cholesterol hemisuccinate (CHS) partly solubilized AtRGS1. Purification of the expressed AtRGS1 protein gave an indication of the yield and efficiency of solubilization, which was slightly better with the C-terminal His tagged AtRGS1 (Figure 2C) than the N-terminal His tagged AtRGS1 (Figure 2B). However, we failed to detect GAP activity by AtRGS1 inserted in liposomes. We conclude that while low levels of AtRGS1 can be synthesized in liposomes and may be useful for some studies, the inefficiency of solubilization and lack of detectable GAP activity precludes further biochemical characterization. Therefore, a different expression and purification scheme was designed.
Figure 2. Incorporation of AtRGS1 into liposomes.
(A) Flow chart detailing the series of centrifugation and solubilization steps to determine incorporation of AtRGS1 into liposomes. Total cell free translation of N-terminal His-tagged AtRGS1 (B) or C-terminal His-tagged AtRGS1 (C), were solubilized in the indicated detergents for 1h at ambient temperature and then centrifuged to separate insoluble and soluble protein (left panels). The soluble fraction of this centrifugation was further purified by using Ni2+ resin in the presence of the solubilization detergent (right panels). The abbreviation and full name of detergents are shown below, followed by their percentage (w/v) used in solubilization and purification: FC10: Fos-choline-10 (2.0, 0.5); FC12: Fos-choline-12 (0.5, 0.05); LDAO: n-dodecyl-N,N-dimethylamine_N-oxide (0.5, 0.05); UDM: n-undecyl-β-D-maltopyranoside (1.0, 0.1); DDM: n-dodecyl-β-D-maltopyranoside (1.0, 0.05); OG: n-octyl-β-D-glucopyranoside (3.0, 1.0); CHAPS: 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (3.0, 0.7); TX-100: polyethylene glycol p-(1,1,3,3-tetramethylbutyl)-phenyl ether (0.05, 0.05); DDM/CHS: DDM/cholesteryl hemi-succinate (1.0/0.2, 0.05/0.01).
Synthesis of MSP1D1 and AtRGS1 from wheat germ cell-free extract
Because detergents solubilization of AtRGS1 failed to preserve GAP activity, we investigated detergent-free solubilization using membrane scaffold proteins[31]. The same in vitro transcription/translation procedures were followed for this method, except that transcriptions of both AtRGS1 and MSP1D1 plasmids were added to the liposome-containing translation reaction, as described below (Figure 3A).
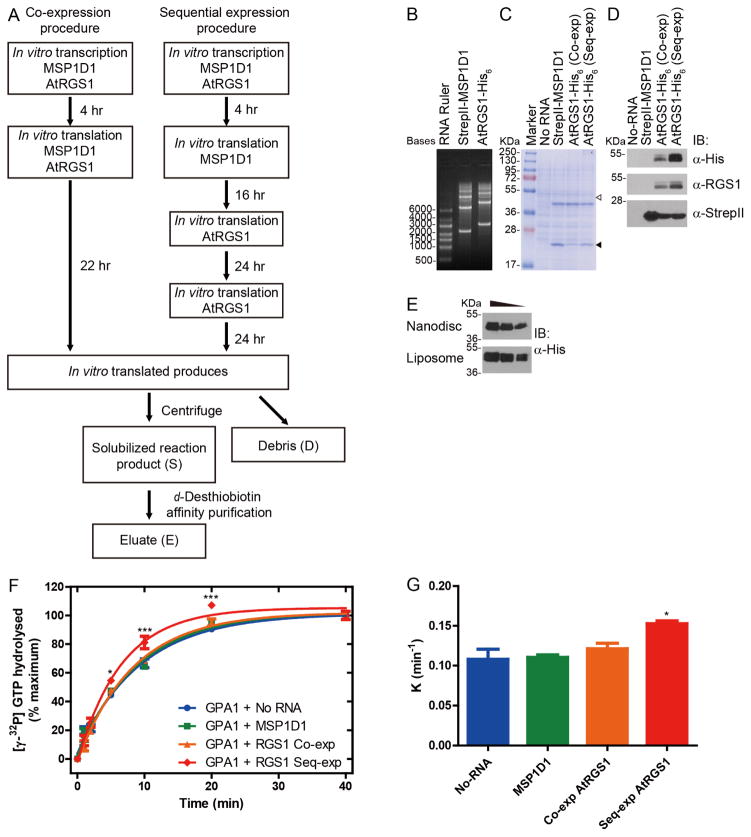
Figure 3. Synthesis of MSP1D1 and AtRGS1 from wheat germ cell-free extract.
(A) Schematic diagram for two procedures of cell-free synthesis and purification. Co-expression of MSP1D1 with AtRGS1 is compared to sequential expression (Seq-exp) of MSP1D1 followed by AtRGS1. (B) In vitro transcription RNA products for MSP1D1 and AtRGS1 were separated by 1.5% agarose gels. (C and D) Crude extract of in vitro translation for No-RNA negative control (No-RNA), MSP1D1 only (StrepII-MSP1D1), co-expressed AtRGS1 (Co-exp) and sequentially-expressed AtRGS1 (Seq-exp) were separated by 12% SDS-PAGE and detected by Coomassie Brilliant Blue Staining (C), immunoblot analysis using anti- His6 (D, upper), anti-RGS1 (D, middle) and anti-StrepII (D, lower) antibodies. The open and closed arrowheads indicate the predicted molecular weights of AtRGS1 and MSP1D1, respectively. (E) Equal volume of crude AtRGS1 extracts in Nanodisc (sequential synthesis) and liposome were loaded in SDS-PAGE and immunoblotted by anti- His6antibody. The gradient indicates 2 fold dilution between lanes. (F) 250 nM of purified AtGPA1 was pre-incubated with [γ-32P] GTP in ice cold buffer. Single turnover GTPase assays were initiated by the addition of GTPγS containing buffer to crude translation extract of No-RNA, MSP1D1, co-expressed AtRGS1 (Co-exp) or Seq-exp AtRGS1. (G) K value represents the rate constant of the reaction with different crude translation extracts. Duplicated reactions were stopped by ice cold quench buffer at the indicated time points. Data were representative of three or more independent experiments. The data were fit to exponential One-phase association functions using GraphPad Prism version 5.0. The quantitative results were expressed as the means ± S.E.M. of at least three experiments. Statistical significance was determined by an analysis of variance (ANOVA). Differences with P values of <0.05 were considered to be statistically significant.
We investigated two protocols for in vitro translation followed by nanodisc construction designated co-expression (Co-exp) and sequential expression (Seq-exp). In the co-expression procedure (Figure 3A, left), RNA encoding MSP1D1 and AtRGS1 were translated simultaneously in the presence of 0.6 mM tetraoleoyl cardiolipin (18:1) for 22 hours at ambient temperature. MSPs can assemble nanodiscs in the presence of lipid[32]. We also tested if expression of MSP1D1 before AtRGS1 increased the solubilization of AtRGS1 into the existing nanodiscs. Based on this hypothesis, a sequential expression procedure was designed (Figure 3A, right). In this procedure, RNA encoding MSP1D1 was translated in the presence of 0.6 mM tetraoleoyl cardiolipin (18:1) for 16 hours at ambient temperature to self-assemble nanodiscs. RNA encoding AtRGS1 was added to the translation system with fresh translation feeding buffer followed by a second addition 24 hours later. Thus, the total translation time for the Seq-exp method was 64 hours.
The translation reactions were examined by both CBB stain and immunoblot analysis (Figure 3C and D). Although no major bands were observed at the predicted position of AtRGS1 (Figure 3C), immunoblot analysis by anti-His and anti-AtRGS1 showed significant bands around 50 kDa in both Co-exp and Seq-exp extracts but not in the negative control (Figure 3D, upper and middle). No other fragments were detected with these antibodies, except a smear between 95~130 kDa, approximately twice the molecular weight for AtRGS1. MSP1D1 migrated at its predicted molecular weight position (17~28 kDa) in the CBB stain gel suggesting that an excess of the nanodisc scaffold was produced (Figure 3C and 3D, Lower). These results indicate that both the co-expression and sequential protocol worked for the expression of AtRGS1 and MSP1D1 but that the yield was greater for the Seq-exp protocol. As shown in Figure 3E, the Co-exp protocol yielded approximately the same as the liposome protocol.
To assess whether the crude in vitro translation extracts contained functional AtRGS1, a single turnover GTPase assay was performed to explore GAP activity of AtRGS1 on its substrate AtGPA1. Seq-exp extract showed a small but statistically-significant GAP activity (Figure 3F and 3G), which suggested synthesized AtRGS1 protein was functional. Neither the No-RNA nor the MSP1D1 extracts showed this effect. However, co-expressed extract showed no GTPase accelerating activity. As an alternative, we tested whether the low activity could be overcome by purification of the AtRGS1 nanodiscs.
Purification and quantification of full length AtRGS1
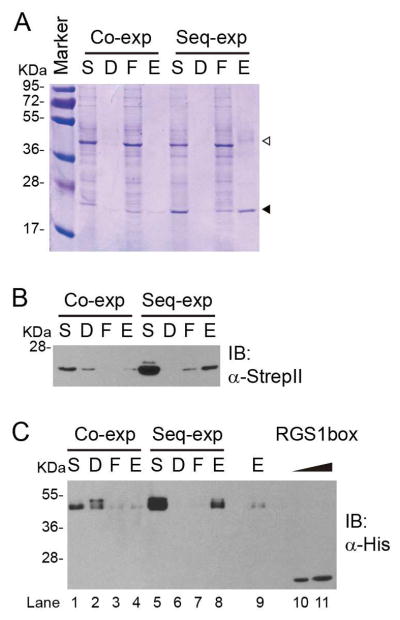
To enrich AtRGS1 incorporated into nanodiscs, the complex was purified using the StrepII tag fused with the C-terminus of MSP1D1. The crude extract was centrifuged to isolate the solubilized reaction product from pellet fractions. The soluble fraction was subjected to StrepTactin purification and eluted with d-desthiobiotin containing elution buffer. Clear bands could be observed in the SDS-PAGE analysis of each fraction of the purification at the predicted molecular weight of MSP1D1 (Figure 4A solid arrowhead). The results of anti-StrepII immunoblotting further confirmed that these bands were StrepII-tagged MSP1D1 (Figure 4B). Two prominent bands corresponding to AtRGS1 and MSP1D1 were observed in the eluate fraction from the Seq-exp AtRGS1 protocol but not the Co-exp protocol (Figure 4A). Immunoblot analysis further confirmed that the upper bands (open arrowhead) were His-tagged AtRGS1 (Figure 4C, lane 8). A weak band was observed in the eluate fraction of the Co-exp protocol (Figure 4C, lane 4), albeit with 4-fold lower intensity than the Seq-exp eluate sample (Figure 4C, lane 9). These results indicated that the Seq-exp protocol was more efficient. AtRGS1 was not detected in the pellet fraction of sequential expression, indicating that the pre-synthesized nanodisc promoted the solubilization of AtRGS1. Doublet bands of AtRGS1 were detected by both anti-His and anti-RGS1 in crude extract from all procedures (Figure 1C, Figure 3D and 3E). During affinity purification, the ratio of these two bands changed in each fraction. The higher molecular weight band was enriched in the debris fraction (Figure 4C, lane 2), in contrast, the lower molecular weight band was prominent in the flow-through and eluate fractions (Figure 4C, 1 and 3, 4 and 8). Thus, we hypothesized that the upper bands represent modified AtRGS1. The eluate contained heterogeneous nanodiscs with or without AtRGS1, therefore the yield of AtRGS1 could not be directly assessed by quantitative assays. To solve this problem, we quantitated full-length AtRGS1 in nanodiscs using immunoblot analysis with the E. coli-purified RGS1 domain as a standard. The antiserum used was raised against this domain. The amount of AtRGS1 was estimated to be between 400 and 800 ng (Figure 4C, lane 10 and 11).
Figure 4. Isolation of MSP1D1-AtRGS1 complex from crude translation extract.
After co-expression or sequential expression, the crude translation extract was fractionated into solubilized reaction product (S) and debris (D) fractions. The solubilized reaction product was incubated with StrepTactin. After discarding the flow-through (F) by centrifugation, StrepII tagged MSP1D1 was eluted by d-desthiobiotin as described in the Materials and Methods. Fractions were isolated by 12% SDS-PAGE and detected by Coomassie Brilliant Blue Staining (A), anti-His (B) and anti-StrepII (C) antibodies. The open and closed arrowheads indicate the predicted molecular weights of AtRGS1 and MSP1D1 respectively. Purified His6tagged RGS1 box (400 ng and 800 ng) was used as the RGS standard (C, lane 10 and 11 respective).
GAP activity of purified AtRGS1 in nanodiscs
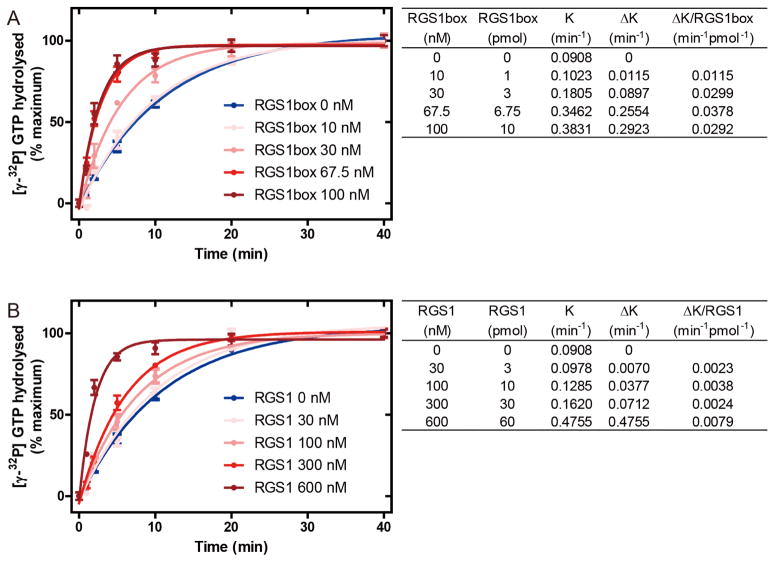
To measure the GAP activity of purified AtRGS1, single turnover GTPase assays were performed (Figure 5). The results of each time points were plotted using the One-phase association model to estimate the rate constant K (min−1). The ΔK of each concentration of RGS1 box domain or AtRGS1 were obtained by subtraction of the intrinsic rate constant of AtGPA1, and the GAP activity of these proteins were compared by ΔK per molecule (min−1 pmol−1). The calculated K value of AtGPA1 was 0.0908 min−1. RGS1 box and full-length AtRGS1 increased the K value in a dose-dependent manner. Saturation of RGS1 box was observed at 67.5 nM, providing a rate constant of 0.3462 min−1 (Figure 5A). Full length AtRGS1 increased the hydrolysis rate to a comparable level (0.4755 min−1) at 600 nM (Figure 5B). Due to the low amount of full-length AtRGS1 nanodisc, saturation of GAP activity could not be confirmed and therefore the GAP activity of AtRGS1 and RGS1 box could not be compared by their EC50. To address this problem, we determined the concentration of each protein that gave a similar reaction rate. At 67.5 nM the GAP activity of RGS1 box domain was comparable to 600 nM AtRGS1 (0.3462 min−1 versus 0.4755 min−1). Therefore, without further purification the GAP activity of the RGS1 box domain was roughly 10 times greater than that of full-length AtRGS1 (0.0378 min−1 pmol−1 versus 0.00024 min−1 pmol−1).
Figure 5. GAP activity of purified MSP1D1-AtRGS1 complex.
250 nM of purified AtGPA1 was pre-incubated with [γ-32P] GTP in ice cold buffer. Single turnover GTPase assay were initiated by the addition of GTPγS containing buffer with the indicated concentration of purified RGS1 box (A) or purified sequential expressed AtRGS1 (B). Duplicated reactions were stopped by ice cold quench buffer at the indicated time points. Each experiment was repeated at least once. The data were fit to exponential One-phase association functions using GraphPad Prism version 5.0 and the calculated rates shown in the tables to the right. K value is the rate constant of the reaction. ΔK is the increase caused by RGS1 box or full-length AtRGS1.
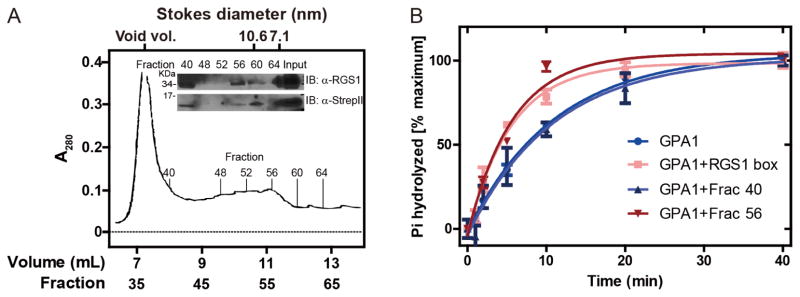
Additional purification by size-exclusion chromatography (SEC) revealed comparable specific activity of the full-length AtRGS1 nanodiscs and the RGS1 box domain. Two peaks in SEC were observed (Figure 6A). The greatest peak occurred between 6.8 to 8.8 mL of eluate volume (fractions between 34 and 44) corresponding to the void volume at 7.2 mL. Another peak was seen at the 8.8 to 12.0 mL elution volume (fractions 44 – 60). The Stokes diameter of MSP1D1 assembled in nanodiscs is calculated to be 9.5 nm [32]. This diameter is slightly affected by the nanodisc cargo such as monomeric β2-adrenergic receptor [18,33] or rhodopsin [21]. Immunoblot analyses were performed to quantitate AtRGS1 in fractions for comparison with GAP activity. Immunoblot analysis revealed that both AtRGS1 and MSP1D1 co-eluted in fraction 40, 56 and 60 (8.0, 11.2 and 12.0 mL of eluate volume) (Figure 6A, inset). Among these fractions, AtRGS1 was enriched in fractions 40 and 56. The concentration of AtRGS1 in these fractions was estimated by comparative immunoblot analysis as described for Figure 4. Single turnover GTPase assays indicated that 8 nM of AtRGS1 from fraction 56 had GAP activity comparable to 30 nM of RGS1box (0.1762 min−1 versus 0.1805 min−1, Figure 6B). These results indicate that the in vitro translated full-length AtRGS1 nanodiscs had comparable GAP activity as the RGS1 box (within 3-fold). In contrast, the eluate from fraction 40 had no effect in GTP hydrolysis, indicating that the peak between fraction 34 and 44 represented aggregated AtRGS1.
Figure 6. Nanodisc-AtRGS1 complex purification by size exclusion chromatography.
(A) Elution profile (OD280) of MSP1D1-AtRGS1 complex performed as described in Materials and Methods. Indicated 0.2-mL fractions were subjected to 12% SDS-PAGE and AtRGS1 and MSD1D1 were detected by anti-RGS1 (inset, upper) and anti-StrepII (inset, lower) antibodies, respectively. The input to the column (eluate from affinity purification) was included. The Stokes radii are provided at the top. (B) The GAP activity of indicated fractions was determined by the single turnover GTPase assay. 10 μL of fraction 40, fraction 50 or RGS1box (final concentration 30 nM) was added into each reaction, respectively. Duplicate reactions were stopped by ice cold quench buffer at indicated time points. Data were fit to exponential One-phase association functions using GraphPad Prism version 5.0.
We described two methods for in vitro AtRGS1 synthesis. Incorporation of AtRGS1 into nanodiscs showed the more promising approach to optimize expression for biochemical studies.
Highlights.
Two protocols are provided for AtRGS1 in vitro synthesis and purification.
AtRGS1 solubilized better in nanodisc than in unilamellar liposomes.
Incorporation into nanodisc conserves the GAP activity of AtRGS1.
Acknowledgments
The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through the grant DE-FG02-05er15671 to A.M.J funded the majority of this work. This work was supported further in part by grants from the NIGMS (R01GM065989) and NSF (MCB-0718202) to A.M.J and a Transmembrane Protein Center grant awarded by the National Institutes of Health (NIH) from the Institute for General Medical Sciences U54GM094584 to B.G.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambert NA. Dissociation of heterotrimeric g proteins in cells. Sci Signal. 2008;1:re5. doi: 10.1126/scisignal.125re5. [DOI] [PubMed] [Google Scholar]
- 2.Zhong H, Wade SM, Woolf PJ, Linderman JJ, Traynor JR, Neubig RR. A spatial focusing model for G protein signals. Regulator of G protein signaling (RGS) protein-mediated kinetic scaffolding. J Biol Chem. 2003;278:7278–84. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]
- 3.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci U S A. 2007;104:17317–22. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, et al. G protein activation without a GEF in the plant kingdom. PLoS Genet. 2012;8:e1002756. doi: 10.1371/journal.pgen.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford W, Buckholz A, Morton J, Price C, Jones AM, Urano D. Eukaryotic G protein signaling evolved to require G protein-coupled receptors for activation. Sci Signal. 2013;6:ra37. doi: 10.1126/scisignal.2003768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urano D, Jones AM. Heterotrimeric G protein-coupled signaling in plants. Annu Rev Plant Biol. 2014;65:365–84. doi: 10.1146/annurev-arplant-050213-040133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, et al. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–31. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 8.Temple BRS, Jones AM. The plant heterotrimeric G-protein complex. Annu Rev Plant Biol. 2007;58:249–66. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- 9.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 30:1185–94. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser L, Graveland-Bikker J, Steuerwald D, Vanberghem M, Herlihy K, Zhang S. Efficient cell-free production of olfactory receptors: detergent optimization, structure, and ligand binding analyses. Proc Natl Acad Sci U S A. 2008;105:15726–15731. doi: 10.1073/pnas.0804766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takai K, Sawasaki T, Endo Y. Practical cell-free protein synthesis system using purified wheat embryos. Nat Protoc. 2010;5:227–238. doi: 10.1038/nprot.2009.207. [DOI] [PubMed] [Google Scholar]
- 12.Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Satoh R, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. [accessed June 9, 2015];Nat Methods. 2008 5:1011–7. doi: 10.1038/nmeth.1273. http://www.ncbi.nlm.nih.gov/pubmed/19054851. [DOI] [PubMed] [Google Scholar]
- 13.Hong J, Krishnamurthi K, Zheng W, Sandberg K. In Vitro Translation of the Angiotensin AT(1) Receptor in Wheat-Germ Extracts. Methods Mol Med. 2001;51:171–92. doi: 10.1385/1-59259-087-X:171. [DOI] [PubMed] [Google Scholar]
- 14.Basu D, Castellano JM, Thomas N, Mishra RK. Cell-free protein synthesis and purification of human dopamine D2 receptor long isoform. Biotechnol Prog. 29:601–8. doi: 10.1002/btpr.1706. [DOI] [PubMed] [Google Scholar]
- 15.Arimitsu E, Ogasawara T, Takeda H, Sawasaki T, Ikeda Y, Hiasa Y, et al. The ligand binding ability of dopamine D1 receptors synthesized using a wheat germ cell-free protein synthesis system with liposomes. Eur J Pharmacol. 2014;745:117–22. doi: 10.1016/j.ejphar.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, et al. Chapter 11 Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Elsevier Masson SAS. 2009 doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2:853–856. doi: 10.1021/nl025623k. [DOI] [Google Scholar]
- 18.Whorton MR, Bokoch MP, Rasmussen SGF, Huang B, Zare RN, Kobilka B, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao XJ, Vélez Ruiz G, Whorton MR, Rasmussen SGF, DeVree BT, Deupi X, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee S, Huber T, Sakmar TP. Rapid Incorporation of Functional Rhodopsin into Nanoscale Apolipoprotein Bound Bilayer (NABB) Particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 21.Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, et al. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayburt TH, Vishnivetskiy Sa, McLean Ma, Morizumi T, Huang CC, Tesmer JJG, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knepp AM, Grunbeck A, Banerjee S, Sakmar TP, Huber T. Direct measurement of thermal stability of expressed CCR5 and stabilization by small molecule ligands. Biochemistry. 2011;50:502–511. doi: 10.1021/bi101059w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denisov IG, Grinkova YV, Lazarides aa, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. Nano Lett. 2004:1–6. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 26.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 27.Sawasaki T, Hasegawa Y, Tsuchimochi M, Kasahara Y, Endo Y. Construction of an efficient expression vector for coupled transcription/translation in a wheat germ cell-free system. [accessed June 9, 2015];Nucleic Acids Symp Ser. 2000 :9–10. doi: 10.1093/nass/44.1.9. http://www.ncbi.nlm.nih.gov/pubmed/12903243. [DOI] [PubMed]
- 28.Makino S, Beebe ET, Markley JL, Fox BG. Cell-free protein synthesis for functional and structural studies. Methods Mol Biol. 2014;1091:161–78. doi: 10.1007/978-1-62703-691-7_11. [DOI] [PubMed] [Google Scholar]
- 29.Goren MA, Nozawa A, Makino S, Wrobel RL, Fox BG. Cell-free translation of integral membrane proteins into unilamelar liposomes. Methods Enzymol. 2009;463:647–73. doi: 10.1016/S0076-6879(09)63037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, et al. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012;14:1079–88. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390:805–14. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 32.Denisov IG, Grinkova YV, Lazarides aa, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 33.Leitz AJ, Bayburt TH, Barnakov AN, Springer Ba, Sligar SG. Functional reconstitution of3??2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601–612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]