Abstract
Objective:
The aim of the current study was to carry out a preliminary validation of devices for standardized collection of whole mouth fluid (WMF) in comparison to the passive drooling method for protein analysis in healthy subjects.
Materials and Methods:
A carefully designed sample collection/pretreatment protocol is crucial to the success of any saliva proteomics project. In this study, WMF was collected from healthy volunteers (n = 10, ages: 18–26 years). Individuals with any oral disease were excluded from the study group. In our study, we evaluated the following collection methods; the classical passive drooling method (unstimulated whole saliva) and standardized tools for saliva collection (Pure·SAL™, and RNAPro·SAL™) from Oasis Diagnostics® Corporation (Vancouver WA, USA). For estimation of protein levels, we used the bicinchoninic acid assay and protein assay kit (Thermo Fisher). The two-dimensional gel electrophoresis sample analysis was carried out for the estimation of proteins in one of the samples.
Results:
When gels were compared, the difference was seen in the resolution of spots. Protein spots were fading from high- to low-molecular weight masses. Hence, advanced devices in comparison to spitting method resulted in much clearer protein spots which in turn prove the validation of devices.
Conclusions:
In this study, we concluded that protein extraction could be possible by both methods such as passive drooling method and through advanced saliva collection devices (Pure·SAL™ and RNAPro·SAL™).
Keywords: Gel electrophoresis and dentistry, proteins, proteomics, saliva
INTRODUCTION
Human saliva produced in the oral cavity having several functions also maintains oral homoeostasis through a broad spectrum of biomolecules.[1] Potential of this biofluid could be a choice for the diagnosis and prognosis of diseases. It contains hundreds of proteins and peptides as its components. The most ubiquitous inorganic components of saliva comprise sodium, potassium chloride, calcium, magnesium, and carbonates, whereas the organic components include amylases, lysozyme, mucins, peroxidase, lipase, lactoferrins, cystatins, hormones, and growth factors.[2] The contents of these proteins and peptides are altered not only in oral disease conditions but also during other disorders of the human body.[3] The advancements in protein sciences not only made it possible to identify the protein components from any biological sample but also the quantification of such proteins has made immense progress in recent years.[4] Transforming scientific results of biomolecules such as nucleic acids, proteins, and metabolites present in biological samples to clinical applications are complicated and challenging. That is the reason why there is a rare chance of reaching to the final destination of implementation of a scientific finding into a clinical test.[5] To do so, the first step of basic research is of utmost importance. Afterward, the technological and methodology develop that ultimate results in the application of research work. Using saliva as liquid biopsy, a biofluid for diagnosis brings its benefits being an inexpensive, fast, easy, and noninvasive collection method.[6] Whereas saliva sample gives no clotting issues, storage and shipping are easy with the physiological and pathological reflection of the state of the individual. Bandhakavi et al. reported 2340 proteins in saliva and highlighted the significance of saliva as a diagnostic medium by identifying 20% of proteins being found in plasma as well.[7] Our group recently reviewed the importance of gingival crevicular fluid as a diagnostic fluid for the proteomics analysis, it contains many inflammatory biomarkers due to surrounding tissue diseases.[8] We have found many studies using saliva proteomics for the detection of diseases such as oral cancer,[9] dental caries,[10,11] periodontitis,[12] gingivitis,[13] oral lichen planus,[14] and Sjogren's syndrome.[15]
Proteomics of human saliva has demonstrated a new approach in the search for protein biomarkers for detection of human diseases.[16] Identification of the proteome of human saliva plays a key role in understanding the pathophysiology of oral and systemic human diseases.[17] Proteins are the physiological effectors, which have a direct relationship with the pathology of the body, that can be highlighted by clinical proteomics. In addition, these proteomic findings may provide the opportunity to monitor remedies, therapeutic outcomes, and disease progression. In spite of much advancements and progress already achieved in saliva proteomics, efforts are going on toward standardizing sample collection method, pretreatment procedure, and extensive range analysis of the whole saliva to be the fluid of choice for diagnostics. Saliva can be collected under two conditions, stimulated and unstimulated way; it can be stimulated through acid, citrus juices, mechanically or chewing on absorbent pad,[18,19] whereas saliva collected at resting condition is preferable. Methods of the collection include spitting, suction, swabbing, passive drooling, and the recent number of commercially available devices are being used such as Oasis Diagnostics, Salimetrics® Oral Swab (SOS), and Greiner Bio-One (GBO), and Saliva Collection System (SCS).[6]
The aim of this study was to analyze the difference (if any) between commercially available saliva collection device (RNAPro·SAL™ and Pure·SAL™) and passive drooling method. For the quantification of protein contents in each group, a method was done using bicinchoninic acid (BCA) kit for protein quantification and two-dimensional (2D) gel electrophoresis in the saliva of healthy individuals.
MATERIALS AND METHODS
Saliva sampling and storage
Saliva sampling was collected at Liaquat College of Medicine and Dentistry (LCMD), Karachi, Pakistan, from 10 healthy volunteers (5 males and 5 females) age group between 22 and 25 years and they have volunteered students of the LCMD from an undergraduate program. Exclusion criteria are any history of oral diseases, malignancy, viral diseases, hormonal diseases, immunodeficiencies, and autoimmune diseases. All the participants signed informed consents after the approval from ethical board committee of the college. A carefully designed sample collection/pretreatment protocol is crucial to the success of any saliva proteomics project. In this study, we evaluated the following collection methods; the classical passive drooling method (unstimulated whole saliva) and standardized tools for saliva collection (Pure·SAL™, and RNAPro·SAL™) from Oasis Diagnostics® Corporation (Vancouver WA, USA) [Figure 1].[20] Subjects were asked to refrain from drinking, eating, or any oral hygiene procedure 2 h before sampling as reported by Topkas et al.[21] Before collection, subjects have been invited to rinse the mouth with drinking water for 15 s to remove any food debris, microorganisms, and desquamated epithelial cells. After rinsing, subjects have been asked to sit straight in a dental chair and wait for 1 min before collection. For all subjects, order of sampling was unstimulated saliva collection through Pure·SAL™ and RNAPro·SAL™ according to manufacturer instructions, and for the passive drooling method, we used 10 mL sterilized tube, and every sample was collected after 24 h on next appointment. After collection, samples were immersed in crushed ice and delivered to National Center for Proteomics for proteomic analysis. The samples were stored at −20°C until further use. For estimation of protein, we used the BCA assay protein assay kit (Thermo Fisher).[22] To check the proteins in all samples, 2D gel electrophoresis was run to see the proteins pattern in all samples.
Figure 1.
Saliva devices by Oasis Diagnostics
RESULTS
Protein concentration measurements
Total protein was estimated by BCA protein assay method of Pierce (Thermo Scientific) by following the instructions of the manufacturer. BCA assay is colorimetric based, and it gives dark purple color when two molecules of BCA chelates with protein and formed a compound of the cuprous ion. The absorbance of the complex was measured at 562 nm. BCA standard reagent A and B mixed freshly in the ratio of 50:1. Bovine serum albumin (2 mg/ml) is used as a standard with 5 working standards 1–5 μg. All the tubes (standards, test samples, and blank) are incubated at 37°C for 30 min. After incubation, absorbance is measured at 562 nm against a reagent blank using a microplate reader (Beckman Coulter Co.). The concentration of test samples was measured with reference to standards.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis or gel electrophoresis
For 2D gel electrophoresis, an equal amount of proteins (80 μg) was dissolved in rehydration buffer (8 M urea, 0.2 M EDTA, 0.5 M dithiothreitol [DTT], glycerol, NP-40, ampholyte solution pH 3–10 in 0.5 M Tris–HCl) for passive rehydration of 7 cm IPG strip 3–10 NL overnight at room temperature. Proteins were focused on the IEF Multiphor II system (GE Healthcare) at 20°C using a total of 7000 V/h. After IEF, IPG strips were equilibrated using equilibration buffer I (6 M urea, 30% glycerol, 2% sodium dodecyl sulfate [SDS], 50 mM Tris–HCl pH 8.8, and 1% DTT) and equilibration buffer II (6 M urea, 30% glycerol, 2% SDS, 50 mM Tris–HCl pH 8.8, and 2.5% iodoacetamide). Following equilibration, the focused proteins were separated on 12.5% SDS-polyacrylamide gel electrophoresis. Gels were visualized by silver staining to detect proteins after electrophoretic separation on 12.5% polyacrylamide gels.
DISCUSSION
There was a difference observed in the gels as shown by the area of gel encircled in Figure 2. The difference was observed in the resolution of spots. The spots circled in locations “a” and “b” were resolved in a better way in saliva sample collected using the passive drool method in comparison to Pure·SAL™ and RNAPro·SAL™ devices. In the case of circle “c,” few of spots that are much clearer in saliva sample collected using the Pure·SAL™ device. The gel showed protein spots from high-molecular weight to low-molecular masses (encircled from top to bottom in the figure). The low-molecular protein spots were not that much resolved. Few of the spots that were differentially resolved could be good candidates for in-gel digestion and mass spectrometric analysis for identification. The gel also indicated that the extracted proteins lie in the range of acidic to slightly basic pH while the basic section (towards pH 10) did not show the presence of resolved spots from top to bottom.
Figure 2.
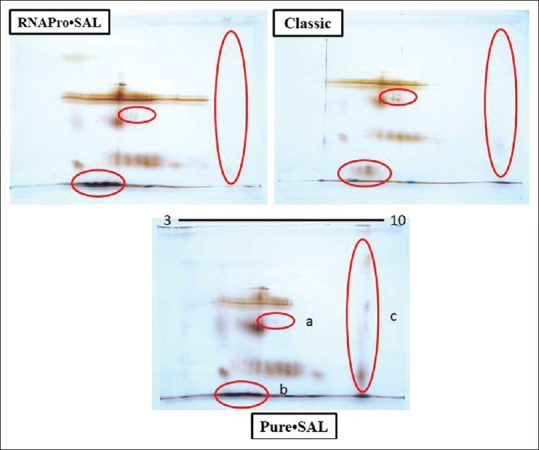
Two-dimensional gel images of the saliva sample. The IEF was run using 3–10 IPG strip from Bio-Rad. The second dimension was run on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis at a constant voltage of 70 volts. The gel was stained by silver staining
Previously, Topkas et al. collected saliva from different commercially available methods such as SOS, Salivette® Cotton and Synthetic (Sarstedt), and GBO SCS® to measure C-reactive protein, IgE, and myoglobin levels in human saliva to analyze the differences between collection methods which showed a relevant variation in the salivary flow rates depending on the collection method.[21] In our study, human saliva was collected by three different approaches, the gold standard passive drool method and by two devices, Pure·SAL™ and RNAPro·SAL™ from Oasis Diagnostics®. 2D gel electrophoresis analysis was performed to profile the proteins. The gel images show a few differences in the protein patterns between the methods. The spots encircled in locations “a” and “b” were well resolved in saliva samples collected using the passive drool method in comparison to the Pure·SAL™ and RNAPro·SAL™ devices. In the case of circle “c,” this showed a few spots that are much clearer in saliva samples collected using the Pure·SAL™ device. The noted differences may be observed due to the intrinsic properties of the RNAPro·SAL™ and Pure·SAL™ technologies, which both use highly absorbent pad materials to collect saliva and act to remove a high percentage of mucinous material (>70%). The recovery of a highly purified sample is readily obtained by compression through a proprietary medium to clear potential interferences likely to cause problems with downstream assays. The devices also feature a unique sample volume adequacy indicator that provides a visual indication that an adequate quantity of sample has been collected for downstream analysis of RNA, proteins, cell-free DNA, and exosomes. Samples may then be applied to techniques such as polymerase chain reaction, genotyping, sequencing, proteomics, and other applications, depending on the desired results. Tomoaki et al.[21] used cotton swab (cotton saliva collection) and passive drooling method for saliva collection, this study nicely revealed the impact of collection on salivary cortisol assays.[23] Our study demonstrated the evaluation of collection method and standardization of device, in comparison to passive drooling. Passive drooling, that requires practice, is less desirable and more time-consuming for participants and provides an unfiltered sample. The advantages of these two commercially available devices provide a readily purified sample directly into a standard collection tube, that is, stabilized independently for immediate use or stored for long-term storage pending analysis. There are a few limitation of this study including lack of advanced characterization using techniques such as high-performance liquid chromatography, mass spectroscopy, and enzyme-linked immunosorbent assay. Due to limited resources, these techniques were not explored in the current study and should be considered for further research.
CONCLUSIONS
This study focused on evaluating the biopsy utility of the RNAPro·SAL and Pure·SAL system for protein content analysis. The RNAPro·SAL and Pure·SAL systems provide a number of advantages over the passive drooling method as they are easy to use and cost-effective tools for protein and peptides evaluation but also to split sample collection. In this way, it could perform as a collection method for a rich source of RNA, miRNA, mRNA, cell-free DNA, and cell-free RNA and exosomes in saliva. These devices are compatible for self-collection and good for speed-up the collection method. This equipments help in the understanding of saliva-based point-of-care (PoC) technology.
Financial support and sponsorship
We are thankful to Sure Bio-diagnostics and Pharmaceuticals for provide us financial support for this project.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We acknowledge Oasis Diagnostics Cooperation for providing us free samples of the RNAPro· SAL and Pure· SAL and Sure Bio-diagnostics & Pharmaceuticlas for providing us Gel Electrophoresis. We would also thank to Pakistan Human Saliva Research Group (PHSRG) and National Center for Proteomics (NCP), Karachi University, for helping in sample collection and processing.
REFERENCES
- 1.Zhang CZ, Cheng XQ, Li JY, Zhang P, Yi P, Xu X, et al. Saliva in the diagnosis of diseases. Int J Oral Sci. 2016;8:133–7. doi: 10.1038/ijos.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khurshid Z, Najeeb S, Mali M, Moin SF, Raza SQ, Zohaib S, et al. Histatin peptides: Pharmacological functions and their applications in dentistry. Saudi Pharm J. 2017;25:25–31. doi: 10.1016/j.jsps.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins Mussi MC, Moffa E, Castro T, Lira Ortega A, Freitas G, Braga M, et al. Salivary parameters and oral health in the moebius syndrome. Spec Care Dentist. 2016;36:265–70. doi: 10.1111/scd.12175. [DOI] [PubMed] [Google Scholar]
- 4.Khurshid Z, Zohaib S, Najeeb S, Zafar MS, Rehman R, Rehman IU, et al. Advances of proteomic sciences in dentistry. Int J Mol Sci. 2016;17:728. doi: 10.3390/ijms17050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller CS, Foley JD, Bailey AL, Campell CL, Humphries RL, Christodoulides N, et al. Current developments in salivary diagnostics. Biomark Med. 2010;4:171–89. doi: 10.2217/bmm.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurshid Z, Zohaib S, Najeeb S, Zafar MS, Slowey PD, Almas K, et al. Human saliva collection devices for proteomics: An update. Int J Mol Sci. 2016;17:846. doi: 10.3390/ijms17060846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8:5590–600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khurshid Z, Mali M, Naseem M, Najeeb S, Zafar MS. Human gingival crevicular fluids (GCF) proteomics: An overview. Dent J. 2017;5:12. doi: 10.3390/dj5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Huang X, Tan X, Si Y, Wang X, Chen F, et al. Salivary peptidome profiling for diagnosis of severe early childhood caries. J Transl Med. 2016;14:240. doi: 10.1186/s12967-016-0996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vacca Smith AM, Scott-Anne KM, Whelehan MT, Berkowitz RJ, Feng C, Bowen WH, et al. Salivary glucosyltransferase B as a possible marker for caries activity. Caries Res. 2007;41:445–50. doi: 10.1159/000107930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant MM, Creese AJ, Barr G, Ling MR, Scott AE, Matthews JB, et al. Proteomic analysis of a noninvasive human model of acute inflammation and its resolution: The twenty-one day gingivitis model. J Proteome Res. 2010;9:4732–44. doi: 10.1021/pr100446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangé H, Léger T, Huchon C, Ciangura C, Diallo D, Poitou C, et al. Salivary proteome modifications associated with periodontitis in obese patients. J Clin Periodontol. 2012;39:799–806. doi: 10.1111/j.1600-051X.2012.01913.x. [DOI] [PubMed] [Google Scholar]
- 14.Cheng YS, Rees T, Jordan L, Oxford L, O’Brien J, Chen HS, et al. Salivary endothelin-1 potential for detecting oral cancer in patients with oral lichen planus or oral cancer in remission. Oral Oncol. 2011;47:1122–6. doi: 10.1016/j.oraloncology.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldini C, Giusti L, Ciregia F, Da Valle Y, Giacomelli C, Donadio E, et al. Proteomic analysis of saliva: A unique tool to distinguish primary Sjögren's syndrome from secondary Sjögren's syndrome and other sicca syndromes. Arthritis Res Ther. 2011;13:R194. doi: 10.1186/ar3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarano E, Fiorita A, Picciotti PM, Passali GC, Calò L, Cabras T, et al. Proteomics of saliva: Personal experience. Acta Otorhinolaryngol Ital. 2010;30:125–30. [PMC free article] [PubMed] [Google Scholar]
- 17.Latterich M, Abramovitz M, Leyland-Jones B. Proteomics: New technologies and clinical applications. Eur J Cancer. 2008;44:2737–41. doi: 10.1016/j.ejca.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Henson BS, Wong DT. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol Biol. 2010;666:21–30. doi: 10.1007/978-1-60761-820-1_2. [DOI] [PubMed] [Google Scholar]
- 19.Malamud D. Saliva as a diagnostic fluid. Dent Clin North Am. 2011;55:159–78. doi: 10.1016/j.cden.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang SH, Thomas GA, Liao W, Grogan T, Buck RL, Fuentes L, et al. RNAPro·SAL: A device for rapid and standardized collection of saliva RNA and proteins. Biotechniques. 2015;58:69–76. doi: 10.2144/000114254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topkas E, Keith P, Dimeski G, Cooper-White J, Punyadeera C. Evaluation of saliva collection devices for the analysis of proteins. Clin Chim Acta. 2012;413:1066–70. doi: 10.1016/j.cca.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Krieg RC, Dong Y, Schwamborn K, Knuechel R. Protein quantification and its tolerance for different interfering reagents using the BCA-method with regard to 2D SDS PAGE. J Biochem Biophys Methods. 2005;65:13–9. doi: 10.1016/j.jbbm.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Kozaki T, Hashiguchi N, Kaji Y, Yasukouchi A, Tochihara Y. Effects of saliva collection using cotton swab on cortisol enzyme immunoassay. Eur J Appl Physiol. 2009;107:743–6. doi: 10.1007/s00421-009-1178-3. [DOI] [PubMed] [Google Scholar]


